Separase and Roads to Disengage Sister Chromatids during Anaphase
Abstract
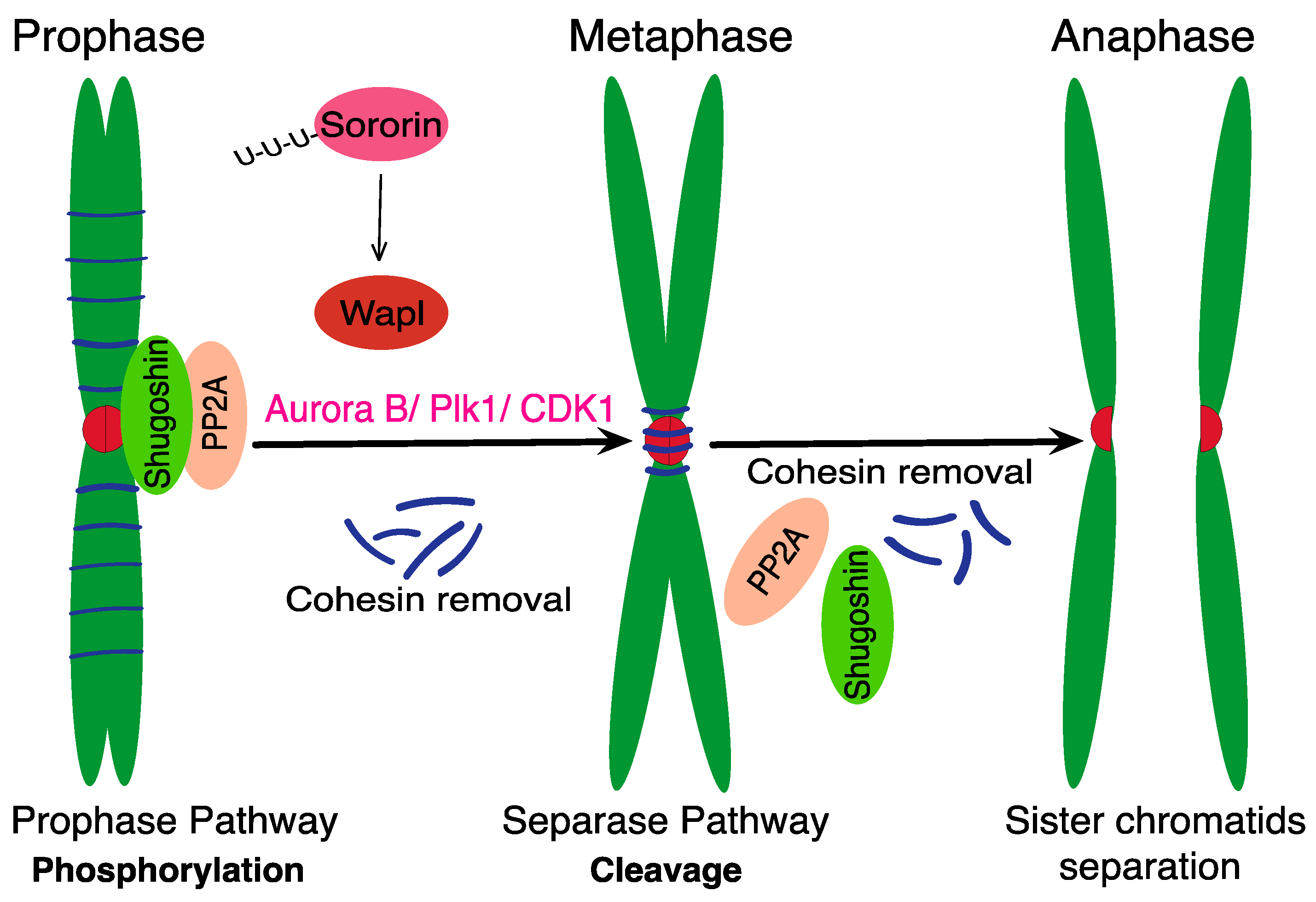
1. Introduction
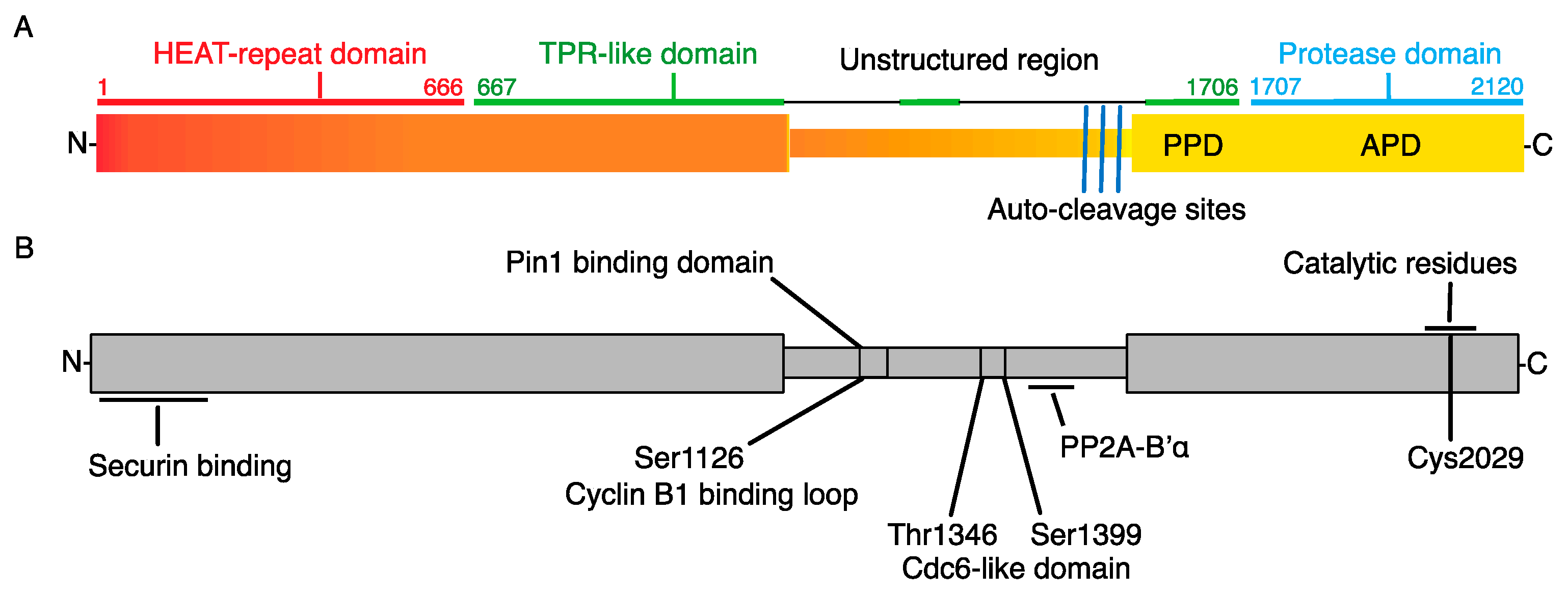
2. Separase Structure and Functional Motifs
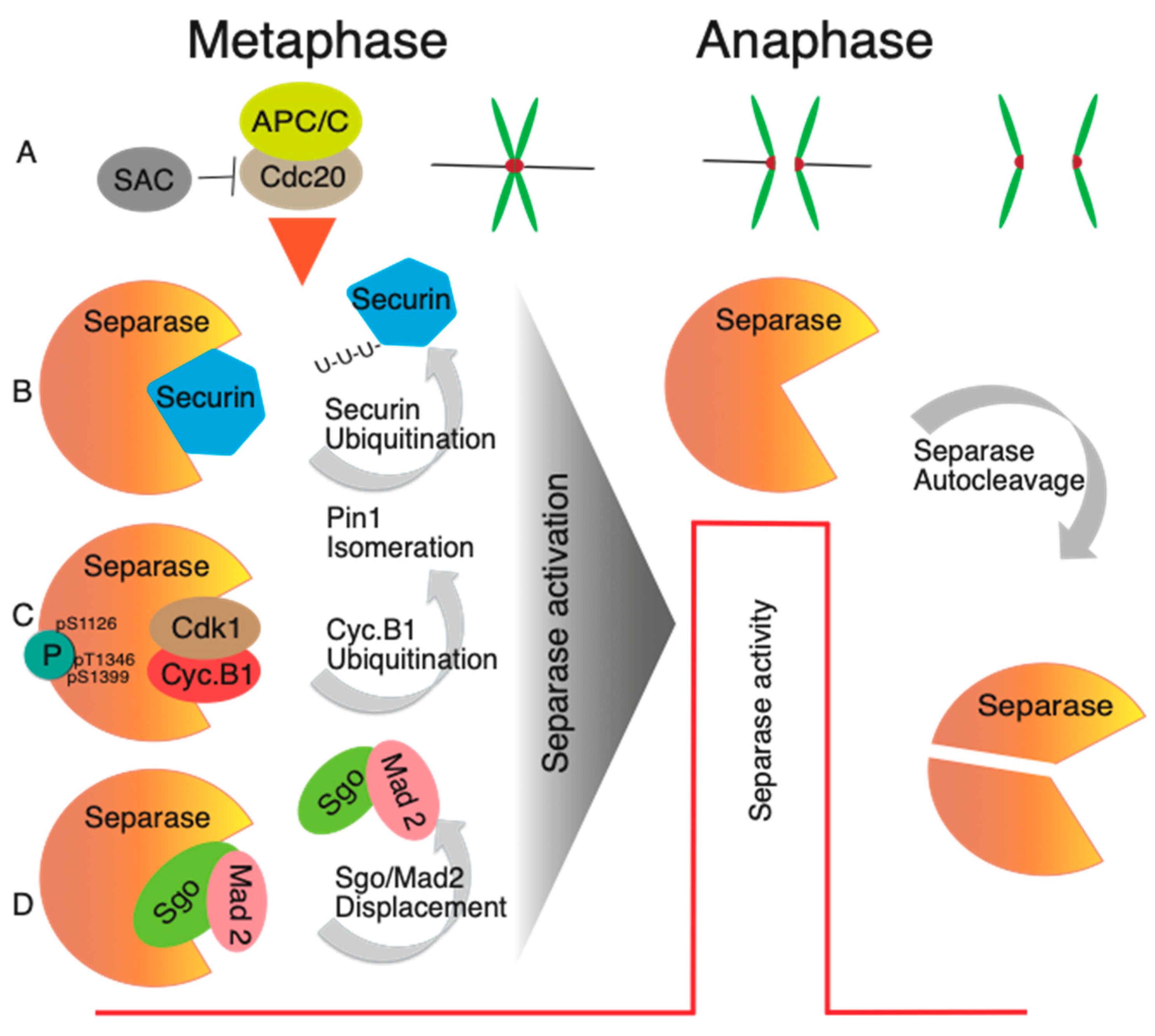
3. How Is the Activation of Separase in Mitosis Controlled?
4. How Is Separase Activation Linked to Assembly of the Spindle?
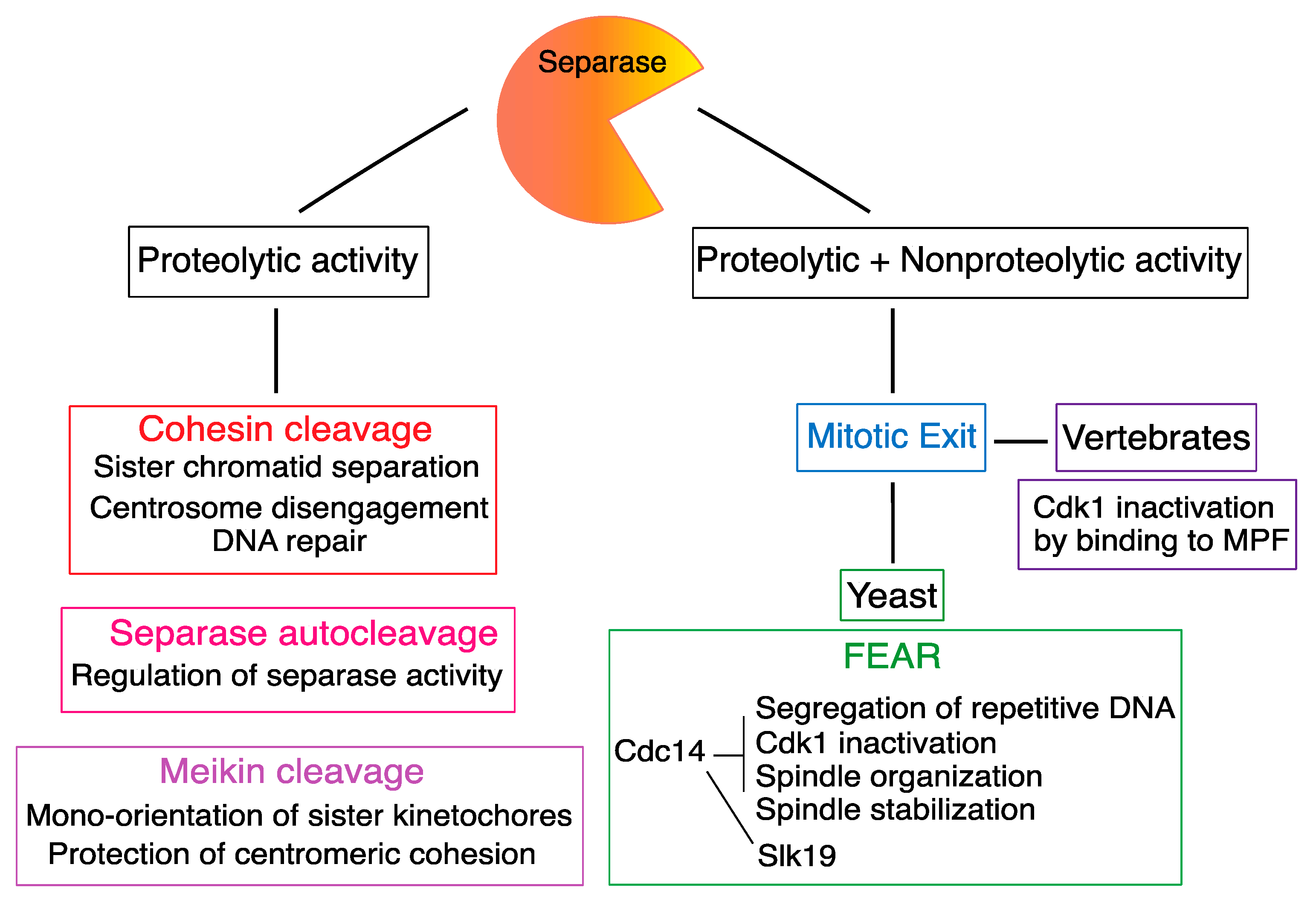
5. Other Targets Than Cohesin?
6. Role and Regulation of Separase during Meiosis and Early Development
7. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| APC/C | Anaphase-promoting complex/cyclosome |
| APD | Active protease domain |
| ARM | Armadillo repeat |
| BUB | Budding uninhibited by benzimidazoles |
| CaMKII | Calcium/calmodulin-dependent protein kinase II |
| Cdc14 | Cell division cycle 14 |
| CDC20 | Cell division cycle 20 |
| Cdc7-dbf4 | Cell division cycle 7-related protein kinase, DBF4-dependent kinase |
| Cdh1 | Cdc20 homolog 1 |
| CDK | Cyclin-dependent kinase |
| CksI | Cyclin-dependent protein kinase regulatory subunit I |
| CSF | Cytostatic factor |
| D-box | Destruction box |
| DNA | Deoxyribonucleic acid |
| ESPl1 | Extra spindle poles-like protein 1 |
| FEAR | Fourteen early anaphase release |
| MAD | Mitotic arrest-deficient protein |
| MAU2 | MAU2 sister chromatid cohesion factor |
| MCC | Mitotic checkpoint complex |
| MEIKIN | Meiosis-specific kinetochore protein |
| MPS1 | Monopolar spindle 1 kinase |
| NES | Nuclear export sequence |
| NIPBL | Nipped-B-like protein |
| Pds | Precocious dissociation of sisters |
| Pin1 | Peptidyl-prolyl cis/trans isomerase |
| Plk1 | Polo-like kinase 1 |
| PP2A | Protein phosphatase 2A |
| PPD | Pseudo protease domain |
| Rec 8 | Meiotic recombination protein Rec8 |
| RNA | Ribonucleic acid |
| SAC | Spindle assembly checkpoint |
| SCC | Sister chromatid cohesion protein |
| SGO | Shugoshin |
| SMC | Structural maintenance of chromosomes |
| Slk19 | Kinetochore-associated protein Slk19 |
| TRIP13 | Thyroid hormone receptor interactor 13 |
| Wapl | Wings apart-like protein |
References
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Guacci, V.; Koshland, D.; Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997, 91, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Haering, C.H.; Farcas, A.M.; Arumugam, P.; Metson, J.; Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 2008, 454, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef]
- Peters, J.M.; Nishiyama, T. Sister chromatid cohesion. Cold Spring Harb. Perspect. Biol. 2012, 4, a011130. [Google Scholar] [CrossRef]
- Morales, C.; Losada, A. Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol. 2018, 52, 51–57. [Google Scholar] [CrossRef]
- Yatskevich, S.; Rhodes, J.; Nasmyth, K. Organization of Chromosomal DNA by SMC Complexes. Annu. Rev. Genet. 2019, 53, 445–482. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Wattendorf, L.; Marzouk, S.; Blower, M.D. Cohesin: Behind Dynamic Genome Topology and Gene Expression Reprogramming. Trends Cell Biol. 2021, 31, 760–773. [Google Scholar] [CrossRef]
- Davidson, I.F.; Peters, J.M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 445–464. [Google Scholar] [CrossRef]
- Higashi, T.L.; Uhlmann, F. SMC complexes: Lifting the lid on loop extrusion. Curr. Opin. Cell. Biol. 2022, 74, 13–22. [Google Scholar] [CrossRef]
- Hartman, T.; Stead, K.; Koshland, D.; Guacci, V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 2000, 151, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Losada, A.; Yokochi, T.; Kobayashi, R.; Hirano, T. Identification and Characterization of Sa/Scc3p Subunits in the Xenopus and Human Cohesin Complexes. J. Cell Biol. 2000, 150, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Kueng, S.; Hegemann, B.; Peters, B.H.; Lipp, J.J.; Schleiffer, A.; Mechtler, K.; Peters, J.M. Wapl controls the dynamic association of cohesin with chromatin. Cell 2006, 127, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Haering, C.H.; Nasmyth, K. Building and breaking bridges between sister chromatids. Bioessays 2003, 25, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Makrantoni, V.; Marston, A.L. Cohesin and chromosome segregation. Curr. Biol. 2018, 28, R688–R693. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; Shirayama, M.; Shevchenko, A.; Tanaka, T.; Toth, A.; Shevchenko, A.; Nasmyth, K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 2000, 5, 243–254. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Hirano, T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004, 14, 1598–1603. [Google Scholar] [CrossRef]
- Tonkin, E.T.; Wang, T.J.; Lisgo, S.; Bamshad, M.J.; Strachan, T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004, 36, 636–641. [Google Scholar] [CrossRef]
- Watrin, E.; Schleiffer, A.; Tanaka, K.; Eisenhaber, F.; Nasmyth, K.; Peters, J.M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006, 16, 863–874. [Google Scholar] [CrossRef]
- Rankin, S.; Ayad, N.G.; Kirschner, M.W. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol. Cell 2005, 18, 185–200. [Google Scholar] [CrossRef]
- Ladurner, R.; Kreidl, E.; Ivanov, M.P.; Ekker, H.; Idarraga-Amado, M.H.; Busslinger, G.A.; Wutz, G.; Cisneros, D.A.; Peters, J.M. Sororin actively maintains sister chromatid cohesion. EMBO J. 2016, 35, 635–653. [Google Scholar] [CrossRef]
- Sundin, O.; Varshavsky, A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: Dissection of the final stages of SV40 DNA replication. Cell 1981, 25, 659–669. [Google Scholar] [CrossRef]
- Murray, A.W.; Schultes, N.P.; Szostak, J.W. Chromosome length controls mitotic chromosome segregation in yeast. Cell 1986, 45, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Uluocak, P.; Helmhart, W.; Nasmyth, K. Cohesin’s concatenation of sister DNAs maintains their intertwining. Mol. Cell 2011, 44, 97–107. [Google Scholar] [CrossRef]
- Coelho, P.A.; Queiroz-Machado, J.; Sunkel, C.E. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 2003, 116, 4763–4776. [Google Scholar] [CrossRef] [PubMed]
- Charbin, A.; Bouchoux, C.; Uhlmann, F. Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic. Acids Res. 2014, 42, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Piskadlo, E.; Oliveira, R.A. A Topology-Centric View on Mitotic Chromosome Architecture. Int. J. Mol. Sci. 2017, 18, 2751. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Zhang, Z.; Mukhina, M.; Zickler, D.; Kleckner, N. Sister chromatids separate during anaphase in a three-stage program as directed by interaxis bridges. Proc. Natl. Acad. Sci. USA 2022, 119, e2123363119. [Google Scholar] [CrossRef]
- Waizenegger, I.C.; Hauf, S.; Meinke, A.; Peters, J.M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 2000, 103, 399–410. [Google Scholar] [CrossRef]
- Haarhuis, J.H.; Elbatsh, A.M.; Rowland, B.D. Cohesin and its regulation: On the logic of X-shaped chromosomes. Dev. Cell 2014, 31, 7–18. [Google Scholar] [CrossRef]
- Tedeschi, A.; Wutz, G.; Huet, S.; Jaritz, M.; Wuensche, A.; Schirghuber, E.; Davidson, I.F.; Tang, W.; Cisneros, D.A.; Bhaskara, V.; et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 2013, 501, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, T.S.; Kawashima, S.A.; Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004, 427, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Rabitsch, K.P.; Gregan, J.; Schleiffer, A.; Javerzat, J.P.; Eisenhaber, F.; Nasmyth, K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004, 14, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Salic, A.; Waters, J.C.; Mitchison, T.J. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 2004, 118, 567–578. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Sakuno, T.; Ishiguro, K.; Iemura, S.; Natsume, T.; Kawashima, S.A.; Watanabe, Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 2006, 441, 46–52. [Google Scholar] [CrossRef]
- Riedel, C.G.; Katis, V.L.; Katou, Y.; Mori, S.; Itoh, T.; Helmhart, W.; Gálová, M.; Petronczki, M.; Gregan, J.; Cetin, B.; et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 2006, 441, 53–61. [Google Scholar] [CrossRef]
- Marston, A.L. Shugoshins: Tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol. Cell Biol. 2015, 35, 634–648. [Google Scholar] [CrossRef]
- Kudo, N.R.; Wassmann, K.; Anger, M.; Schuh, M.; Wirth, K.G.; Xu, H.; Helmhart, W.; Kudo, H.; McKay, M.; Maro, B.; et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 2006, 126, 135–146. [Google Scholar] [CrossRef]
- Silva, M.C.C.; Powell, S.; Ladstätter, S.; Gassler, J.; Stocsits, R.; Tedeschi, A.; Peters, J.M.; Tachibana, K. Wapl releases Scc1-cohesin and regulates chromosome structure and segregation in mouse oocytes. J. Cell Biol. 2020, 219, e201906100. [Google Scholar] [CrossRef]
- Uhlmann, F.; Lottspeich, F.; Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 1999, 400, 37–42. [Google Scholar] [CrossRef]
- Uhlmann, F.; Wernic, D.; Poupart, M.A.; Koonin, E.V.; Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 2000, 103, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Hauf, S.; Waizenegger, I.C.; Peters, J.M. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 2001, 293, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Wirth, K.G.; Wutz, G.; Kudo, N.R.; Desdouets, C.; Zetterberg, A.; Taghybeeglu, S.; Seznec, J.; Ducos, G.M.; Ricci, R.; Firnberg, N.; et al. Separase: A universal trigger for sister chromatid disjunction but not chromosome cycle progression. J. Cell Biol. 2006, 172, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Baum, P.; Yip, C.; Goetsch, L.; Byers, B. A yeast gene essential for regulation of spindle pole duplication. Mol. Cell Biol. 1988, 8, 5386–5397. [Google Scholar]
- Uzawa, S.; Samejima, I.; Hirano, T.; Tanaka, K.; Yanagida, M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell 1990, 62, 913–925. [Google Scholar] [CrossRef]
- Buonomo, S.B.; Clyne, R.K.; Fuchs, J.; Loidl, J.; Uhlmann, F.; Nasmyth, K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 2000, 103, 387–398. [Google Scholar] [CrossRef]
- Alexandru, G.; Uhlmann, F.; Mechtler, K.; Poupart, M.A.; Nasmyth, K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 2001, 105, 459–472. [Google Scholar] [CrossRef]
- Hauf, S.; Roitinger, E.; Koch, B.; Dittrich, C.M.; Mechtler, K.; Peters, J.M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005, 3, e69. [Google Scholar] [CrossRef]
- Katis, V.L.; Lipp, J.J.; Imre, R.; Bogdanova, A.; Okaz, E.; Habermann, B.; Mechtler, K.; Nasmyth, K.; Zachariae, W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell 2010, 18, 397–409. [Google Scholar] [CrossRef]
- Viadiu, H.; Stemmann, O.; Kirschner, M.W.; Walz, T. Domain structure of separase and its binding to securin as determined by EM. Nat. Struct. Mol. Biol. 2005, 12, 552–553. [Google Scholar] [CrossRef]
- Luo, S.; Tong, L. Molecular mechanism for the regulation of yeast separase by securin. Nature 2017, 542, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Boland, A.; Martin, T.G.; Zhang, Z.; Yang, J.; Bai, X.C.; Chang, L.; Scheres, S.H.; Barford, D. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2017, 24, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Luo, X.; Yu, H. Structural basis of cohesin cleavage by separase. Nature 2016, 532, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kucej, M.; Fan, H.Y.; Yu, H.; Sun, Q.Y.; Zou, H. Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner. Cell 2009, 137, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Hellmuth, S.; Gutiérrez-Caballero, C.; Llano, E.; Pendás, A.M.; Stemmann, O. Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Hellmuth, S.; Rata, S.; Brown, A.; Heidmann, S.; Novak, B.; Stemmann, O. Human chromosome segregation involves multi-layered regulation of separase by the peptidyl-prolyl-isomerase Pin1. Mol. Cell 2015, 58, 495–506. [Google Scholar] [CrossRef]
- Stemmann, O.; Zou, H.; Gerber, S.A.; Gygi, S.P.; Kirschner, M.W. Dual inhibition of sister chromatid separation at metaphase. Cell 2001, 107, 715–726. [Google Scholar] [CrossRef]
- Yu, J.; Raia, P.; Ghent, C.M.; Raisch, T.; Sadian, Y.; Cavadini, S.; Sabale, P.M.; Barford, D.; Raunser, S.; Morgan, D.O.; et al. Structural basis of human separase regulation by securin and CDK1-cyclin B1. Nature 2021, 596, 138–142. [Google Scholar] [CrossRef]
- Holland, A.J.; Taylor, S.S. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J. Cell Sci. 2006, 119, 3325–3336. [Google Scholar] [CrossRef]
- Boos, D.; Kuffer, C.; Lenobel, R.; Körner, R.; Stemmann, O. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J. Biol. Chem. 2008, 283, 816–823. [Google Scholar] [CrossRef]
- Gorr, I.H.; Boos, D.; Stemmann, O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol. Cell 2005, 19, 135–141. [Google Scholar] [CrossRef]
- Holland, A.J.; Böttger, F.; Stemmann, O.; Taylor, S.S. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J. Biol. Chem. 2007, 282, 24623–24632. [Google Scholar] [CrossRef] [PubMed]
- Waizenegger, I.; Giménez-Abián, J.F.; Wernic, D.; Peters, J.M. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 2002, 12, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Stemman, O.; Anderson, J.S.; Mann, M.; Kirschner, M.W. Anaphase specific auto-cleavage of separase. FEBS Lett. 2002, 528, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Funabiki, H.; Kumada, K.; Yanagida, M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996, 15, 6617–6628. [Google Scholar] [CrossRef] [PubMed]
- Funabiki, H.; Yamano, H.; Kumada, K.; Nagao, K.; Hunt, T.; Yanagida, M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature 1996, 381, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Guacci, V.; Koshland, D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 1996, 133, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Fix, O.; Peters, J.M.; Kirschner, M.W.; Koshland, D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996, 10, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; Zachariae, W.; Michaelis, C.; Shevchenko, A.; Mann, M.; Nasmyth, K. An ESP1/PDS1 Complex Regulates Loss of Sister Chromatid Cohesion at the Metaphase to Anaphase Transition in Yeast. Cell 1998, 93, 1067–1076. [Google Scholar] [CrossRef]
- Zou, H.; McGarry, T.J.; Bernal, T.; Kirschner, M.W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 1999, 285, 418–422. [Google Scholar] [CrossRef]
- Rosen, L.E.; Klebba, J.E.; Asfaha, J.B.; Ghent, C.M.; Campbell, M.G.; Cheng, Y.; Morgan, D.O. Cohesin cleavage by separase is enhanced by a substrate motif distinct from the cleavage site. Nat. Commun. 2019, 10, 5189. [Google Scholar] [CrossRef] [PubMed]
- Shindo, N.; Kumada, K.; Hirota, T. Separase sensor reveals dual roles for separase coordinating cohesin cleavage and cdk1 inhibition. Dev. Cell 2012, 23, 112–123. [Google Scholar] [CrossRef]
- Hellmuth, S.; Böttger, F.; Pan, C.; Mann, M.; Stemmann, O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014, 33, 1134–1147. [Google Scholar] [CrossRef]
- Thomas, C.; Wetherall, B.; Levasseur, M.D.; Harris, R.J.; Kerridge, S.T.; Higgins, J.M.G.; Davies, O.R.; Madgwick, S. A prometaphase mechanism of securin destruction is essential for meiotic progression in mouse oocytes. Nat. Commun. 2021, 12, 4322. [Google Scholar] [CrossRef]
- Kishimoto, T. MPF-based meiotic cell cycle control: Half a century of lessons from starfish oocytes. Proc. Jpn. Acad. Ser. B 2018, 94, 180–203. [Google Scholar] [CrossRef]
- Crncec, A.; Hochegger, H. Triggering mitosis. FEBS Lett. 2019, 593, 2868–2888. [Google Scholar] [CrossRef]
- Holder, J.; Poser, E.; Barr, F.A. Getting out of mitosis: Spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett. 2019, 593, 2908–2924. [Google Scholar] [CrossRef] [PubMed]
- Radonova, L.; Pauerova, T.; Jansova, D.; Danadova, J.; Skultety, M.; Kubelka, M.; Anger, M. Cyclin A1 in Oocytes Prevents Chromosome Segregation and Anaphase Entry. Sci. Rep. 2020, 10, 7455. [Google Scholar] [CrossRef] [PubMed]
- Papi, M.; Berdougo, E.; Randall, C.L.; Ganguly, S.; Jallepalli, P.V. Multiple roles for separase auto-cleavage during the G2/M transition. Nat. Cell Biol. 2005, 7, 1029–1035. [Google Scholar] [CrossRef]
- Shindo, N.; Kumada, K.; Iemura, K.; Yasuda, J.; Fujimori, H.; Mochizuki, M.; Tamai, K.; Tanaka, K.; Hirota, T. Autocleavage of separase suppresses its premature activation by promoting binding to cyclin B1. Cell Rep. 2022, 41, 111723. [Google Scholar] [CrossRef] [PubMed]
- Hellmuth, S.; Gómez-H, L.; Pendás, A.M.; Stemmann, O. Securin-independent regulation of separase by checkpoint-induced shugoshin-MAD2. Nature 2020, 580, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.; Schultz, R.M.; Lampson, M.A. Age-dependent susceptibility of chromosome cohesion to premature separase activation in mouse oocytes. Biol. Reprod. 2011, 85, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, H.; Zou, H. Nuclear exclusion of separase prevents cohesin cleavage in interphase cells. Cell Cycle 2006, 5, 2537–2542. [Google Scholar] [CrossRef] [PubMed]
- Hornig, N.C.; Knowles, P.P.; McDonald, N.Q.; Uhlmann, F. The dual mechanism of separase regulation by securin. Curr. Biol. 2002, 12, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Pines, J.; Desai, A. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 2021, 117, 86–98. [Google Scholar] [CrossRef]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef]
- McVey, S.L.; Cosby, J.K.; Nannas, N.J. Aurora B Tension Sensing Mechanisms in the Kinetochore Ensure Accurate Chromosome Segregation. Int. J. Mol. Sci. 2021, 22, 8818. [Google Scholar] [CrossRef]
- Sudakin, V.; Ganoth, D.; Dahan, A.; Heller, H.; Hershko, J.; Luca, F.C.; Ruderman, J.V.; Hershko, A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 1995, 6, 185–197. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 1970, 66, 352–359. [Google Scholar] [CrossRef]
- King, R.W.; Peters, J.M.; Tugendreich, S.; Rolfe, M.; Hieter, P.; Kirschner, M.W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995, 81, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef]
- King, R.W.; Glotzer, M.; Kirschner, M.W. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell 1996, 7, 1343–1357. [Google Scholar] [CrossRef]
- Gregan, J.; Polakova, S.; Zhang, L.; Tolić-Nørrelykke, I.M.; Cimini, D. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 2011, 21, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Adachi, Y.; Yanagida, M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 2004, 430, 1044–1048. [Google Scholar] [CrossRef]
- McAleenan, A.; Clemente-Blanco, A.; Cordon-Preciado, V.; Sen, N.; Esteras, M.; Jarmuz, A.; Aragón, L. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature 2013, 493, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Stegmeier, F.; Visintin, R.; Amon, A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 2002, 108, 207–220. [Google Scholar] [CrossRef]
- D’Amours, D.; Amon, A. At the interface between signaling and executing anaphase--Cdc14 and the FEAR network. Genes Dev. 2004, 18, 2581–2595. [Google Scholar] [CrossRef]
- Sullivan, M.; Lehane, C.; Uhlmann, F. Orchestrating anaphase and mitotic exit: Separase cleavage and localization of Slk19. Nat. Cell Biol. 2001, 3, 771–777. [Google Scholar] [CrossRef]
- Wurzenberger, C.; Gerlich, D.W. Phosphatases: Providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 2011, 12, 469–482. [Google Scholar] [CrossRef]
- Kim, J.; Ishiguro, K.; Nambu, A.; Akiyoshi, B.; Yokobayashi, S.; Kagami, A.; Ishiguro, T.; Pendas, A.M.; Takeda, N.; Sakakibara, Y.; et al. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 2015, 517, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Maier, N.K.; Ma, J.; Lampson, M.A.; Cheeseman, I.M. Separase cleaves the kinetochore protein Meikin at the meiosis I/II transition. Dev. Cell 2021, 56, 2192–2206.e8. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Centrosome duplication: Of rules and licenses. Trends Cell Biol. 2007, 17, 215–221. [Google Scholar] [CrossRef]
- Tsou, M.F.; Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006, 442, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Tsou, M.F.; Wang, W.J.; George, K.A.; Uryu, K.; Stearns, T.; Jallepalli, P.V. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 2009, 17, 344–354. [Google Scholar] [CrossRef]
- Schöckel, L.; Möckel, M.; Mayer, B.; Boos, D.; Stemmann, O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 2011, 13, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Ohsumi, K.; Iwabuchi, M.; Kawamata, T.; Ono, Y.; Takahashi, M. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr. Biol. 2012, 22, 915–921. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Nasmyth, K. Cohesin cleavage is insufficient for centriole disengagement in Drosophila. Curr. Biol. 2013, 23, R601–R603. [Google Scholar] [CrossRef]
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef]
- Petronczki, M.; Siomos, M.F.; Nasmyth, K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell 2003, 112, 423–440. [Google Scholar] [CrossRef]
- Terret, M.E.; Wassmann, K.; Waizenegger, I.; Maro, B.; Peters, J.-M.; Verlhac, M.-H. The Meiosis I-to-Meiosis II Transition in Mouse Oocytes Requires Separase Activity. Curr. Biol. 2003, 13, 1797–1802. [Google Scholar] [CrossRef] [PubMed]
- Herbert, M.; Levasseur, M.; Homer, H.; Yallop, K.; Murdoch, A.; McDougall, A. Homologue disjunction in mouse oocytes requires proteolysis of securin and cyclin B1. Nat. Cell Biol. 2003, 5, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Wassmann, K. Separase Control and Cohesin Cleavage in Oocytes: Should I Stay or Should I Go. Cells 2022, 11, 3399. [Google Scholar] [CrossRef]
- Nabti, I.; Reis, A.; Levasseur, M.; Stemmann, O.; Jones, K.T. Securin and not CDK1/cyclin B1 regulates sister chromatid disjunction during meiosis II in mouse eggs. Dev. Biol. 2008, 321, 379–386. [Google Scholar] [CrossRef]
- Nabti, I.; Grimes, R.; Sarna, H.; Marangos, P.; Carroll, J. Maternal age-dependent APC/C-mediated decrease in securin causes premature sister chromatid separation in meiosis II. Nat. Commun. 2017, 8, 15346. [Google Scholar] [CrossRef]
- Huang, X.; Andreu-Vieyra, C.V.; Wang, M.; Cooney, A.J.; Matzuk, M.M.; Zhang, P. Preimplantation mouse embryos depend on inhibitory phosphorylation of separase to prevent chromosome missegregation. Mol. Cell Biol. 2009, 29, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018, 32, 620–638. [Google Scholar] [CrossRef]
- Mei, J.; Huang, X.; Zhang, P. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr. Biol. 2001, 11, 1197–1201. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, R.; Melmed, S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 2001, 15, 1870–1879. [Google Scholar] [CrossRef]
- Henschke, L.; Frese, M.; Hellmuth, S.; Marx, A.; Stemmann, O.; Mayer, T.U. Identification of Bioactive Small Molecule Inhibitors of Separase. ACS Chem. Biol. 2019, 14, 2155–2159. [Google Scholar] [CrossRef]
- Spiess, B.; Kleiner, H.; Flach, J.; Fabarius, A.; Saussele, S.; Hofmann, W.K.; Seifarth, W. Separase activity distribution can be a marker of major molecular response and proliferation of CD34+ cells in TKI-treated chronic myeloid leukemia patients. Ann. Hematol. 2020, 99, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.; Kusano, Y.; Hirota, T. Unraveling pathologies underlying chromosomal instability in cancers. Cancer Sci. 2021, 112, 2975–2983. [Google Scholar] [CrossRef] [PubMed]
- Gurvits, N.; Löyttyniemi, E.; Nykänen, M.; Kuopio, T.; Kronqvist, P.; Talvinen, K. Separase is a marker for prognosis and mitotic activity in breast cancer. Br. J. Cancer 2017, 117, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Scorsone, K.; Ge, G.; Kaffes, C.C.; Dobrolecki, L.E.; Mukherjee, M.; Lewis, M.T.; Berg, S.; Stephan, C.C.; Pati, D. Identification and Characterization of Separase Inhibitors (Sepins) for Cancer Therapy. J. Biomol. Screen 2014, 19, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Pati, D. Separase Inhibitor Sepin-1 Inhibits Foxm1 Expression and Breast Cancer Cell Growth. J. Cancer Sci. Ther. 2018, 10, 517. [Google Scholar] [CrossRef]
| Inhibitor | Effect | References |
|---|---|---|
| Sepin-1 | Identified by Rad21 cleavage in vitro, good in vitro inhibitory effect and selective in vivo inhibition, molecular mechanisms of inhibition involve transcription factor FoxM1. | [124,125] |
| SIC1, 3, 5 | Identified by Rad21 cleavage in vitro, the in vivo activity requires lower Separase levels. | [120] |
| SIC5−6 | Improved in vitro inhibitory effect. | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konecna, M.; Abbasi Sani, S.; Anger, M. Separase and Roads to Disengage Sister Chromatids during Anaphase. Int. J. Mol. Sci. 2023, 24, 4604. https://doi.org/10.3390/ijms24054604
Konecna M, Abbasi Sani S, Anger M. Separase and Roads to Disengage Sister Chromatids during Anaphase. International Journal of Molecular Sciences. 2023; 24(5):4604. https://doi.org/10.3390/ijms24054604
Chicago/Turabian StyleKonecna, Marketa, Soodabeh Abbasi Sani, and Martin Anger. 2023. "Separase and Roads to Disengage Sister Chromatids during Anaphase" International Journal of Molecular Sciences 24, no. 5: 4604. https://doi.org/10.3390/ijms24054604
APA StyleKonecna, M., Abbasi Sani, S., & Anger, M. (2023). Separase and Roads to Disengage Sister Chromatids during Anaphase. International Journal of Molecular Sciences, 24(5), 4604. https://doi.org/10.3390/ijms24054604




