Gut-Microbiota Dysbiosis in Stroke-Prone Spontaneously Hypertensive Rats with Diet-Induced Steatohepatitis
Abstract
1. Introduction
2. Results
2.1. Quantitative Analysis of 16S Ribosomal RNA Genes of Bacteria of Microbiota
2.2. Next-Generation Sequencing of the V4–V5 Region of 16S rRNA Genes of Gut-Microbiota
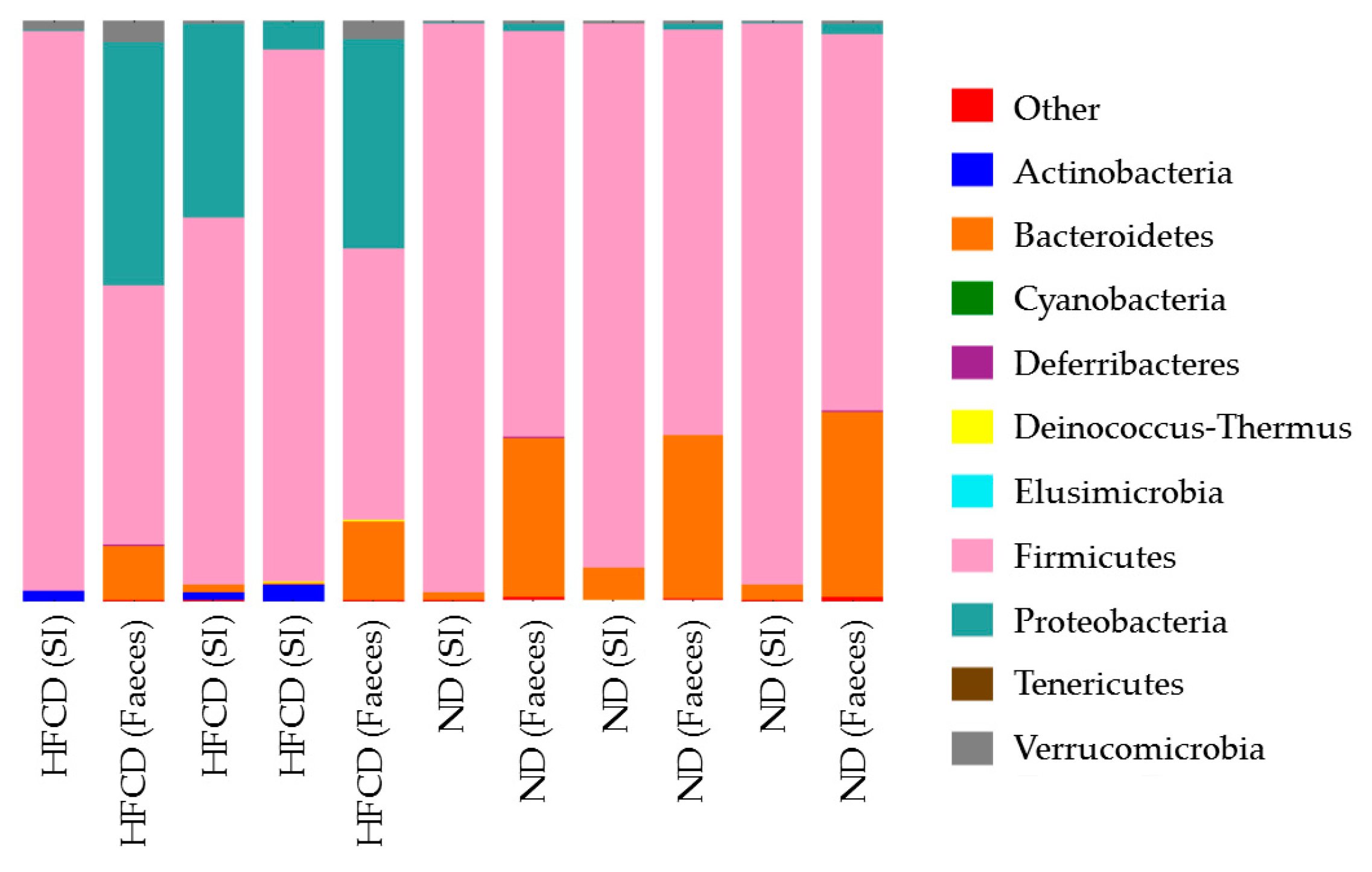
2.3. Microbiota of Small Intestine in SHRSP5 Rats Fed ND Are More Similar to Those of Small Intestine or Feces in SHRSP5 Rats Fed HFCD Than to Those of Feces in SHRSP5 Rats Fed ND
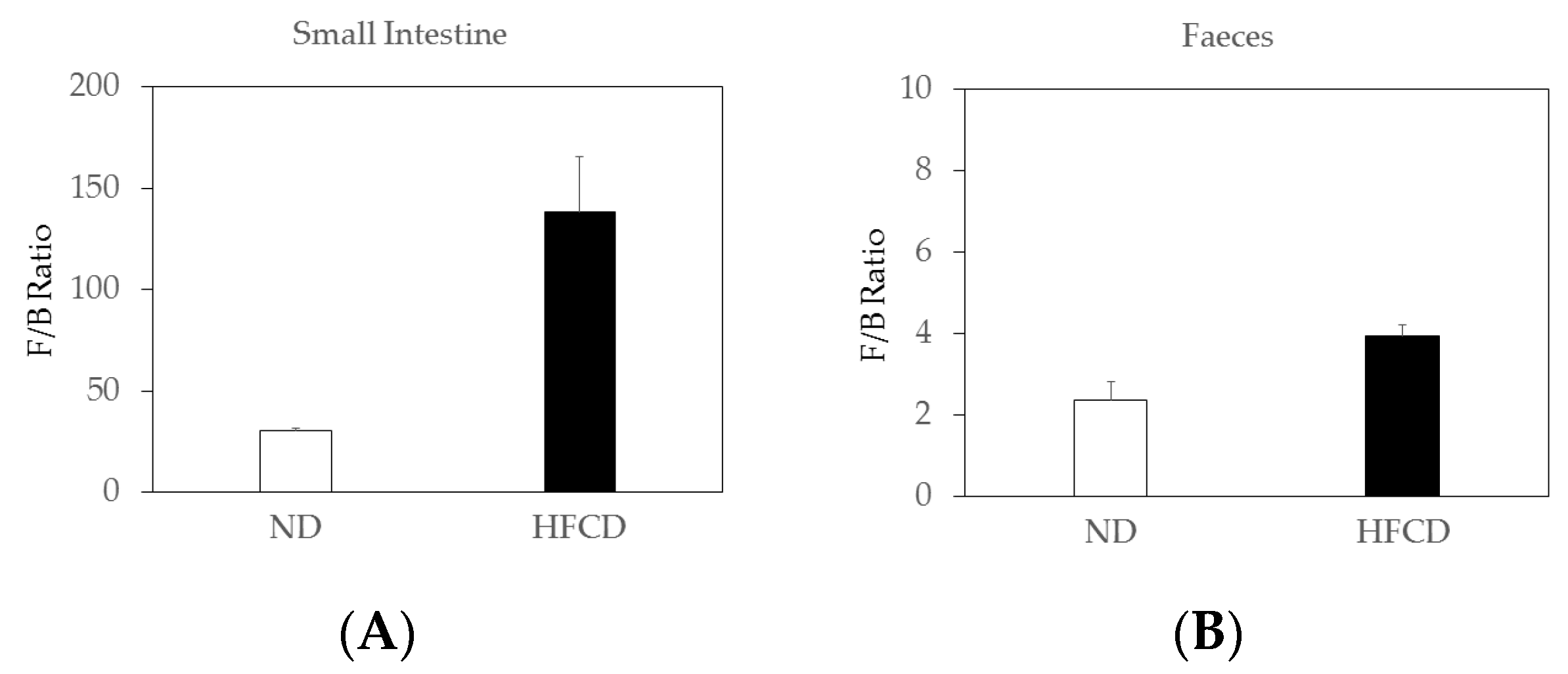
2.4. The Firmicutes/Bacteroidetes (F/B) Ratio Increased in the Small Intestines of SHRSP5 Rats Fed HFCD Compared to That in SHRSP5 Rats Fed ND
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Dietary Intervention
4.3. Sample Collection
4.4. Quantification of 16S rRNA Genes by Real-Time PCR
4.5. Next-Generation Sequencing 16S rRNA Genes
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henry, L.; Paik, J.; Younossi, Z.M. Review article: The epidemiologic burden of non-alcoholic fatty liver disease across the world. Aliment. Pharmacol. Ther. 2022, 56, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Matsuoka, S.; Yamazaki, M.; Shibata, T.; Nirei, K.; Takahashi, H.; Kaneko, T.; Fujisawa, M.; Higuchi, T.; Nakamura, H.; et al. Apoptosis and non-alcoholic fatty liver diseases. World J. Gastroenterol. 2018, 24, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Goto, T.; Hirotsu, Y.; Masuzaki, R.; Moriyama, M.; Omata, M. Molecular Mechanisms: Connections between Nonalcoholic Fatty Liver Disease, Steatohepatitis and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 1525. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sanyal, A.J.; George, J.; International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sarin, S.K.; Wong, V.W.; Fan, J.G.; Kawaguchi, T.; Ahn, S.H.; Zheng, M.H.; Shiha, G.; Yilmaz, Y.; Gani, R.; et al. The Asian Pacific Association for the Study of the Liver clinical practice guidelines for the diagnosis and management of metabolic associated fatty liver disease. Hepatol. Int. 2020, 14, 889–919. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Downes, M.; Evans, R.; Witztum, J.; Glass, C.; Loomba, R. Shared mechanisms between cardiovascular disease and NAFLD. Semin. Liver Dis. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Wigg, A.J.; Roberts-Thomson, I.C.; Dymock, R.B.; McCarthy, P.J.; Grose, R.H.; Cummins, A.G. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of non-alcoholic steatohepatitis. Gut 2001, 48, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Miele, L.; Valenza, V.; La Torre, G.; Montalto, M.; Cammarota, G.; Ricci, R.; Mascianà, R.; Forgione, A.; Gabrieli, M.L.; Perotti, G.; et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009, 49, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Santisteban, M.M.; Rodriguez, V.; Li, E.; Ahmari, N.; Carvajal, J.M.; Zadeh, M.; Gong, M.; Qi, Y.; Zubcevic, J.; et al. Gut dysbiosis is linked to hypertension. Hypertension 2015, 65, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Sharma, V.; Shahjouei, S.; Li, J.; Chaudhary, D.; Khan, A.; Wolk, D.M.; Zand, R.; Abedi, V. At the Intersection of Gut Microbiome and Stroke: A Systematic Review of the Literature. Front Neurol. 2021, 12, 729399. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, S.B.; Rathipriya, A.G.; Mahalakshmi, A.M.; Sharma, S.; Hediyal, T.A.; Ray, B.; Sunanda, T.; Rungratanawanich, W.; Kashyap, R.S.; Qoronfleh, M.W.; et al. The Influence of Gut Dysbiosis in the Pathogenesis and Management of Ischemic Stroke. Cells 2022, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Fukuda, N.; Nagase, H.; Tsunemi, A.; Tahira, K.; Matsumoto, T.; Hiraoka-Yamamoto, J.; Ikeda, K.; Mitsumata, M.; Sato, Y.; et al. Atherogenic dyslipidemia and altered hepatic gene expression in SHRSP.Z-Leprfa/IzmDmcr rats. Int. J. Mol. Med. 2009, 23, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Fukuda, N.; Maeshima, A.; Yamamoto, C.; Matsumoto, T.; Ueno, T.; Nojima, Y.; Matsumoto, K.; Soma, M. Treatment with valsartan stimulates endothelial progenitor cells and renal label-retaining cells in hypertensive rats. J. Hypertens. 2011, 29, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Moriyama, M.; Fukushima, A.; Matsumura, H.; Matsuoka, S.; Kanda, T.; Sugitani, M.; Tsunemi, A.; Ueno, T.; Fukuda, N. Association of mRNA expression of iron metabolism-associated genes and progression of non-alcoholic steatohepatitis in rats. Oncotarget 2018, 9, 26183–26194. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, F.; Huang, Y.; Liu, B. The Positive Effects of Grifola frondosa Heteropolysaccharide on NAFLD and Regulation of the Gut Microbiota. Int. J. Mol. Sci. 2019, 20, 5302. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Panasevich, M.R.; Schuster, C.M.; Phillips, K.E.; Meers, G.M.; Chintapalli, S.V.; Wankhade, U.D.; Shankar, K.; Butteiger, D.N.; Krul, E.S.; Thyfault, J.P.; et al. Soy compared with milk protein in a Western diet changes fecal microbiota and decreases hepatic steatosis in obese OLETF rats. J. Nutr. Biochem. 2017, 46, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yao, X.; Xia, F.; Yang, M.; Chen, Z.; Zhou, B.; Liu, Q. Modulation of the Gut Microbiota in Rats by Hugan Qingzhi Tablets during the Treatment of High-Fat-Diet-Induced Nonalcoholic Fatty Liver Disease. Oxid. Med. Cell Longev. 2018, 2018, 7261619. [Google Scholar] [CrossRef] [PubMed]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2014, 63, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Adenote, A.; Dumic, I.; Madrid, C.; Barusya, C.; Nordstrom, C.W.; Rueda Prada, L. NAFLD and Infection, a Nuanced Relationship. Can. J. Gastroenterol. Hepatol. 2021, 2021, 5556354. [Google Scholar] [CrossRef] [PubMed]
- Papić, N.; Jelovčić, F.; Karlović, M.; Marić, L.S.; Vince, A. Nonalcoholic fatty liver disease as a risk factor for Clostridioides difficile infection. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.C.; Zhao, W.; Li, S. Small intestinal bacteria overgrowth decreases small intestinal motility in the NASH rats. World J. Gastroenterol. 2008, 14, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kapil, S.; Duseja, A.; Sharma, B.K.; Singla, B.; Chakraborti, A.; Das, A.; Ray, P.; Dhiman, R.K.; Chawla, Y. Small intestinal bacterial overgrowth and toll-like receptor signaling in patients with non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2016, 31, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- De Faria Ghetti, F.; Oliveira, D.G.; de Oliveira, J.M.; de Castro Ferreira, L.E.V.V.; Cesar, D.E.; Moreira, A.P.B. Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur. J. Nutr. 2018, 57, 861–876. [Google Scholar] [CrossRef] [PubMed]
- Lira, M.M.P.; de Medeiros Filho, J.E.M.; Baccin Martins, V.J.; da Silva, G.; de Oliveira Junior, F.A.; de Almeida Filho, É.J.B.; Silva, A.S.; Henrique da Costa-Silva, J.; de Brito Alves, J.L. Association of worsening of nonalcoholic fatty liver disease with cardiometabolic function and intestinal bacterial overgrowth: A cross-sectional study. PLoS ONE 2020, 15, e0237360. [Google Scholar] [CrossRef] [PubMed]
- Mikolasevic, I.; Delija, B.; Mijic, A.; Stevanovic, T.; Skenderevic, N.; Sosa, I.; Krznaric-Zrnic, I.; Abram, M.; Krznaric, Z.; Domislovic, V.; et al. Small intestinal bacterial overgrowth and non-alcoholic fatty liver disease diagnosed by transient elastography and liver biopsy. Int. J. Clin. Pract. 2021, 75, e13947. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Yokosuka, O.; Suzuki, Y. Prolonged hepatitis caused by cytomegalovirus and non-alcoholic steatohepatitis in 16-year-old obese boy. Eur. J. Pediatr. 2005, 164, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Stepanov, Y.M.; Zavhorodnia, N.Y.; Yagmur, V.B.; Lukianenko, O.Y.; Zygalo, E.V. Association of nonalcoholic fatty liver disease with small intestine bacterial overgrowth in obese children. Wiad. Lek. 2019, 72, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Zhou, W.; Jiang, Y. Association of small intestinal bacterial overgrowth with nonalcoholic fatty liver disease in children: A meta-analysis. PLoS ONE 2021, 16, e0260479. [Google Scholar] [CrossRef] [PubMed]
- Domper Bardají, F.; Gil Rendo, A.; Illescas Fernández-Bermejo, S.; Patón Arenas, R.; Hernández Albújar, A.; Martín Dávila, F.; Murillo Lázaro, C.; Sánchez Alonso, M.; Serrano Dueñas, M.; Sobrino López, A.; et al. An assessment of bacterial overgrowth and translocation in the non-alcoholic fatty liver of patients with morbid obesity. Rev. Esp. Enferm. Dig. 2019, 111, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.S.; Jiang, F. Cisapride decreasing orocecal transit time in patients with nonalcoholic steatohepatitis. Hepatobiliary Pancreat. Dis. Int. 2006, 5, 534–537. [Google Scholar] [PubMed]
- Bures, J.; Cyrany, J.; Kohoutova, D.; Förstl, M.; Rejchrt, S.; Kvetina, J.; Vorisek, V.; Kopacova, M. Small intestinal bacterial overgrowth syndrome. World J. Gastroenterol. 2010, 16, 2978–2990. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ding, P.H.; Liu, L.J.; Shi, L.; Mao, T.Y.; Li, J.X.; Wang, Y.L. Gegen Qinlian Decoction Attenuates High-Fat Diet-Induced Steatohepatitis in Rats via Gut Microbiota. Evid. Based Complement Alternat. Med. 2018, 2018, 7370891. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yao, Y.; Gao, P.; Bu, S. The Therapeutic Efficacy of Curcumin vs. Metformin in Modulating the Gut Microbiota in NAFLD Rats: A Comparative Study. Front Microbiol. 2021, 11, 555293. [Google Scholar] [CrossRef] [PubMed]
- Di Ciaula, A.; Bonfrate, L.; Portincasa, P. The role of microbiota in nonalcoholic fatty liver disease. Eur. J. Clin. Investig. 2022, 52, e13768. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Kassam, Z.; Mullish, B.H.; Chiang, A.; Carrellas, M.; Hurtado, J.; Marchesi, J.R.; McDonald, J.A.K.; Pechlivanis, A.; Barker, G.F.; et al. Effects of Fecal Microbiota Transplantation With Oral Capsules in Obese Patients. Clin. Gastroenterol. Hepatol. 2020, 18, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Vancells Lujan, P.; Viñas Esmel, E.; Sacanella Meseguer, E. Overview of Non-Alcoholic Fatty Liver Disease (NAFLD) and the Role of Sugary Food Consumption and Other Dietary Components in Its Development. Nutrients 2021, 13, 1442. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J.; Dull, T.J.; Sleeter, D.D.; Noller, H.F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 1981, 148, 107–127. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Inoue, S.I.; Matsuhisa, A.; Iwata, Y.; Aizawa, N.; Sakai, Y.; Takata, R.; Ikeda, N.; Hasegawa, K.; Nakano, C.; et al. Amplification of bacterial genomic DNA from all ascitic fluids with a highly sensitive polymerase chain reaction. Mol. Med. Rep. 2018, 18, 2117–2123. [Google Scholar] [CrossRef] [PubMed]
- Pataky, Z.; Genton, L.; Spahr, L.; Lazarevic, V.; Terraz, S.; Gaïa, N.; Rubbia-Brandt, L.; Golay, A.; Schrenzel, J.; Pichard, C. Impact of Hypocaloric Hyperproteic Diet on Gut Microbiota in Overweight or Obese Patients with Nonalcoholic Fatty Liver Disease: A Pilot Study. Dig. Dis. Sci. 2016, 61, 2721–2731. [Google Scholar] [CrossRef] [PubMed]
- Delik, A.; Dinçer, S.; Ülger, Y.; Akkız, H.; Karaoğullarından, Ü. Metagenomic identification of gut microbiota distribution on the colonic mucosal biopsy samples in patients with non-alcoholic fatty liver disease. Gene 2022, 833, 146587. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D. Amplicon Sequence Variants Artificially Split Bacterial Genomes into Separate Clusters. mSphere 2021, 6, e0019121. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.T.; Xu, F.; Ng, R.T.; Hogg, J.C. Mian: Interactive web-based microbiome data table visualization and machine learning platform. Bioinformatics 2022, 38, 1176–1178. [Google Scholar] [CrossRef] [PubMed]
- Chiarello, M.; McCauley, M.; Villéger, S.; Jackson, C.R. Ranking the biases: The choice of OTUs vs. ASVs in 16S rRNA amplicon data analysis has stronger effects on diversity measures than rarefaction and OTU identity threshold. PLoS ONE 2022, 17, e0264443. [Google Scholar] [CrossRef] [PubMed]
- Serrana, J.M.; Watanabe, K. Sediment-associated microbial community profiling: Sample pre-processing through sequential membrane filtration for 16S rRNA amplicon sequencing. BMC Microbiol. 2022, 22, 33. [Google Scholar] [CrossRef] [PubMed]
- Kitamori, K.; Naito, H.; Tamada, H.; Kobayashi, M.; Miyazawa, D.; Yasui, Y.; Sonoda, K.; Tsuchikura, S.; Yasui, N.; Ikeda, K.; et al. Development of novel rat model for high-fat and high-cholesterol diet-induced steatohepatitis and severe fibrosis progression in SHRSP5/Dmcr. Environ. Health Prev. Med. 2012, 17, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Moriya, T.; Kitamori, K.; Naito, H.; Yanagiba, Y.; Ito, Y.; Yamagishi, N.; Tamada, H.; Jia, X.; Tsuchikura, S.; Ikeda, K.; et al. Simultaneous changes in high-fat and high-cholesterol diet-induced steatohepatitis and severe fibrosis and those underlying molecular mechanisms in novel SHRSP5/Dmcr rat. Environ. Health Prev. Med. 2012, 17, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Ritalahti, K.M.; Amos, B.K.; Sung, Y.; Wu, Q.; Koenigsberg, S.S.; Löffler, F.E. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 2006, 72, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, Y.L.; Yang, T.; Sahay, B.; Zadeh, M.; Cheng, S.X.; Wang, G.P.; Owen, J.L.; Mohamadzadeh, M. Colonic immune suppression, barrier dysfunction, and dysbiosis by gastrointestinal bacillus anthracis Infection. PLoS ONE 2014, 9, e100532. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal dysbiosis and depletion of butyrogenic bacteria in clostridium difficile infection and nos-ocomial diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [PubMed]

| Materials | 16S rRNA Genes (Copies/g) |
|---|---|
| ND (SI) | 4.8 × 1010 |
| ND (SI) | 5.1 × 1010 |
| ND (SI) | 8.0 × 1010 |
| ND (Feces) | 5.0 × 1011 |
| ND (Feces) | 5.2 × 1011 |
| ND (Feces) | 6.4 × 1011 |
| HFCD (SI) | 8.9 × 106 |
| HFCD (SI) | 4.3 × 107 |
| HFCD (SI) | 2.0 × 108 |
| HFCD (Feces) | 2.3 × 1010 |
| HFCD (Feces) | 5.2 × 1010 |
| Materials | Sequence-Read Number |
|---|---|
| ND (SI) | 31,756 |
| ND (SI) | 27,989 |
| ND (SI) | 28,957 |
| ND (Feces) | 18,255 |
| ND (Feces) | 24,888 |
| ND (Feces) | 21,820 |
| HFCD (SI) | 29,786 |
| HFCD (SI) | 26,581 |
| HFCD (SI) | 30,091 |
| HFCD (Feces) | 29,040 |
| HFCD (Feces) | 27,399 |
| Primer/Probe | Sequence | Target Gene |
|---|---|---|
| Bac1055YF * | 5′-ATGGYTGTCGTCAGCT-3 | Bacteria |
| Bac1392R * | 5′-ACGGGCGGTGTGTAC-3 | Bacteria |
| Bac1115Probe * | 5′-FAM-CAACGAGCGCAACCC-TAMRA | Bacteria |
| U515F | 5′-GTGYCAGCMGCCGCGGTA-3′ | V4–V5 region of the 16S rRNA |
| 926R | 5′-CCGYCAATTCMTTTRAGTT-3′ | V4–V5 region of the 16S rRNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanezawa, S.; Moriyama, M.; Kanda, T.; Fukushima, A.; Masuzaki, R.; Sasaki-Tanaka, R.; Tsunemi, A.; Ueno, T.; Fukuda, N.; Kogure, H. Gut-Microbiota Dysbiosis in Stroke-Prone Spontaneously Hypertensive Rats with Diet-Induced Steatohepatitis. Int. J. Mol. Sci. 2023, 24, 4603. https://doi.org/10.3390/ijms24054603
Kanezawa S, Moriyama M, Kanda T, Fukushima A, Masuzaki R, Sasaki-Tanaka R, Tsunemi A, Ueno T, Fukuda N, Kogure H. Gut-Microbiota Dysbiosis in Stroke-Prone Spontaneously Hypertensive Rats with Diet-Induced Steatohepatitis. International Journal of Molecular Sciences. 2023; 24(5):4603. https://doi.org/10.3390/ijms24054603
Chicago/Turabian StyleKanezawa, Shini, Mitsuhiko Moriyama, Tatsuo Kanda, Akiko Fukushima, Ryota Masuzaki, Reina Sasaki-Tanaka, Akiko Tsunemi, Takahiro Ueno, Noboru Fukuda, and Hirofumi Kogure. 2023. "Gut-Microbiota Dysbiosis in Stroke-Prone Spontaneously Hypertensive Rats with Diet-Induced Steatohepatitis" International Journal of Molecular Sciences 24, no. 5: 4603. https://doi.org/10.3390/ijms24054603
APA StyleKanezawa, S., Moriyama, M., Kanda, T., Fukushima, A., Masuzaki, R., Sasaki-Tanaka, R., Tsunemi, A., Ueno, T., Fukuda, N., & Kogure, H. (2023). Gut-Microbiota Dysbiosis in Stroke-Prone Spontaneously Hypertensive Rats with Diet-Induced Steatohepatitis. International Journal of Molecular Sciences, 24(5), 4603. https://doi.org/10.3390/ijms24054603










