Assessment of Lab4P Probiotic Effects on Cognition in 3xTg-AD Alzheimer’s Disease Model Mice and the SH-SY5Y Neuronal Cell Line
Abstract
1. Introduction
2. Results
2.1. Lab4P Maintains Cognitive Performance and Preserves Hippocampal Spine Densities in 3xTg-AD Mice (Study A)
2.2. Lab4P Maintains Cognitive Performance and Preserves Hippocampal Spine Densities in HFD-Fed 3xTg-AD Mice (STUDY B)
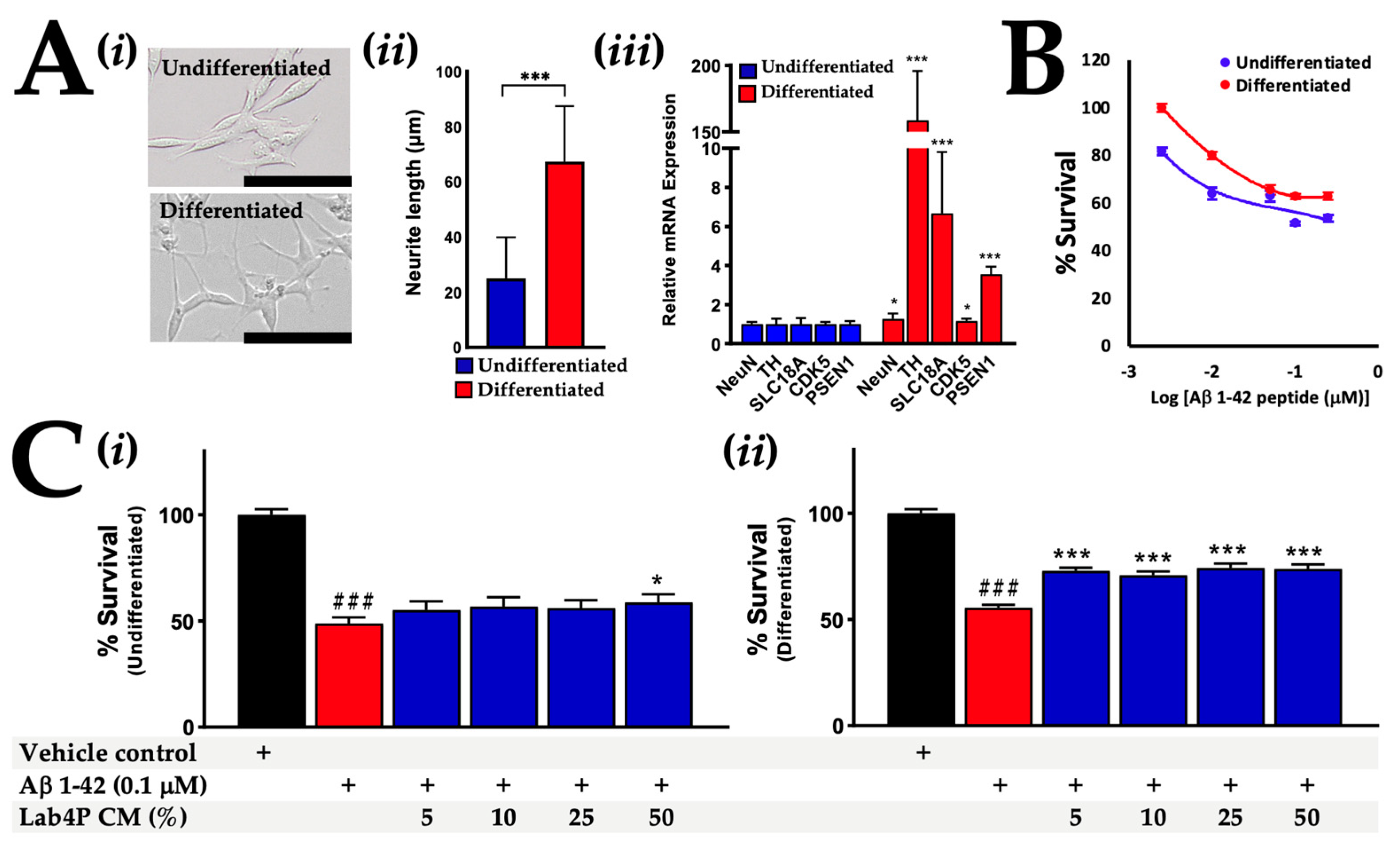
2.3. Lab4P CM Promotes Survival in Human SH-SY5Y Cells Challenged with β-Amyloid
3. Discussion
4. Materials and Methods
4.1. Experiments in 3xTg-AD Mice
4.1.1. Probiotic Intervention
4.1.2. Mouse Husbandry and Study Designs
4.1.3. Mouse Behavioural Testing
4.1.4. Staining, Imaging and Morphological Classification of Neuronal Dendritic Spines in the Hippocampus
4.1.5. mRNA Expression Analysis of Hippocampus
4.2. Experiments In Vitro in the Human SH-SY5Y Neuronal Cell Line
4.2.1. Maintenance and Differentiation of SH-SY5Y Cell Cultures
4.2.2. Quantification of Neurite Length
4.2.3. mRNA Expression Analysis in SH-SY5Y Cells
4.2.4. Generation of Lab4P Conditioned Media (CM) and SH-SY5Y Cell Stimulation
4.2.5. Assessment of SH-SY5Y Cell Viability
4.3. Statistical Analysis
Studies In Vivo and In Vitro
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2020, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Olaimat, A.N.; Aolymat, I.; Al-Holy, M.; Ayyash, M.; Abu Ghoush, M.; Al-Nabulsi, A.A.; Osaili, T.; Apostolopoulos, V.; Liu, S.Q.; Shah, N.P. The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Sci. Food 2020, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Agusti, A.; Garcia-Pardo, M.P.; Lopez-Almela, I.; Campillo, I.; Maes, M.; Romani-Perez, M.; Sanz, Y. Interplay Between the Gut-Brain Axis, Obesity and Cognitive Function. Front. Neurosci. 2018, 12, 155. [Google Scholar] [CrossRef]
- Zhuang, Z.Q.; Shen, L.L.; Li, W.W.; Fu, X.; Zeng, F.; Gui, L.; Lu, Y.; Cai, M.; Zhu, C.; Tan, Y.L.; et al. Gut Microbiota is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yue, L.; Fang, X.; Wang, G.; Li, C.; Sun, X.; Jia, X.; Yang, J.; Song, J.; Zhang, Y.; et al. Altered gut microbiota in Parkinson’s disease patients/healthy spouses and its association with clinical features. Park. Relat. Disord. 2020, 81, 84–88. [Google Scholar] [CrossRef]
- Alzheimer’s Society’s View on Demography; Alzheimer’s Society: London, UK, 2019.
- World Health Organisation (WHO). Fact Sheet on Dementia; World Health Organisation: Geneva, Switzerland, 2020. [Google Scholar]
- Reza-Zaldivar, E.E.; Hernandez-Sapiens, M.A.; Minjarez, B.; Gomez-Pinedo, U.; Sanchez-Gonzalez, V.J.; Marquez-Aguirre, A.L.; Canales-Aguirre, A.A. Dendritic Spine and Synaptic Plasticity in Alzheimer’s Disease: A Focus on MicroRNA. Front. Cell Dev. Biol. 2020, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Jahn, H. Memory loss in Alzheimer’s disease. Dialogues Clin. Neurosci. 2013, 15, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Aisen, P.; Lemere, C.; Atri, A.; Sabbagh, M.; Salloway, S. Aducanumab produced a clinically meaningful benefit in association with amyloid lowering. Alzheimers Res. 2021, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Yiannopoulou, K.G.; Papageorgiou, S.G. Current and Future Treatments in Alzheimer Disease: An Update. J. Cent. Nerv. Syst. Dis. 2020, 12, 1179573520907397. [Google Scholar] [CrossRef]
- Anstey, K.J.; Cherbuin, N.; Budge, M.; Young, J. Body mass index in midlife and late-life as a risk factor for dementia: A meta-analysis of prospective studies. Obes. Rev. 2011, 12, e426–e437. [Google Scholar] [CrossRef]
- Julien, C.; Tremblay, C.; Phivilay, A.; Berthiaume, L.; Emond, V.; Julien, P.; Calon, F. High-fat diet aggravates amyloid-beta and tau pathologies in the 3xTg-AD mouse model. Neurobiol. Aging 2010, 31, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Knight, E.M.; Martins, I.V.; Gumusgoz, S.; Allan, S.M.; Lawrence, C.B. High-fat diet-induced memory impairment in triple-transgenic Alzheimer’s disease (3xTgAD) mice is independent of changes in amyloid and tau pathology. Neurobiol. Aging 2014, 35, 1821–1832. [Google Scholar] [CrossRef]
- Kim, D.; Cho, J.; Lee, I.; Jin, Y.; Kang, H. Exercise Attenuates High-Fat Diet-induced Disease Progression in 3xTg-AD Mice. Med. Sci. Sport. Exerc. 2017, 49, 676–686. [Google Scholar] [CrossRef]
- World Health Organisation (WHO). Fact Sheet No 311: Obesity and Overweight; World Health Organisation: Geneva, Switzerland, 2018. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Tenorio-Jimenez, C.; Martinez-Ramirez, M.J.; Gil, A.; Gomez-Llorente, C. Effects of Probiotics on Metabolic Syndrome: A Systematic Review of Randomized Clinical Trials. Nutrients 2020, 12, 124. [Google Scholar] [CrossRef]
- Bonfili, L.; Cecarini, V.; Gogoi, O.; Gong, C.; Cuccioloni, M.; Angeletti, M.; Rossi, G.; Eleuteri, A.M. Microbiota modulation as preventative and therapeutic approach in Alzheimer’s disease. FEBS J. 2020, 288, 2836–2855. [Google Scholar] [CrossRef] [PubMed]
- Bonfili, L.; Cecarini, V.; Berardi, S.; Scarpona, S.; Suchodolski, J.S.; Nasuti, C.; Fiorini, D.; Boarelli, M.C.; Rossi, G.; Eleuteri, A.M. Microbiota modulation counteracts Alzheimer’s disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci. Rep. 2017, 7, 2426. [Google Scholar] [CrossRef] [PubMed]
- Bonfili, L.; Cecarini, V.; Cuccioloni, M.; Angeletti, M.; Berardi, S.; Scarpona, S.; Rossi, G.; Eleuteri, A.M. SLAB51 Probiotic Formulation Activates SIRT1 Pathway Promoting Antioxidant and Neuroprotective Effects in an AD Mouse Model. Mol. Neurobiol. 2018, 55, 7987–8000. [Google Scholar] [CrossRef]
- Webberley, T.S.; Masetti, G.; Bevan, R.J.; Kerry-Smith, J.; Jack, A.A.; Michael, D.R.; Thomas, S.; Glymenaki, M.; Li, J.; McDonald, J.A.K.; et al. The Impact of Probiotic Supplementation on Cognitive, Pathological and Metabolic Markers in a Transgenic Mouse Model of Alzheimer’s Disease. Front. Neurosci. 2022, 16, 843105. [Google Scholar] [CrossRef]
- Marx, W.; Scholey, A.; Firth, J.; D’Cunha, N.M.; Lane, M.; Hockey, M.; Ashton, M.M.; Cryan, J.F.; O’Neil, A.; Naumovski, N.; et al. Prebiotics, probiotics, fermented foods and cognitive outcomes: A meta-analysis of randomized controlled trials. Neurosci. Biobehav. Rev. 2020, 118, 472–484. [Google Scholar] [CrossRef]
- Lv, T.; Ye, M.; Luo, F.; Hu, B.; Wang, A.; Chen, J.; Yan, J.; He, Z.; Chen, F.; Qian, C.; et al. Probiotics treatment improves cognitive impairment in patients and animals: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2021, 120, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.; Davies, T.; Loxley, K.; Allen, M.; Good, M.; Hughes, T.; Plummer, S.J.B. In vitro neuroprotective activities of two distinct probiotic consortia. Benef. Microbes. 2019, 10, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.R.; Jack, A.A.; Masetti, G.; Davies, T.S.; Loxley, K.E.; Kerry-Smith, J.; Plummer, J.F.; Marchesi, J.R.; Mullish, B.H.; McDonald, J.A.K.; et al. A randomised controlled study shows supplementation of overweight and obese adults with lactobacilli and bifidobacteria reduces bodyweight and improves well-being. Sci. Rep. 2020, 10, 4183. [Google Scholar] [CrossRef]
- Michael, D.R.; Davies, T.S.; Moss, J.W.E.; Calvente, D.L.; Ramji, D.P.; Marchesi, J.R.; Pechlivanis, A.; Plummer, S.F.; Hughes, T.R. The anti-cholesterolaemic effect of a consortium of probiotics: An acute study in C57BL/6J mice. Sci. Rep. 2017, 7, 2883. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.R.; Davies, T.S.; Jack, A.A.; Masetti, G.; Marchesi, J.R.; Wang, D.; Mullish, B.H.; Plummer, S.F. Daily supplementation with the Lab4P probiotic consortium induces significant weight loss in overweight adults. Sci. Rep. 2021, 11, 5. [Google Scholar] [CrossRef]
- Mullish, B.H.; Michael, D.R.; McDonald, J.A.; Masetti, G.; Plummer, S.F.; Marchesi, J.R.J.G. Identifying the factors influencing outcome in probiotic studies in overweight and obese patients: Host or microbiome? Gut 2021, 70, 225–226. [Google Scholar] [CrossRef]
- Mullish, B.H.; Marchesi, J.R.; McDonald, J.A.K.; Pass, D.A.; Masetti, G.; Michael, D.R.; Plummer, S.; Jack, A.A.; Davies, T.S.; Hughes, T.R.; et al. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: Should we be considering probiotics during viral pandemics? Gut Microbes 2021, 13, 1–9. [Google Scholar] [CrossRef]
- O’Morain, V.L.; Chan, Y.-H.; Williams, J.O.; Alotibi, R.; Alahmadi, A.; Rodrigues, N.P.; Plummer, S.F.; Hughes, T.R.; Michael, D.R.; Ramji, D.P. The Lab4P Consortium of Probiotics Attenuates Atherosclerosis in LDL Receptor Deficient Mice Fed a High Fat Diet and Causes Plaque Stabilization by Inhibiting Inflammation and Several Pro-atherogenic Processes. Mol. Nutr. Food Res. 2021, 65, 2100214. [Google Scholar] [CrossRef]
- De Medeiros, L.M.; De Bastiani, M.A.; Rico, E.P.; Schonhofen, P.; Pfaffenseller, B.; Wollenhaupt-Aguiar, B.; Grun, L.; Barbe-Tuana, F.; Zimmer, E.R.; Castro, M.A.A.; et al. Cholinergic Differentiation of Human Neuroblastoma SH-SY5Y Cell Line and Its Potential Use as an In vitro Model for Alzheimer’s Disease Studies. Mol. Neurobiol. 2019, 56, 7355–7367. [Google Scholar] [CrossRef]
- Teppola, H.; Sarkanen, J.R.; Jalonen, T.O.; Linne, M.L. Morphological Differentiation Towards Neuronal Phenotype of SH-SY5Y Neuroblastoma Cells by Estradiol, Retinoic Acid and Cholesterol. Neurochem. Res. 2016, 41, 731–747. [Google Scholar] [CrossRef]
- Kovalevich, J.; Langford, D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. [Google Scholar] [PubMed]
- Lopes, F.M.; Schröder, R.; da Frota, M.L.; Zanotto-Filho, A.; Müller, C.B.; Pires, A.S.; Meurer, R.T.; Colpo, G.D.; Gelain, D.P.; Kapczinski, F.; et al. Comparison between proliferative and neuron-like SH-SY5Y cells as an in vitro model for Parkinson disease studies. Brain Res. 2010, 1337, 85–94. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, C.; Li, J.V.; Marchesi, J.R.; Plummer, S.; Garaiova, I.; Good, M.A. Long-term multi-species Lactobacillus and Bifidobacterium dietary supplement enhances memory and changes regional brain metabolites in middle-aged rats. Neurobiol. Learn Mem. 2017, 144, 36–47. [Google Scholar] [CrossRef]
- Sun, M.; Bao, W.; Huang, C.; Xia, Z.; Zhang, C.; Wang, G.; Wang, R.; Li, J.; Roux, S.; Li, Q.; et al. A Novel Probiotic Formula, BIOCG, Protects Against Alzheimer’s-Related Cognitive Deficits via Regulation of Dendritic Spine Dynamics. Curr. Alzheimer Res. 2021, 18, 558–572. [Google Scholar] [CrossRef]
- Tan, C.; Liu, Y.; Zhang, H.; Di, C.; Xu, D.; Liang, C.; Zhang, N.; Han, B.; Lang, W. Neuroprotective Effects of Probiotic-Supplemented Diet on Cognitive Behavior of 3xTg-AD Mice. J. Health Eng. 2022, 2022, 4602428. [Google Scholar] [CrossRef] [PubMed]
- Abraham, D.; Feher, J.; Scuderi, G.L.; Szabo, D.; Dobolyi, A.; Cservenak, M.; Juhasz, J.; Ligeti, B.; Pongor, S.; Gomez-Cabrera, M.C.; et al. Exercise and probiotics attenuate the development of Alzheimer’s disease in transgenic mice: Role of microbiome. Exp. Gerontol. 2019, 115, 122–131. [Google Scholar] [CrossRef]
- Kaur, H.; Nagamoto-Combs, K.; Combs, C.K. Impact of modulating gut bacteria using antibiotic, probiotic, and prebiotic interventions in the APPNL-G-F mouse model of Alzheimer disease. Alzheimer’s Dement. 2020, 16, e042122. [Google Scholar] [CrossRef]
- Filali, M.; Lalonde, R.; Theriault, P.; Julien, C.; Calon, F.; Planel, E. Cognitive and non-cognitive behaviors in the triple transgenic mouse model of Alzheimer’s disease expressing mutated APP, PS1, and Mapt (3xTg-AD). Behav. Brain Res. 2012, 234, 334–342. [Google Scholar] [CrossRef]
- Roda, A.R.; Esquerda-Canals, G.; Marti-Clua, J.; Villegas, S. Cognitive Impairment in the 3xTg-AD Mouse Model of Alzheimer’s Disease is Affected by Abeta-ImmunoTherapy and Cognitive Stimulation. Pharmaceutics 2020, 12, 944. [Google Scholar] [CrossRef]
- Rollins, C.P.E.; Gallino, D.; Kong, V.; Ayranci, G.; Devenyi, G.A.; Germann, J.; Chakravarty, M.M. Contributions of a high-fat diet to Alzheimer’s disease-related decline: A longitudinal behavioural and structural neuroimaging study in mouse models. Neuroimage Clin. 2019, 21, 101606. [Google Scholar] [CrossRef]
- Robison, L.S.; Gannon, O.J.; Thomas, M.A.; Salinero, A.E.; Abi-Ghanem, C.; Poitelon, Y.; Belin, S.; Zuloaga, K.L. Role of sex and high-fat diet in metabolic and hypothalamic disturbances in the 3xTg-AD mouse model of Alzheimer’s disease. J. Neuroinflammation 2020, 17, 285. [Google Scholar] [CrossRef] [PubMed]
- Castelli, V.; Benedetti, E.; Antonosante, A.; Catanesi, M.; Pitari, G.; Ippoliti, R.; Cimini, A.; d’Angelo, M. Neuronal Cells Rearrangement During Aging and Neurodegenerative Disease: Metabolism, Oxidative Stress and Organelles Dynamic. Front. Mol. Neurosci. 2019, 12, 132. [Google Scholar] [CrossRef] [PubMed]
- Trevino, S.; Vazquez-Roque, R.A.; Lopez-Lopez, G.; Perez-Cruz, C.; Moran, C.; Handal-Silva, A.; Gonzalez-Vergara, E.; Flores, G.; Guevara, J.; Diaz, A. Metabolic syndrome causes recognition impairments and reduced hippocampal neuronal plasticity in rats. J. Chem. Neuroanat. 2017, 82, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Bloss, E.; Morrison, J.; Hof, P.; Dickstein, D. Influence of aging and neurodegeneration on dendritic spine morphology. Transl. Neurosci. 2011, 2, 49–60. [Google Scholar] [CrossRef]
- Bevan, R.J.; Hughes, T.R.; Williams, P.A.; Good, M.A.; Morgan, B.P.; Morgan, J.E. Retinal ganglion cell degeneration correlates with hippocampal spine loss in experimental Alzheimer’s disease. Acta Neuropathol. Commun. 2020, 8, 216. [Google Scholar] [CrossRef]
- Tackenberg, C.; Ghori, A.; Brandt, R. Thin, stubby or mushroom: Spine pathology in Alzheimer’s disease. Curr. Alzheimer Res. 2009, 6, 261–268. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2020, 17, 157–172. [Google Scholar] [CrossRef]
- Barrientos, R.M.; Kitt, M.M.; Watkins, L.R.; Maier, S.F. Neuroinflammation in the normal aging hippocampus. Neuroscience 2015, 309, 84–99. [Google Scholar] [CrossRef]
- Van Dyken, P.; Lacoste, B. Impact of Metabolic Syndrome on Neuroinflammation and the Blood-Brain Barrier. Front. Neurosci. 2018, 12, 930. [Google Scholar] [CrossRef]
- Kwilasz, A.J.; Grace, P.M.; Serbedzija, P.; Maier, S.F.; Watkins, L.R. The therapeutic potential of interleukin-10 in neuroimmune diseases. Neuropharmacology 2015, 96 Pt A, 55–69. [Google Scholar] [CrossRef]
- Mishra, A.; Kim, H.J.; Shin, A.H.; Thayer, S.A. Synapse loss induced by interleukin-1beta requires pre- and post-synaptic mechanisms. J. Neuroimmune Pharm. 2012, 7, 571–578. [Google Scholar] [CrossRef]
- Quintanilla, R.A.; Orellana, D.I.; González-Billault, C.; Maccioni, R.B. Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp. Cell Res. 2004, 295, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O.; Tschopp, J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef]
- Chen, C.; Ahn, E.H.; Kang, S.S.; Liu, X.; Alam, A.; Ye, K. Gut dysbiosis contributes to amyloid pathology, associated with C/EBPβ/AEP signaling activation in Alzheimer’s disease mouse model. Sci. Adv. 2020, 6, eaba0466. [Google Scholar] [CrossRef]
- Xu, W.L.; Atti, A.R.; Gatz, M.; Pedersen, N.L.; Johansson, B.; Fratiglioni, L. Midlife overweight and obesity increase late-life dementia risk: A population-based twin study. Neurology 2011, 76, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, S.; Misrani, A.; Yang, L. Exploiting Common Aspects of Obesity and Alzheimer’s Disease. Front. Hum. Neurosci. 2020, 14, 602360. [Google Scholar] [CrossRef]
- Ratter, J.M.; Rooijackers, H.M.M.; Hooiveld, G.J.; Hijmans, A.G.M.; de Galan, B.E.; Tack, C.J.; Stienstra, R. In vitro and in vivo Effects of Lactate on Metabolism and Cytokine Production of Human Primary PBMCs and Monocytes. Front. Immunol. 2018, 9, 2564. [Google Scholar] [CrossRef]
- Baker, L.M.; Davies, T.S.; Masetti, G.; Hughes, T.R.; Marchesi, J.R.; Jack, A.A.; Joyce, T.S.C.; Allen, M.D.; Plummer, S.F.; Michael, D.R.; et al. A genome guided evaluation of the Lab4 probiotic consortium. Genomics 2021, 113, 4028–4038. [Google Scholar] [CrossRef] [PubMed]
- Webberley, T.S.; Masetti, G.; Baker, L.M.; Dally, J.; Hughes, T.R.; Marchesi, J.R.; Jack, A.A.; Plummer, S.F.; Ramanathan, G.; Facey, P.D.; et al. The Impact of Lab4 Probiotic Supplementation in a 90-Day Study in Wistar Rats. Front. Nutr. 2021, 8, 778289. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- Belfiore, R.; Rodin, A.; Ferreira, E.; Velazquez, R.; Branca, C.; Caccamo, A.; Oddo, S. Temporal and regional progression of Alzheimer’s disease-like pathology in 3xTg-AD mice. Aging Cell 2019, 18, e12873. [Google Scholar] [CrossRef]
- Javonillo, D.I.; Tran, K.M.; Phan, J.; Hingco, E.; Kramár, E.A.; da Cunha, C.; Forner, S.; Kawauchi, S.; Milinkeviciute, G.; Gomez-Arboledas, A.; et al. Systematic Phenotyping and Characterization of the 3xTg-AD Mouse Model of Alzheimer’s Disease. Front. Neurosci. 2021, 15, 785276. [Google Scholar] [CrossRef]
- Bell, K.F.; de Kort, G.J.; Steggerda, S.; Shigemoto, R.; Ribeiro-da-Silva, A.; Cuello, A.C. Structural involvement of the glutamatergic presynaptic boutons in a transgenic mouse model expressing early onset amyloid pathology. Neurosci. Lett. 2003, 353, 143–147. [Google Scholar] [CrossRef]
- Haam, J.; Yakel, J.L. Cholinergic modulation of the hippocampal region and memory function. J. Neurochem. 2017, 142 (Suppl. 2), 111–121. [Google Scholar] [CrossRef] [PubMed]
- Sirin, S.; Aslim, B. Characterization of lactic acid bacteria derived exopolysaccharides for use as a defined neuroprotective agent against amyloid beta1-42-induced apoptosis in SH-SY5Y cells. Sci. Rep. 2020, 10, 8124. [Google Scholar] [CrossRef] [PubMed]
- Castelli, V.; d’Angelo, M.; Lombardi, F.; Alfonsetti, M.; Antonosante, A.; Catanesi, M.; Benedetti, E.; Palumbo, P.; Cifone, M.G.; Giordano, A.; et al. Effects of the probiotic formulation SLAB51 in in vitro and in vivo Parkinson’s disease models. Aging 2020, 12, 4641–4659. [Google Scholar] [CrossRef] [PubMed]
- Cheon, M.J.; Lim, S.M.; Lee, N.K.; Paik, H.D. Probiotic Properties and Neuroprotective Effects of Lactobacillus buchneri KU200793 Isolated from Korean Fermented Foods. Int. J. Mol. Sci. 2020, 21, 1227. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Lee, N.K.; Paik, H.D. Potential neuroprotective effects of heat-killed Lactococcus lactis KC24 using SH-SY5Y cells against oxidative stress induced by hydrogen peroxide. Food Sci. Biotechnol. 2020, 29, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.; Yeom, Z.; Heo, D.; Lim, Y.H. Neuroprotective effect of Ruminococcus albus on oxidatively stressed SH-SY5Y cells and animals. Sci. Rep. 2017, 7, 14520. [Google Scholar] [CrossRef]
- Lee, N.K.; Lim, S.M.; Cheon, M.J.; Paik, H.D. Physicochemical Analysis of Yogurt Produced by Leuconostoc mesenteroides H40 and Its Effects on Oxidative Stress in Neuronal Cells. Food Sci. Anim. Resour. 2021, 41, 261–273. [Google Scholar] [CrossRef]
- Giordano, S.; Lee, J.; Darley-Usmar, V.M.; Zhang, J. Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS ONE 2012, 7, e44610. [Google Scholar] [CrossRef] [PubMed]
- Macleod, M.R.; Allsopp, T.E.; McLuckie, J.; Kelly, J.S. Serum withdrawal causes apoptosis in SHSY 5Y cells. Brain Res. 2001, 889, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, V.B.; Askari, V.R.; Mousavi, S.H. Ellagic acid reveals promising anti-aging effects against d-galactose-induced aging on human neuroblastoma cell line, SH-SY5Y: A mechanistic study. Biomed. Pharm. 2018, 108, 1712–1724. [Google Scholar] [CrossRef] [PubMed]
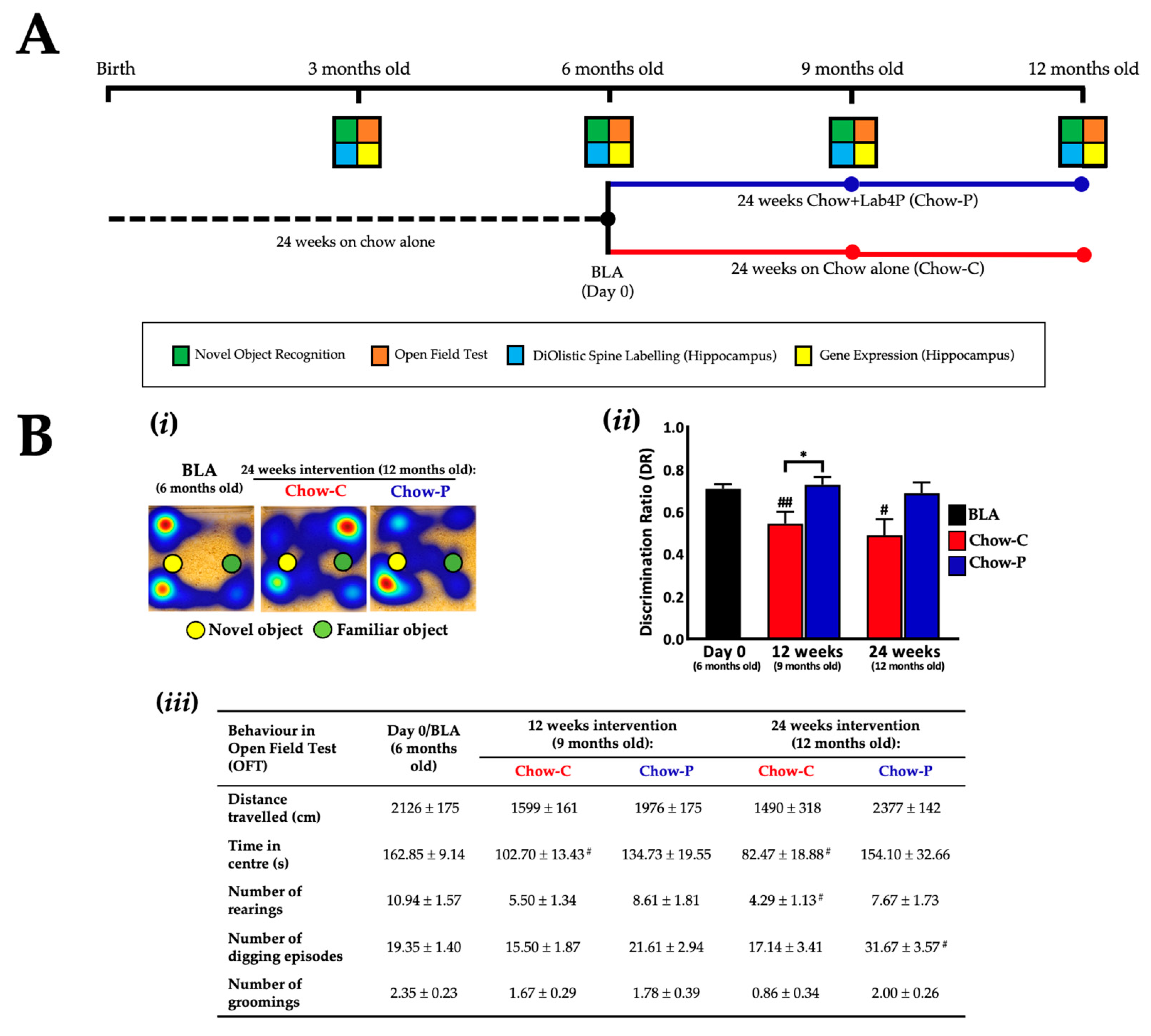
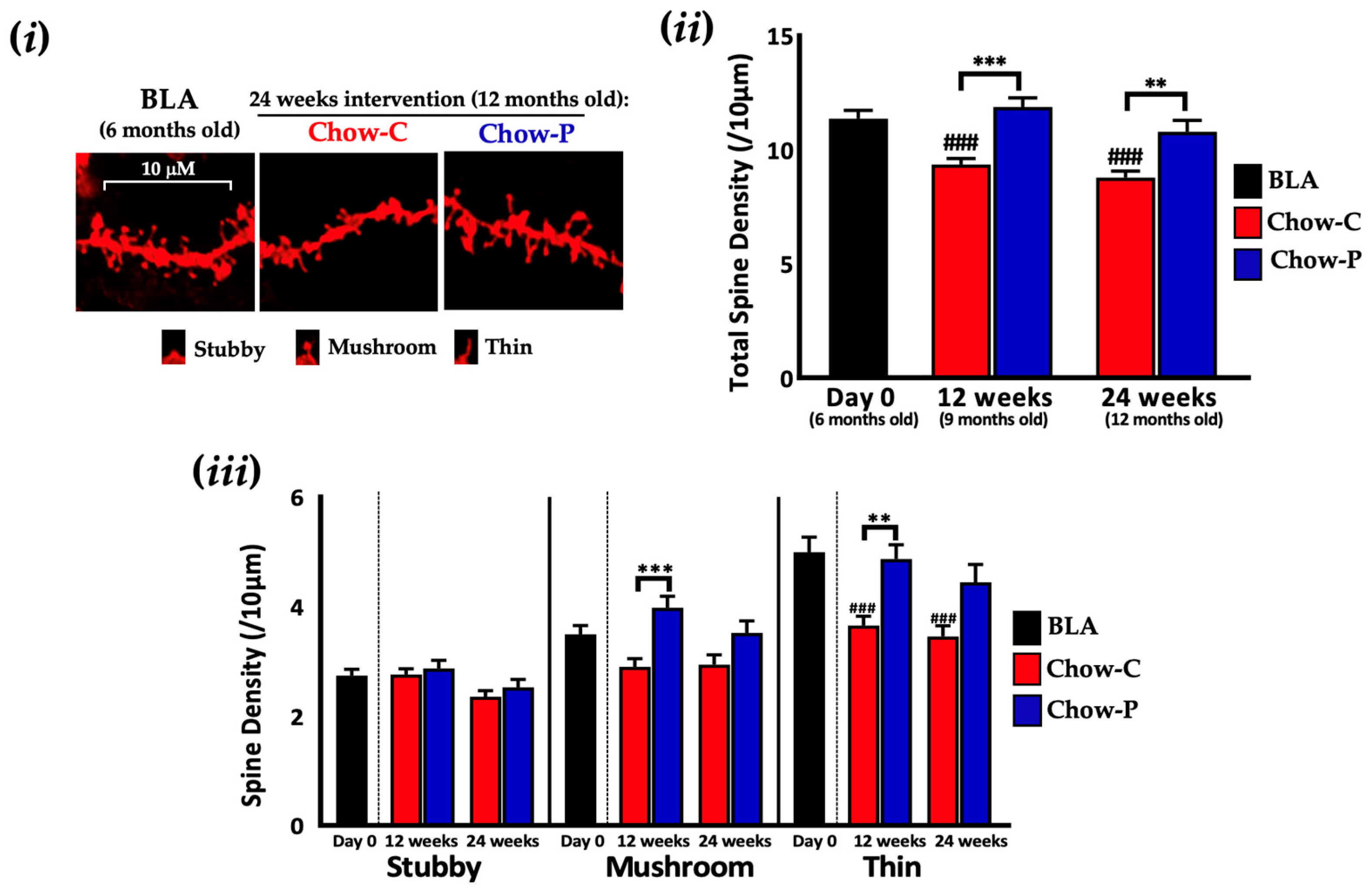
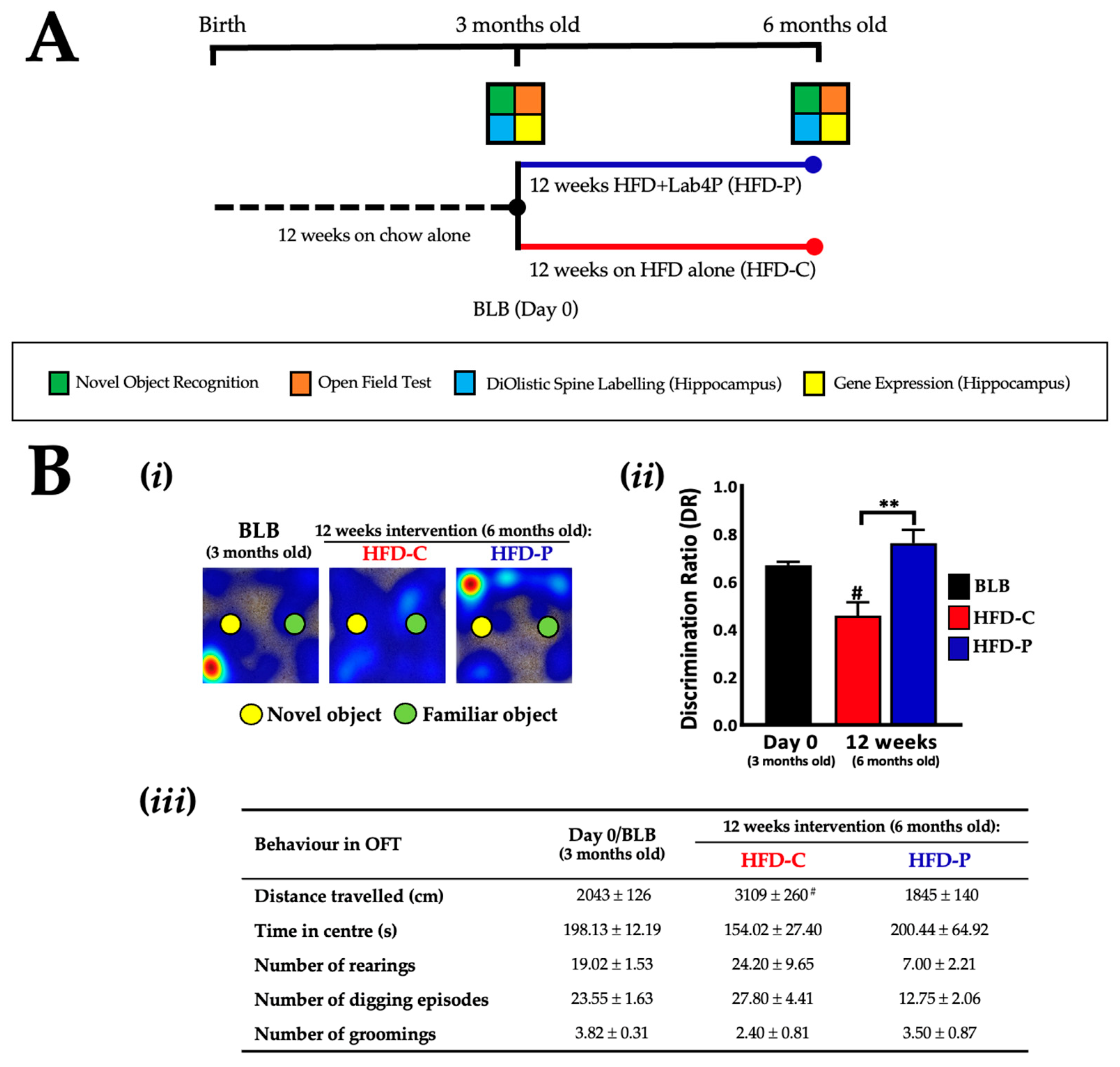
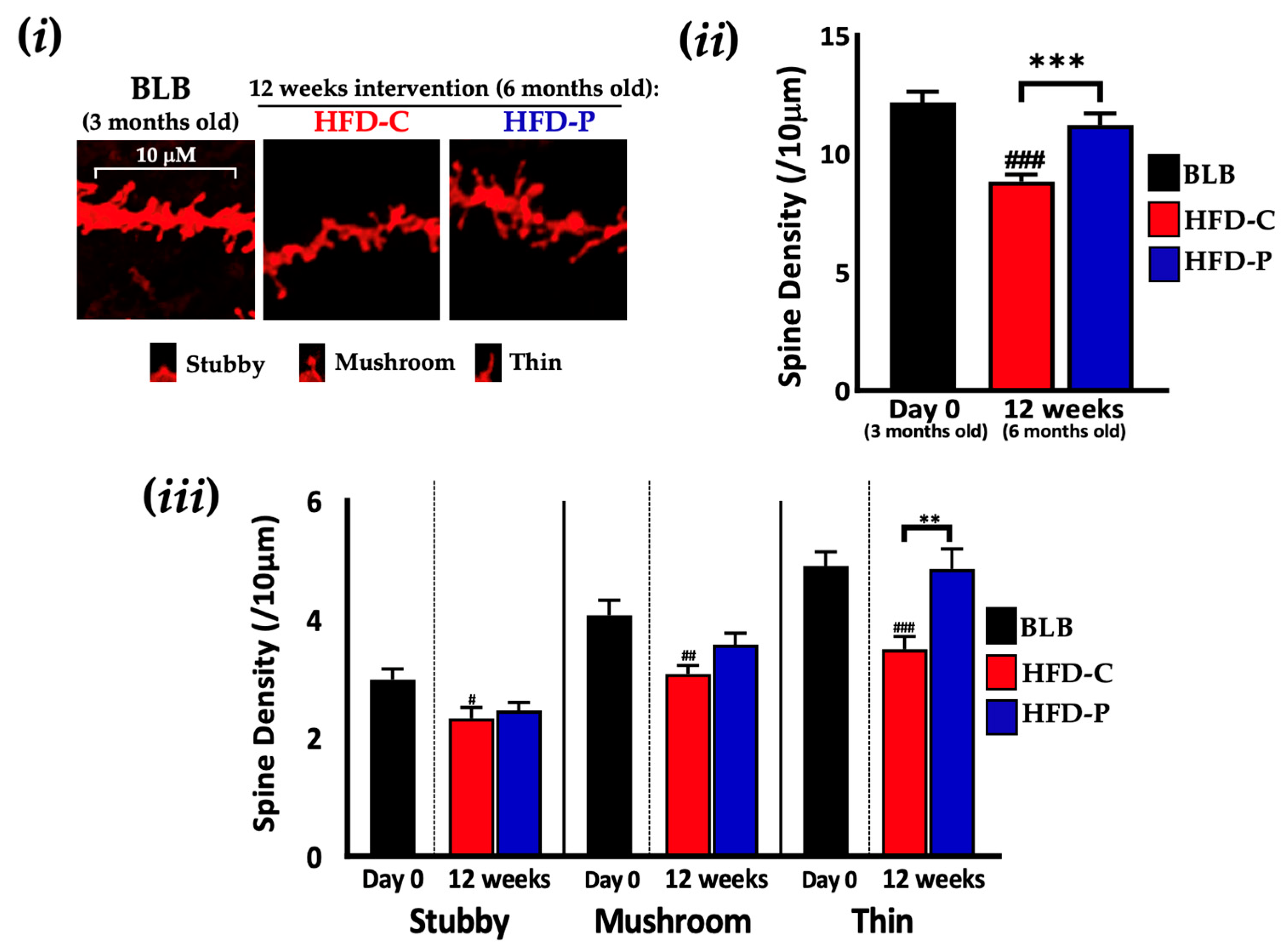
| Gene | Baseline (6 Months Old) | 12 Weeks Intervention (9 Months Old) | 24 Weeks Intervention (12 Months Old) | ||
|---|---|---|---|---|---|
| Chow-C | Chow-P | Chow-C | Chow-P | ||
| Iba-1 | 1.00 ± 0.23 | 1.56 ± 0.34 | 1.22 ± 0.12 | 1.45 ± 0.18 | 1.16 ± 0.35 |
| IL-1β | 1.00 ± 0.25 | 0.77 ± 0.10 | 0.84 ± 0.10 | 1.19 ± 0.22 | 1.01 ± 0.33 |
| IL-6 | 1.00 ± 0.13 | 1.08 ± 0.18 | 0.98 ± 0.19 | 0.71 ± 0.23 | 0.67 ± 0.14 |
| IL-8 | 1.00 ± 0.34 | 0.42 ± 0.05 | 1.01 ± 0.22 | 0.56 ± 0.09 | 0.58 ± 0.13 |
| IL-10 | 1.00 ± 0.10 | 1.06 ± 0.08 | 1.18 ± 0.9 | 1.43 ± 0.26 | 0.48 ± 0.07 *** |
| TNF-α | 1.00 ± 0.18 | 1.38 ± 0.65 | 1.11 ± 0.15 | 2.25 ± 1.05 | 1.51 ± 0.32 |
| Bax:Bcl-2 | 1.00 ± 0.11 | 1.06 ± 0.06 | 1.01 ± 0.04 | 1.16 ± 0.12 | 1.30 ± 0.15 |
| Gene | Baseline (3 Months Old) | 12 Weeks Intervention (6 Months Old) | |
|---|---|---|---|
| HFD-C | HFD-P | ||
| Iba-1 | 1.00 ± 0.17 | 0.79 ± 0.19 | 0.70 ± 0.17 |
| IL-1β | 1.00 ± 0.15 | 5.78 ± 1.42 ### | 1.61 ± 0.36 ** |
| IL-6 | 1.00 ± 0.17 | 2.09 ± 0.56 # | 1.38 ± 0.20 |
| IL-8 | 1.00 ± 0.10 | 1.45 ± 0.28 | 1.19 ± 0.13 |
| IL-10 | 1.00 ± 0.08 | 0.93 ± 0.15 | 0.67 ± 0.14 |
| TNF-α | 1.00 ± 0.16 | 5.58 ± 0.86 ### | 3.16 ± 0.87 *# |
| Bax:Bcl-2 | 1.00 ± 0.06 | 1.04 ± 0.08 | 1.06 ± 0.08 |
| Gene | Vehicle Control | β-Amyloid 1-42 (0.1 μM) | β-Amyloid 1–42 (0.1 μM) + Lab4P CM (50%) Pre-Stimulation |
|---|---|---|---|
| IL-1β | ND | ND | ND |
| IL-6 | 1.00 ± 0.67 | 1.46 ± 0.91 | 0.28 ± 0.35 # |
| IL-8 | 1.00 ± 0.19 | 1.01 ± 0.29 | 3.04 ± 1.01 |
| IL-10 | ND | ND | ND |
| TNF-α | 1.00 ± 0.24 | 0.99 ± 0.71 | 0.35 ± 0.10 |
| Bax:Bcl-2 | 1.00 ± 0.04 | 0.97 ± 0.05 | 1.39 ±0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Webberley, T.S.; Bevan, R.J.; Kerry-Smith, J.; Dally, J.; Michael, D.R.; Thomas, S.; Rees, M.; Morgan, J.E.; Marchesi, J.R.; Good, M.A.; et al. Assessment of Lab4P Probiotic Effects on Cognition in 3xTg-AD Alzheimer’s Disease Model Mice and the SH-SY5Y Neuronal Cell Line. Int. J. Mol. Sci. 2023, 24, 4683. https://doi.org/10.3390/ijms24054683
Webberley TS, Bevan RJ, Kerry-Smith J, Dally J, Michael DR, Thomas S, Rees M, Morgan JE, Marchesi JR, Good MA, et al. Assessment of Lab4P Probiotic Effects on Cognition in 3xTg-AD Alzheimer’s Disease Model Mice and the SH-SY5Y Neuronal Cell Line. International Journal of Molecular Sciences. 2023; 24(5):4683. https://doi.org/10.3390/ijms24054683
Chicago/Turabian StyleWebberley, Thomas S., Ryan J. Bevan, Joshua Kerry-Smith, Jordanna Dally, Daryn R. Michael, Sophie Thomas, Meg Rees, James E. Morgan, Julian R. Marchesi, Mark A. Good, and et al. 2023. "Assessment of Lab4P Probiotic Effects on Cognition in 3xTg-AD Alzheimer’s Disease Model Mice and the SH-SY5Y Neuronal Cell Line" International Journal of Molecular Sciences 24, no. 5: 4683. https://doi.org/10.3390/ijms24054683
APA StyleWebberley, T. S., Bevan, R. J., Kerry-Smith, J., Dally, J., Michael, D. R., Thomas, S., Rees, M., Morgan, J. E., Marchesi, J. R., Good, M. A., Plummer, S. F., Wang, D., & Hughes, T. R. (2023). Assessment of Lab4P Probiotic Effects on Cognition in 3xTg-AD Alzheimer’s Disease Model Mice and the SH-SY5Y Neuronal Cell Line. International Journal of Molecular Sciences, 24(5), 4683. https://doi.org/10.3390/ijms24054683






