Abstract
The landscape of pervasive transcription in eukaryotic genomes has made space for the identification of thousands of transcripts that are difficult to frame in a specific functional category. A new class has been broadly named as long non-coding RNAs (lncRNAs) and shortly defined as transcripts that are longer than 200 nucleotides with no or limited coding potential. So far, about 19,000 lncRNAs genes have been annotated in the human genome (Gencode 41), nearly matching the number of protein-coding genes. A key scientific priority is the functional characterization of lncRNAs, a major challenge in molecular biology that has encouraged many high-throughput efforts. LncRNA studies have been stimulated by the enormous clinical potential that these molecules promise and have been based on the characterization of their expression and functional mechanisms. In this review, we illustrate some of these mechanisms as they have been pictured in the context of breast cancer.
1. Introduction
The next-generation sequencing era has strongly increased the number of annotated non-canonical transcripts, such as lncRNAs. There are several factors that make the current annotation of these non-coding transcripts challenging compared to protein-coding genes, since lncRNAs are (i) weakly conserved at the sequence levels during evolution, (ii) generally low-expressed and (iii) strongly context-dependent, meaning that their levels of expression can differ greatly between tissues or even different cell types within a tissue. So, it is not surprising that several databases such as LNCipedia, lncRNADisease 2.0, LncATLAS, LncRNAdb and Lnc2Cancer are not coherently concordant and need to be updated timely and be supported by biological validation [1,2,3,4,5]. The use of high-throughput data (including CAGE, RNA-seq and polyA site-seq) from available consortia such as ENCODE (https://www.encodeproject.org, accessed on 20 February 2023) or FANTOM (https://fantom.gsc.riken.jp, accessed on 20 February 2023) can be useful for characterizing the expression of a given candidate in a specific tissue/cell type. Multiple studies have highlighted the role of lncRNAs in diseases [6]. Expression studies comparing normal vs. cancer tissues have revealed many lncRNAs to be regulated in cancer, often with a very high cancer specificity. In addition, cancer pathways have been found to be regulated by intricated networks of coding and non-coding transcripts. As for coding transcripts, lncRNAs can have either tumor-suppressing or oncogenic functions [7].
Among cancers, breast cancer is a life-threatening disease that mirrors the complexity and heterogeneity of the mammary gland. In this context, investigations on lncRNAs have been frequently aimed at the identification of novel or more accurate biomarkers for diagnosis and, above all, the search of novel therapeutic targets that are potentially useful in treating the most-lethal forms of the disease. Importantly, several molecular mechanisms involving lncRNAs have emerged in breast cancer studies and can be considered prototypical for their mode of action. In this review, we aimed at illustrating the complexity of lncRNA mechanisms and their mode of action, focusing on breast cancer as a unifying model system, which is useful for illustrating the biological role and, at the same time, the therapeutic potential of lncRNAs. A comprehensive list of lncRNAs with their mechanisms involved in breast cancer is summarized in Table 1. Below, we will discuss in detail the main mechanisms of lncRNAs, distinguishing those that occur in the nucleus from those that require cytoplasmic localization, as depicted in Figure 1. For each mechanism, we focused on a few lncRNAs with large support from the literature and evidence in breast cancer, which can be used as a prototypical example.

Table 1.
Long non-coding RNAs and their mechanisms.
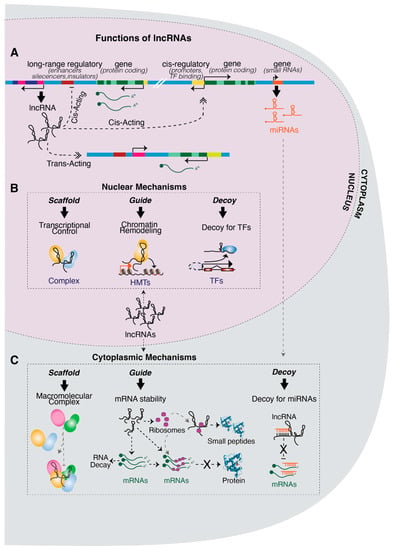
Figure 1.
Genomic activity and molecular functions of lncRNAs: (A) the panel summarizes the possible modes of action of lncRNA, either in cis or in trans. The activity in cis is exerted on elements in close genomic proximity, while trans-acting lncRNAs regulate genes in genomic loci distant from their site of transcription. (B) Nuclear lncRNAs can regulate epigenetic and/or gene transcription by acting as: (i) scaffolds for macromolecular complexes; (ii) guides for chromatin-remodeling complexes to specific regulatory regions and (iii) decoys for transcription factors. (C) Cytoplasmic lncRNAs can either interact with mRNAs and/or proteins, acting as: (i) scaffolds for promoting the assembly of RNA-binding protein complexes; (ii) guides for modulating mRNA stability, either operating on RNA decay or RNA translation or, alternatively, by being translated in small peptides and associating with proteins and (iii) decoys for miRNAs through MREs.
2. Functions of lncRNAs in the Nucleus
A large fraction of lncRNAs is expressed almost exclusively in the nucleus [60,61] and, hence, exhibits functions related to nuclear processes, such as the regulation of RNA transcription and RNA splicing or the organization of functionally distinct nuclear domains. Various mechanisms contribute to the nuclear localization of lncRNAs. In general, we can distinguish “passive” mechanisms, favoring the nuclear accumulation of lncRNAs, such as inefficient transcription and low-yield RNA processing (i.e., splicing) [62], from “active” mechanisms based on nuclear retention signals that allow interaction with protein complexes and ribonucleoproteins localized in the nucleus, [63]. Overall, the nuclear functions of lncRNAs are related to the control of gene expression and, hence, fall into two main categories, cis- or trans-acting, depending on whether the lncRNA influences nearby genes or acts on long-distance regions [64]. Similarly, the activity of lncRNAs can be also categorized as sequence-dependent or -independent, as it may or may not depend on their exact nucleotide sequence.
2.1. lncRNAs as Regulators of Chromatin Status
A recurring theme of nuclear functions is the regulation of the chromatin status. LncRNAs have been shown to influence chromatin organization at different levels. Indeed, the mere act of transcription can modulate the chromatin accessibility of a locus and, thus, lncRNA transcription can function as a cis-acting mechanism, influencing the expression of nearby protein-coding genes in a sequence-independent manner [65]. Alternatively, lncRNAs can interact with chromatin modifiers through the recognition of specific binding sites or secondary structures in the lncRNA transcript. Their interaction with proteins can have multiple readouts: lncRNAs can act as a molecular scaffold, bridging multiple proteins in a single macromolecular complex, or as a molecular decoy, coordinating the regulatory activity in a locus. In both cases, there are examples of cis- and trans-acting lncRNAs (some of them are reviewed in [66]).
HOTAIR
HOX transcript antisense intergenic RNA (HOTAIR) is one of the most strikingly cancer-associated lncRNAs [67] and is a typical example of a nuclear lncRNA acting both as a molecular guide and as a scaffold. HOX genes are a group of conserved protein-coding genes used in the control of the correct body patterning and are organized into different clusters in the genome. HOX genes are tightly regulated during their development and are frequently over-expressed in cancer [68]. The lncRNA HOTAIR is a conserved 2.1 kb transcript produced from the HOXC locus on chromosome twelve and is composed of six exons, which are actively spliced and polyadenylated [69]. Initially, HOTAIR has been suggested to regulate chromatin in trans at the distal HOXD cluster. Indeed, the knock-down of HOTAIR by siRNAs induces a de-repression of the locus and a reduction in the repressive histone modification H3K27me3 [70]. Further studies have suggested that HOTAIR acts as a scaffold, coordinating two different chromatin-modifying activities: the deposition of H3K27me3 mediated by Polycomb-repressive complex (PRC2) and the simultaneous demethylation of H3K4me3 by lysine-specific demethylase 1 (LSD1). Two loops in HOTAIR’s secondary structure, at the 5′ and 3′ ends, have been proposed to mediate its interaction with PRC2 and LSD1, respectively [70]. This lncRNA is frequently found as dysregulated (mostly over-expressed) in different cancer types. As it relates to breast cancer, HOTAIR has been reported to aberrantly target genomic regions other than the HOXD cluster, mediating chromatin dysregulation and promoting breast tumor metastasis [8,71].
2.2. Enhancer-Like Functions
Enhancers are regions of open chromatins which act as hubs for different transcription-factor-binding sites and operate in a cell-type-specific fashion to activate the expression of target genes (reviewed in [72,73]). Enhancers function on nearby genes (cis-acting), which can even be placed at several kb distances thanks to the formation of long-range chromatin interactions. As enhancers are actively transcribed, they also generate non-coding transcripts, which can either be shortly and rapidly degraded (eRNAs) or longer and more frequently processed [74]. These non-coding RNAs may participate in an enhancer function by several types of mechanisms. Here, we describe two lncRNAs involved in breast cancer with their reported enhancer-like functions.
CCAT1-L
Colon-cancer-associated transcript-1-long isoform (CCAT1-L) is a lncRNA gene, so named as it was found to be highly expressed in colorectal cancer (CRC) samples [75]. CCAT1-L is a 5.2 kb long RNA that is enriched at its site of transcription and is chromatin-bound. It is expressed from the 8q24 genomic region, 500 kb upstream of the myelocytomatosis (MYC) locus. The CCAT1-L locus has been associated with several chromatin marks, which are typical of enhancer regions, such as high levels of H3K27Ac and H3K4Me1, low levels of H3K4Me3 and the presence of DNase-I-hypersensitive sites [72,76]. According to these epigenetic marks, the 150 kb long region encompassing CCAT1-L has been proposed to act as a putative super-enhancer responsible for controlling MYC expression via a regulatory element embedded in a lncRNA promoter (MYC-515) and a downstream regulatory element (MYC-335) [77,78]. Three-dimensional conformation capture data have supported the molecular interaction occurring among MYC-515, MYC-335 and MYC promoter. Strikingly, the downregulation of the CCAT1-L transcript by antisense oligonucleotides (ASOs) corresponds to a reduction in MYC expression and a decreased contact frequency among MYC, MYC-335 and MYC-515. These findings highlight the importance of the lncRNA transcript in mediating enhancer activities, other than DNA features occurring at the locus of its transcription. Overall, the proposed model suggests that the CCAT1-L transcript operates in cis and participates in establishing long-range contacts that bring the MYC locus into proximity with its enhancers thanks to its direct interaction with CCCTC-binding factor (CTCF) [79]. Given the wide oncogenic role of MYC, it is not surprising that CCAT1-L is frequently expressed at elevated levels in many cancer types. In breast cancer, CCAT1-L is a promising prognostic biomarker, as its expression correlates with a decreased overall survival and progression-free survival independently from the receptor status of the disease [80].
A-ROD
In some cases, processed (mature) lncRNAs, rather than primary unprocessed transcripts, may directly contribute to enhancer function control. This is the case of A-ROD, a lncRNA involved in the regulation of a downstream gene, namely a negative regulator of the Wnt pathway named Dickkopf-1 (DKK1) [81]. A-ROD is a non-coding transcript originating from a locus acting as an enhancer and is located 130 kb upstream of the DKK1 locus. In breast cancer cell lines (MCF-7) and samples from breast cancer patients, the two loci showed a correlated expression and were found to be involved in chromatin looping [17]. Interestingly, ASOs targeting the nascent A-ROD transcript had no effect on DKK1 mRNA levels, while siRNAs targeting the mature transcript were able to reduce the expression level of DKK1 and, at the same time, increase the pausing of RNA polymerase 2 at the DKK1 transcription start site. The proposed model suggests that A-ROD is not involved in chromosomal looping, conversely to CCAT1-L. A pre-existing conformation maintains the A-ROD locus in proximity to DKK1, and the A-ROD mature transcript recruits the transcriptional activator EBP4 to enhance DKK1 transcription. Experiments on the splicing inhibition and transcriptional termination of this lncRNA both supported the fact that the enhancer-like function of A-ROD is mediated by the mature transcript. Interestingly, Ntini et al. [17] provided data in support of many other lncRNAs with similar features, as they are transcribed from regions involved in chromatin loops and with a poor association with chromatin, suggesting that the mature form of the lncRNA plays a functional role in gene expression control. In support of this, bioinformatic analyses have proposed that the activity of enhancers is correlated with both the transcription and splicing of their encoded lncRNAs [82,83].
2.3. Regulation of Splicing
Recently, lncRNAs were shown to exploit another mechanism of gene expression control by affecting gene splicing. Splicing is a fundamental step in mRNA maturation that allows the excision of introns from transcripts, and the usage of alternative splice sites can affect the production of multiple isoforms subjected to differential regulation in physiology and disease [84]. An emblematic case is the stress-induced lncRNA known as lncRNA associated with SART3 regulation of splicing (LASTR), which is induced by c-JUN together with other survival genes during hypoxia and DNA damage. As c-JUN is frequently overexpressed in epithelial tumors, LASTR was found to be highly expressed in most breast cancer subtypes, as reported in The Cancer Genome Atlas—TCGA [21]. This lncRNA is a 714 nt transcript composed of two exons expressed mainly in the nucleus and has been found to interact with SART3, a splicing protein involved in the assembly of the U4/U6 ribonucleic complex, by RNA pulldown assays followed by mass spectrometry [85]. LASTR seems to have an impact on splicing control at a global level, as shown by knock-down experiments with ASOs, which resulted in the impairment of the SART3 disassembly from the U4 snRNA and the prevention of the recycling of spliceosome components, increasing intron retention, exon skipping and the non-sense-mediated decay of mRNAs [86].
In normal mammary epithelial cells, the expression of LASTR induced by hypoxia facilitates the dissociation of SART3 from the U4/U6 snRNP, preserving cell physiology when stress conditions are present [87]. Similarly, the constitutive overexpression of LASTR helps cancer cells to increase their cell fitness by avoiding splicing defects. Strikingly, the knockdown of LASTR can sensitize the triple-negative breast cancer cell line MDA-MB-231 to irradiation and impair the tumor growth in mice xenografts, suggesting that this lncRNA is a potential therapeutic target. This work exemplifies how the dynamic regulation of one single lncRNA can largely impact fundamental physiological cellular processes and how cancer cells favorably exploit these simple but effective mechanisms.
2.4. Organization of Nuclear Architecture
Some lncRNAs that are abundantly expressed in the nucleus can function in coordinating the organization and activity of functionally distinct nuclear compartments [88]. This is the case of two well-known lncRNAs, named nuclear enriched abundant transcript 1 (NEAT1) and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) (also known as NEAT2). These lncRNAs are transcribed from proximal loci but are then localized in separate compartments.
NEAT1 is localized and is essential for the assembly of paraspeckles, a dynamic compartment responsible for transcription and RNA processing. NEAT1 is associated with actively transcribed genes [89]. Thanks to the different protein interacting domains in its sequence, NEAT1 realizes the precise localization of proteins in this compartment [90]. NEAT1 expression is dysregulated in many cancer types and is found in the peripheral blood of breast cancer patients [24]. The increased expression of NEAT1 is associated with a poor prognosis and overall survival [91]. The expression of NEAT1 is regulated by several tumorigenic transcription factors such as STAT3 or NFkB [92] and contributes to the regulation of the genes responsible for invasion, metastasis and chemoresistance [91].
MALAT1 is a conserved lncRNA [93] that localizes in nuclear speckles, a subnuclear domain where components of the spliceosome concentrate [94]. The current model suggests that MALAT1 functions in the periphery of nuclear speckles and participates in the positioning of the speckles towards actively transcribed genes, contributing to pre-mRNA splicing [89,95]. Consistent with this model, MALAT1 knockdown does not impair the formation of nuclear speckles but changes their composition and functionality. The down-modulation of the phosphorylation levels of serine/arginine-rich proteins, which are key splicing components, has been observed upon MALAT1 silencing, which may explain the impact it has on splicing. MALAT1 expression has been frequently associated with a poor prognosis, metastasis [26] and chemo- and radio-resistance in several human tumors and in breast cancer [96]. Tandem duplications of the MALAT1 gene have also been reported [97]. However, Kim et al. provided evidence of a tumor-suppressive role of MALAT1 in breast cancer cells and primary mammary tumors [25]. According to this work, MALAT1 interacts with TEAD, preventing the association with the YAP co-activator and the expression of pro-metastatic target genes. These opposite views may not be mutually exclusive but may co-exist in the interpretation of a complex and abundant lncRNA such as MALAT1.
3. Functions of lncRNAs in the Cytoplasm
A large portion of lncRNAs can exert their functions on the cytoplasm after being transported from the nucleus. A very recent study highlighted that long transcripts that are A/U-rich and with few exons, such as the vast majority of lncRNAs, are dependent on the NXF1 factor for export [98]. Upon arrival, lncRNAs can be sorted into different organelles [99] (i.e., mitochondria or exosomes) or be associated with proteins/nucleic acids. One representative example is the nuclear-encoded lncRNA SAMMSON, which is expressed in melanoma cells and is re-located into mitochondria and plays a fundamental role in the regulation of mitochondrial metabolism through the interaction with p32 [57]. How the sorting occurs into different cellular compartments still needs better elucidation; however, it is commonly accepted that it depends on the interaction of lncRNAs with RNA-binding proteins (RBPs) and/or with other RNA species, such as for most (if not all) of the cytosolic functions of lncRNAs. An important caveat for lncRNA/target interaction is represented by the critical stoichiometric ratio between the lncRNA and its target molecule, either a protein or an RNA molecule. In fact, in the case of cis-acting lncRNAs, few copies are sufficient to exert a biologically relevant function on a single target gene located in proximity. However, for trans-acting lncRNAs, such as all cytoplasmic lncRNAs, it is essential to start with a clear copy number quantification of both the target and the lncRNA in order to verify if a proposed biological mechanism is plausible [64,65]. This especially applies to those lncRNAs acting as competing endogenous RNAs (ceRNAs). This is a mechanism where an lncRNA acts as sponge, soaking up many molecules of a given miRNA and, hence, competing for the interaction of the miRNA with its own set of mRNA targets. A seminal work by Bartel’s group focused on miR-122 as a proof-of-principle in order to challenge the contention that a change in the copy number of a single miRNA target could compete with other shared targets in such a way as to result in a variation in the miRNA’s effects. This work suggests that the miRNA:ceRNA stoichiometry should be better investigated, as the upregulation/downregulation of a single ceRNA may not be sufficient to exert a measurable effect on miRNA activity [100]. Moreover, to further complicate this scenario, lncRNA expression could be associated to a specific tissue (normal vs. cancer) or even restricted to a specific cell type (i.e., rare cell types as stem cells). Thus, to understand the biological effects mediated by the lncRNA, a careful stoichiometric quantification is advisable, accounting for sample purity rather than using a bulk population, which can contain different cell types.
3.1. LncRNAs Acting as miRNA Sponges
Linc-ROR
LncRNA-regulator of reprogramming, a.k.a., Linc-ROR, was initially identified as a competing endogenous RNA (ceRNA) involved in hESC self-renewal. Linc-ROR levels are high in hESC and drop down during differentiation [101]. The ceRNA activity was dependent on the microRNA responsive elements (MREs) within Linc-ROR sequence and matching with miR-145. This resulted in the sequestering of miR-145 molecules and maintaining the hESC’s pluripotent state by avoiding the miRNA-mediated repression of several embryonic transcription factors (ETFs) [101]. In the breast, Linc-ROR was reported to have an impact on the regulation of the epithelial-to-mesenchymal transition (EMT) and the acquisition of chemoresistance and cancer stem cell traits. One of the first pieces of evidence came from a study using MCF10A, a normal breast epithelial cell line, which described Linc-ROR as a ceRNA that was able to sponge miR-205, thus causing an increase in ZEB2 levels, a TF-promoting EMT and a well-known target for miR-205. This mechanism can explain the increase in the expression of mesenchymal markers and the effects on proliferation, cell migration and the acquisition of some traits observed upon Linc-ROR expression, such as the increased CD44high/CD24low population and its capability of forming non-adherent spheroids (mammosphere) [36]. A more recent study on MCF7 cells suggests that Linc-ROR acts as ceRNA for miR-194-3, causing an increase in MECP2 levels and, thus, promoting the proliferation, invasion and resistance to rapamycin treatment of breast cancer cells [37]. Linc-ROR was also described with another type of mechanism, supporting the estrogen-independent growth of ER+ breast tumors and their resistance to tamoxifen [102]. The proposed mechanism involves the direct inhibition of the phosphatase DUSP7 and the consequent activation of MAPK/ERK signaling, which, in turn, phosphorylates the estrogen receptor and fosters the growth of breast cancer cells and resistance to hormonal chemotherapy [102].
H19
The transcript from the H19 locus was one of the first lncRNAs to be acknowledged. It belongs to a conserved imprinted region located on the human chromosome 11, encoding for a 2.3 kb long cytosolic transcript associated with the epigenetic silencing of the IGF2 locus [103]. In mouse embryonic and extra-embryonic cell lines, H19 has also been described as a precursor for miR-675 and capable of regulating placental development by regulating the abundance of IGF2 at two levels through the imprinting of the gene locus and the silencing of the IGF1R receptor via miR-675 [104].
In breast cancer cells, H19/miR-675 expression has been associated with increased proliferation, tumor growth aggressiveness and metastases in vivo. Mechanistically, this was suggested to be dependent on miR-675 activity in b-Cbl and c-Cbl mRNAs, which, in turn, leads to the hyperactivation of EGFR and c-Met and the consequent activation of Akt and Erk signaling [38]. In addition, H19 has also been described as an miRNA sponge for Let-7, highlighting a prototypical ceRNA mechanism active during muscle differentiation, as shown by Kallen et al. [105]. The ceRNA mechanism was also reported to occur in breast tumors in several reports. For instance, the interaction of H19 with Let-7c was shown to influence the type of division (symmetric or asymmetric) of cancer stem cells (CSC) by controlling WNT signaling [39]. In another report, H19 was associated to Let-7a/b and the core pluripotency factor LIN28, a transcription factor critical for stem cells. A positive feedback loop mechanism was described, with H19 competing with LIN28 for Let-7a/b binding, leading to an increase in LIN28 levels, which, in turn, inhibits the generation of mature Let-7a/b molecules from precursors, derepressing all the target genes for Let-7 miRNAs [40]. Lastly, under hypoxic conditions, H19 was reported to sequester Let-7 miRNAs and to relieve HIF1α mRNA levels. In this report, the H19/Let-7/HIF-1α axis was shown to act as metabolic gatekeeper under hypoxia, controlling the switch from OXPHOS to glycolysis [106].
3.2. LncRNAs Acting as Guide
NORAD
Besides acting as sponges for miRNAs, lncRNAs in the cytosol can also affect mRNA stability, by acting as guide controlling mRNA degradation or mRNA translation. One representative example is the non-coding RNA activated by DNA damage (NORAD). This lncRNA is very abundant in the cytoplasm, where it is bound to PUMILIO1/2 proteins and acts as a decoy. This mechanism depends on the sequence of NORAD, which contains several PUMILIO response elements (PRE), a stretch 8 nt long, which is typically located in the in 3′ UTR of PUMILIO target mRNAs. Upon genotoxic stress, NORAD acts as reservoir of PUMILIO1/2 proteins and controls genomic stability. In fact, the loss or downregulation of NORAD causes a sudden release of PUMILIO1/2 proteins, which bind and accelerate the mRNA turnover of targets involved in DNA repair and DNA replication, thus driving chromosome instability [51].
In colon cancer cells, by combining RNA antisense purification (RAP) and quantitative MS, NORAD was identified as a necessary component for the assembly of a ribonucleic complex (NORAD-activated ribonucleoprotein complex 1, NARC1) involved in genome stability maintenance. Cells depleted for NORAD have increased defects in chromosome segregation, reduced replication fork speed and an altered cell cycle [107].
In breast cancer, NORAD was suggested to act as tumor suppressor. A report showed NORAD under transcriptional repression by the YAP/TAZ and NuRD complexes, which usually act as oncogenic factors. In addition, NORAD was shown to act as a decoy for S100P, counteracting its pro-migratory and pro-invasive activity [52].
3.3. LncRNA-Encoding Polypeptides
According to their definition, lncRNAs should not have coding functions. However, some studies suggested that small polypeptides can be synthetized from small open-reading frames (ORFs) and can participate in lncRNA-regulatory functions. For instance, in breast cells, LINC00665 was shown to encode for a micropeptide of 5.5 kDa named CIP2A-BP, which binds CIP2A and competes with the subunit PP2A, an oncogene that promotes tumor progression [31]. Of note is the fact that the translation of LINC00665 is under the control of the TGFβ and SMAD pathways, while the overall levels are not affected. Consistent with this model, the migration and invasion properties of triple-negative breast cancer cells can be inhibited both in vitro and in vivo by the overexpression of the CIP2A-BP protein but not by LINC00665 expression [31].
LncRNA EPR (epithelial cell program regulator) was found to be a typical breast epithelial lncRNA, whose expression is inhibited by TGFβ treatment. LncRNA EPR was shown to encode for an ~8 kDa small peptide, which localizes at the epithelial cell junctions of mammary glands together with junctional proteins such as ZO-1, CGNL1 and Cortactin [32]. LncRNA EPR was suggested to have a dual mechanism. At the RNA level, it can interact with the Cdkn1a gene on chromatin and can sustain the expression and stability of Cdkn1a mRNA, thus promoting epithelial phenotype and cell cycle arrest [32].
PVT1: One lncRNA, Many Functions
Many lncRNAs display multiple regulatory functions that are associated with complex and sometimes conflicting phenotypes. In this category, one representative example is plasmacytoma variant translocation 1 (PVT1), a long non-coding RNA. The human PVT1 gene shows a high level of homology with mouse and rat genomes [108,109].
Six different transcription start sites (TSS) can drive the expression of PVT1 lncRNA and are distributed in a region of 300 kb and are located downstream of the promoter of the MYC oncogene [110]. MYC and PVT1 belong to the 8q24 genomic region, which is frequently altered in cancer. Specifically, this locus is mostly susceptible to amplifications and other structural alterations that lead to the co-amplification of the two genes [111]. Similarly to MYC, high expression levels of PVT1 have been associated with a poor prognosis in breast cancer and other human malignancies [112,113]. The pro-tumorigenic function of PVT1 can be explained by the activity miRNAs embedded in the lncRNA transcript. PVT1, indeed, hosts a miRNA cluster composed of miR-1204, -1205, -1206, -1207-5p and -3p and -1208, which can act as oncomiRs, promoting cell proliferation [114], increasing glycolytic metabolism [115] and suppressing stress-induced apoptosis [116]. However, other studies reported tumor-suppressive functions for the same miRNAs [117,118], suggesting that tissue-specific targets and effects may be involved. In addition, the PVT1 sequence holds several sites matching miRNAs (MREs), invoking a potential molecular sponge mechanism for the transcript [119]. Indeed, different reports support the possibility that the PVT1 transcript may sequester miRNAs with tumor-suppressive functions, thus leading to the activation of proliferative and survival pathways [120,121] and the acquisition of metastatic traits.
Moreover, the PVT1 transcript was shown to interact with the MYC protein in trans, regulating MYC stability by interfering with Threonine 58 phosphorylation and proteosome-mediated degradation. MYC, in turn, binds the PVT1 promoter in two E-boxes sites, creating a positive feedback loop that sustains MYC expression and MYC-induced proliferation [122,123].
In breast cancer, the amplification of the 8q24 region leads to both a gain in the copy number of the PVT1 gene and the accumulation of genetic alterations at the level of the promoter region of PVT1, which abrogates its expression [124]. Starting from this observation, Cho et al. showed that the most-upstream promoter of PVT1 has a tumor-suppressive function that is independent from the transcription of the lncRNA and aids in tightly regulating the expression of MYC [110]. In breast cancer cell lines, the promoters of PVT1 and MYC compete for the binding of intragenic enhancers that are located within the gene body of PVT1. When the promoter of PVT1 is functional, it interacts with the enhancers, which are closer, sustaining the expression of PVT1 transcript only. When the promoter of PVT1 is non-functional, the intragenic enhancers rewire it towards the promoter of MYC with a topological rearrangement in the 3D genome that boosts the oncogenic expression of MYC, thus enhancing cancer cell proliferation [110].
This mechanism of enhancer retargeting seems to be not restricted to the case of the PVT1-MYC pair. Oh et al. [125] showed that cancer cells often accumulate mutations in the promoters of genes owing to topological rearrangements of the genome that are able to reinforce the expression of oncogenes. In addition to this, a recent work by Oliviero et al. [126] described an alternative type of relationship occurring between the Pvt1-Myc pair in mouse embryonic fibroblasts (MEFs). In this work, a p53-responsive element was found on a downstream TSS for Pvt1 that, in stress conditions and upon p53 binding, could elicit the expression of the Pvt1b isoform and the concurrent repression of Myc transcription. The authors suggested that this mechanism is RNA-dependent, as the repression of Myc occurs in cis in the absence of topological rearrangements and that it is abrogated in the presence of antisense oligonucleotides targeting Pvt1b [126]. This mechanism still needs to be clarified in human malignancies, but it highlights a role for lncRNA in adjuvating key stress response pathways such as the one coordinated by p53.
More than 30 years of study have just scratched the complexity of the PVT1 locus and show the contextual presence of enhancer-like functions of lncRNAs, the trans-activity of the PVT1 transcript in regulating the stability of MYC protein, the contribution of DNA-regulatory elements within a non-coding locus as well as RNA-dependent functions occurring in cis. Moreover, this illustrates how cells regulate non-coding RNAs to coordinate multiple physiological or oncogenic activities and realize the fine dosing of key factors.
4. Final Remarks
The molecular mechanisms of lncRNAs are manifold, as are their implications in cellular and tumor biology. In this review, we summarized some of the most representative examples that have emerged in recent years and illustrated, in the context of breast cancer, one of the most-studied tumor types. Although it is a common belief that lncRNAs contribute to the key steps of gene expression regulation at either the global level or by optimizing target gene dosage, addressing the complexity of their function or biological activity still represents a major challenge in lncRNA research. Recent technological advances that allow the endogenous investigation of the regulatory function of RNAs together with the development of new sequencing approaches other than short-reading sequencing promise to give new life to this research field and contribute to the development of new fundamental discoveries. In addition to participating in the definition of cancer phenotypes, lncRNAs currently represent promising biomarkers of pathological states and promising therapeutic opportunities. RNA is, by its nature, easier to target and degrade (as compared to proteins or DNA), and the high tissue- and cell-type specificity of lncRNA expression is compatible with highly targeted approaches. All these characteristics make the study of lncRNAs in pathology extremely valuable.
Author Contributions
These authors contributed equally to the work: B.G. and C.T. Conceptualization, writing, reviewing: B.G., C.T. and F.N. Funding acquisition: F.N., B.G. All authors have read and agreed to the published version of the manuscript.
Funding
Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG22851 to F.N.]; B.G. was supported by a FIRC-AIRC fellowship for Italy [25405]. Funding for open access charge: core funding from IIT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef] [PubMed]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Pulido, T.H.; Guigo, R.; Johnson, R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Turnbull, C.; Ahmed, S.; Morrison, J.; Pernet, D.; Renwick, A.; Maranian, M.; Seal, S.; Ghoussaini, M.; Hines, S.; Healey, C.S.; et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010, 42, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Xiu, B.; Chi, Y.; Liu, L.; Chi, W.; Zhang, Q.; Chen, J.; Guo, R.; Si, J.; Li, L.; Xue, J.; et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 2019, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Wijesinghe, S.; Wilson, C.; Halsall, J.; Liloglou, T.; Kanhere, A. A long intergenic non-coding RNA regulates nuclear localization of DNA methyl transferase-1. Iscience 2021, 24, 102273. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Mitra, S.; Subhash, S.; Hertwig, F.; Kanduri, M.; Mishra, K.; Fransson, S.; Ganeshram, A.; Mondal, T.; Bandaru, S.; et al. The Risk-Associated Long Noncoding RNA NBAT-1 Controls Neuroblastoma Progression by Regulating Cell Proliferation and Neuronal Differentiation. Cancer Cell 2014, 26, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Chu, J.; Wu, Y.; Sun, L.; Lv, X.; Zhu, Y.; Li, J.; Guo, Q.; Gong, C.; Liu, B.; et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget 2015, 6, 32410–32425. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Guo, C.; Xia, T.; Zhang, R.; Zen, K.; Pan, Y.; Jin, L. LncCCAT1 Promotes Breast Cancer Stem Cell Function through Activating WNT/beta-catenin Signaling. Theranostics 2019, 9, 7384–7402. [Google Scholar] [CrossRef] [PubMed]
- Ntini, E.; Louloupi, A.; Liz, J.; Muino, J.M.; Marsico, A.; Ørom, U.A.V. Long ncRNA A-ROD activates its target gene DKK1 at its release from chromatin. Nat. Commun. 2018, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.O.A.; Yamamoto, T.; Maehara, K.; Nogami, J.; Ohkawa, Y.; Miura, H.; Poonperm, R.; Hiratani, I.; Nakayama, H.; Nakao, M.; et al. The Eleanor ncRNAs activate the topological domain of the ESR1 locus to balance against apoptosis. Nat. Commun. 2019, 10, 3778. [Google Scholar] [CrossRef]
- Fukuoka, M.; Ichikawa, Y.; Osako, T.; Fujita, T.; Baba, S.; Takeuchi, K.; Tsunoda, N.; Ebata, T.; Ueno, T.; Ohno, S.; et al. The ELEANOR noncoding RNA expression contributes to cancer dormancy and predicts late recurrence of estrogen receptor-positive breast cancer. Cancer Sci. 2022, 113, 2336–2351. [Google Scholar] [CrossRef]
- Rossi, T.; Pistoni, M.; Sancisi, V.; Gobbi, G.; Torricelli, F.; Donati, B.; Ribisi, S.; Gugnoni, M.; Ciarrocchi, A. RAIN Is a Novel Enhancer-Associated lncRNA That Controls RUNX2 Expression and Promotes Breast and Thyroid Cancer. Mol. Cancer Res. 2020, 18, 140–152. [Google Scholar] [CrossRef]
- De Troyer, L.; Zhao, P.; Pastor, T.; Baietti, M.F.; Barra, J.; Vendramin, R.; Dok, R.; Lechat, B.; Najm, P.; Van Haver, D.; et al. Stress-induced lncRNA LASTR fosters cancer cell fitness by regulating the activity of the U4/U6 recycling factor SART3. Nucleic Acids Res. 2020, 48, 2502–2517. [Google Scholar] [CrossRef] [PubMed]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Álvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Gupta, S.C.; Peng, W.-X.; Zhou, N.; Pochampally, R.; Atfi, A.; Watabe, K.; Lu, Z.; Mo, Y.-Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016, 7, e2262. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.-Y.; Siu, M.-T.; Ho, C.-W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Piao, H.-L.; Kim, B.-J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.-C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Richart, L.; Picod-Chedotel, M.-L.; Wassef, M.; Macario, M.; Aflaki, S.; Salvador, M.A.; Héry, T.; Dauphin, A.; Wicinski, J.; Chevrier, V.; et al. XIST loss impairs mammary stem cell differentiation and increases tumorigenicity through Mediator hyperactivation. Cell 2022, 185, 2164–2183.e25. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.-H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNARMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020, 217, e20190950. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP 2A-BP encoded by LINC 00665 inhibits triple-negative breast cancer progression. EMBO J. 2019, 39, e102190. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Bucci, G.; Rizzotto, D.; Bordo, D.; Marzi, M.J.; Puppo, M.; Flinois, A.; Spadaro, D.; Citi, S.; Emionite, L.; et al. LncRNA EPR controls epithelial proliferation by coordinating Cdkn1a transcription and mRNA decay response to TGF-beta. Nat. Commun. 2019, 10, 1969. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-Z.; Chen, M.; Chen, D.; Gao, X.-C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.-R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nature 2017, 19, 1105–1115. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell. Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Zhou, Q.; Guo, J.; Huang, W.; Yu, X.; Xu, C.; Long, X. Linc-ROR promotes the progression of breast cancer and decreases the sensitivity to rapamycin through miR-194-3p targeting MECP2. Mol. Oncol. 2020, 14, 2231–2250. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Y.; Xiao, G.-D.; Zheng, X.-Q.; Wang, J.-C.; Xu, C.-W.; Qin, S.; Ren, H.; Tang, S.-C.; Sun, X. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019, 52, e12534. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Li, R.-H.; Chen, M.; Liu, J.; Shao, C.-C.; Guo, C.-P.; Wei, X.-L.; Li, Y.-C.; Huang, W.-H.; Zhang, G.-J. Long noncoding RNA ATB promotes the epithelial−mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018, 9, 1171. [Google Scholar] [CrossRef]
- Wang, Q.; Li, G.; Ma, X.; Liu, L.; Liu, J.; Yin, Y.; Li, H.; Chen, Y.; Zhang, X.; Zhang, L.; et al. LncRNA TINCR impairs the efficacy of immunotherapy against breast cancer by recruiting DNMT1 and downregulating MiR-199a-5p via the STAT1–TINCR-USP20-PD-L1 axis. Cell Death Dis. 2023, 14, 76. [Google Scholar] [CrossRef]
- Han, L.; Yan, Y.; Zhao, L.; Liu, Y.; Lv, X.; Zhang, L.; Zhao, Y.; Zhao, H.; He, M.; Wei, M. LncRNA HOTTIP facilitates the stemness of breast cancer via regulation of miR-148a-3p/WNT1 pathway. J. Cell. Mol. Med. 2020, 24, 6242–6252. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Jin, T.; Guan, S.; Cheng, S.; Wen, S.; Zeng, H.; Zhao, M.; Yang, L.; Wan, X.; Qiu, Y.; et al. Long non-coding RNA Lnc-408 promotes invasion and metastasis of breast cancer cell by regulating LIMK1. Oncogene 2021, 40, 4198–4213. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Chen, X.; Wang, J.; Bin Liu, B. Glycolysis-related lncRNA TMEM105 upregulates LDHA to facilitate breast cancer liver metastasis via sponging miR-1208. Cell Death Dis. 2023, 14, 80. [Google Scholar] [CrossRef]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLOS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Chen, J.; Yang, L.; Ouyang, Q.; Li, J.; Lao, L.; Zhao, J.; Liu, J.; Lu, Y.; Xing, Y.; et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol. 2018, 19, 1112–1125. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2012, 493, 231–235. [Google Scholar] [CrossRef]
- Sharma, U.; Barwal, T.S.; Malhotra, A.; Pant, N.; Vivek; Dey, D.; Gautam, A.; Tuli, H.S.; Vasquez, K.M.; Jain, A. Long non-coding RNA TINCR as potential biomarker and therapeutic target for cancer. Life Sci. 2020, 257, 118035. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.-C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2015, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.-S.; Yang, M.-C.; Singh, S.; Chou, Y.-C.; Chen, H.-Y.; Wang, M.-Y.; Wang, Y.-C.; Chen, R.-H. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 2019, 38, 5612–5626. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lao, L.; Chen, J.; Li, J.; Zeng, W.; Zhu, X.; Li, J.; Chen, X.; Yang, L.; Xing, Y.; et al. The IRENA lncRNA converts chemotherapy-polarized tumor-suppressing macrophages to tumor-promoting phenotypes in breast cancer. Nat. Cancer 2021, 2, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Li, J.; Huang, H.; Fu, F.; Lin, Y.; Wang, C. LncRNA PCIR Is an Oncogenic Driver via Strengthen the Binding of TAB3 and PABPC4 in Triple Negative Breast Cancer. Front. Oncol. 2021, 11, 630300. [Google Scholar] [CrossRef]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nature 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Lin, X.; Dinglin, X.; Cao, S.; Zheng, S.; Wu, C.; Chen, W.; Li, Q.; Hu, Q.; Zheng, F.; Wu, Z.; et al. Enhancer-Driven lncRNA BDNF-AS Induces Endocrine Resistance and Malignant Progression of Breast Cancer through the RNH1/TRIM21/mTOR Cascade. Cell Rep. 2020, 31, 107753. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Vendramin, R.; Verheyden, Y.; Ishikawa, H.; Goedert, L.; Nicolas, E.; Saraf, K.; Armaos, A.; Ponti, R.D.; Izumikawa, K.; Mestdagh, P.; et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018, 25, 1035–1046. [Google Scholar] [CrossRef]
- Sang, L.; Ju, H.-Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 2021, 3, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-J.; Ma, X.-K.; Xing, Y.-H.; Zheng, C.-C.; Xu, Y.-F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636.e22. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liu, S.; Zhu, H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol. Biol. 2011, 11, 102. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Wang, L.; Zeng, X.; Chen, S.; Ding, L.; Zhong, J.; Zhao, J.C.; Wang, L.; Sarver, A.; Koller, A.; Zhi, J.; et al. BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 2013, 32, 1584–1597. [Google Scholar] [CrossRef]
- Gasperini, M.; Tome, J.M.; Shendure, J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat. Rev. Genet. 2020, 21, 292–310. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, W.L.D.N.M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Vučićević, D.; Corradin, O.; Ntini, E.; Scacheri, P.C.; Ørom, U.A. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle 2015, 14, 253–260. [Google Scholar] [CrossRef]
- Pomerantz, M.M.; Ahmadiyeh, N.; Jia, L.; Herman, P.; Verzi, M.P.; Doddapaneni, H.; Beckwith, C.A.; Chan, J.A.; Hills, A.; Davis, M.; et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009, 41, 882–884. [Google Scholar] [CrossRef] [PubMed]
- Tuupanen, S.; Turunen, M.; Lehtonen, R.; Hallikas, O.; Vanharanta, S.; Kivioja, T.; Björklund, M.; Wei, G.; Yan, J.; Niittymäki, I.; et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009, 41, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yu, H. Shaping of the 3D genome by the ATPase machine cohesin. Exp. Mol. Med. 2020, 52, 1891–1897. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Chen, Y.; Lin, Y.; Ye, L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol. Int. 2018, 42, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Xu, Z.-L.; Zhao, T.-J.; Ye, L.-H.; Zhang, X.-D. Dkk-1 secreted by mesenchymal stem cells inhibits growth of breast cancer cells via depression of Wnt signalling. Cancer Lett. 2008, 269, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Y.; Biasini, A.; Young, R.S.; Marques, A.C. Splicing of enhancer-associated lincRNAs contributes to enhancer activity. Life Sci. Alliance 2020, 3, e202000663. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Ulitsky, I. Production of Spliced Long Noncoding RNAs Specifies Regions with Increased Enhancer Activity. Cell Syst. 2018, 7, 537–547.e3. [Google Scholar] [CrossRef]
- Kitamura, K.; Nimura, K. Regulation of RNA Splicing: Aberrant Splicing Regulation and Therapeutic Targets in Cancer. Cells 2021, 10, 923. [Google Scholar] [CrossRef]
- Rader, S.D.; Guthrie, C. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 2002, 8, 1378–1392. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.G.; Smith, C.W. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Memon, D.; Dawson, K.; Smowton, C.S.; Xing, W.; Dive, C.; Miller, C.J. Hypoxia-driven splicing into noncoding isoforms regulates the DNA damage response. NPJ Genom. Med. 2016, 1, 16020. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790.e30. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Kobayashi, N.; Todo, Y.; Watari, H. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front. Genet. 2018, 9, 471. [Google Scholar] [CrossRef]
- Pang, Y.; Wu, J.; Li, X.; Wang, C.; Wang, M.; Liu, J.; Yang, G. NEAT1/miR-124/STAT3 feedback loop promotes breast cancer progression. Int. J. Oncol. 2019, 55, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. et Biophys. Acta (BBA) -Gen. Subj. 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Yu, J.; Jin, T.; Zhang, T. Suppression of Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Potentiates Cell Apoptosis and Drug Sensitivity to Taxanes and Adriamycin in Breast Cancer. Experiment 2020, 26, e922672. [Google Scholar] [CrossRef]
- Menghi, F.; Barthel, F.P.; Yadav, V.; Tang, M.; Ji, B.; Tang, Z.; Carter, G.W.; Ruan, Y.; Scully, R.; Verhaak, R.G.; et al. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell 2018, 34, 197–210.e5. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ron, M.; Mikl, M.; Segal, E.; Ulitsky, I. Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol. Cell 2020, 79, 251–267.e6. [Google Scholar] [CrossRef]
- Szostak, N.; Royo, F.; Rybarczyk, A.; Szachniuk, M.; Blazewicz, J.; Del Sol, A.; Falcon-Perez, J.M. Sorting signal targeting mRNA into hepatic extracellular vesicles. RNA Biol. 2014, 11, 836–844. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Huang, J.G.; Yang, L.; Gong, A.H.; Mo, Y.Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer 2017, 16, 161. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Ripoche, M.-A.; Le Digarcher, A.; Watrin, F.; Ziyyat, A.; Forné, T.; Jammes, H.; Ainscough, J.F.X.; Surani, M.A.; Journot, L.; et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.-B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.-S.; Zhang, H.; et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.-H.; Fan, W.-J.; Meng, Y.-T.; Li, M.-M.; Li, T.-T.; Cui, B.; Wang, H.-F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018, 37, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Munschauer, M.; Nguyen, C.T.; Sirokman, K.; Hartigan, C.R.; Hogstrom, L.; Engreitz, J.M.; Ulirsch, J.C.; Fulco, C.P.; Subramanian, V.; Chen, J.; et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 2018, 561, 132–136. [Google Scholar] [CrossRef]
- Shtivelman, E.; Bishop, J.M. Effects of translocations on transcription from PVT. Mol. Cell. Biol. 1990, 10, 1835–1839. [Google Scholar] [CrossRef]
- Huppi, K.; Siwarski, D. Chimeric transcripts with an open reading frame are generated as a result of translocation to thePvt-1 region in mouse B-cell tumors. Int. J. Cancer 1994, 59, 848–851. [Google Scholar] [CrossRef]
- Cho, S.W.; Xu, J.; Sun, R.; Mumbach, M.R.; Carter, A.C.; Chen, Y.G.; Yost, K.E.; Kim, J.; He, J.; Nevins, S.A.; et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018, 173, 1398–1412.e22. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Palacio, O.R.; Dunning, M.; Speed, D.; Lynch, A.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2020, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, W.; Hu, H.; Zhang, T.; Wu, T.; Li, X.; Li, Y.; Kong, Q.; Lu, H.; Lu, Z. Long noncoding RNA PVT1 promotes breast cancer proliferation and metastasis by binding miR-128-3p and UPF1. Breast Cancer Res. 2021, 23, 115. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Kuo, W.-L.; Stilwell, J.L.; Takano, H.; Lapuk, A.V.; Fridlyand, J.; Mao, J.-H.; Yu, M.; Miller, M.A.; Santos, J.L.; et al. Amplification of PVT1 Contributes to the Pathophysiology of Ovarian and Breast Cancer. Clin. Cancer Res. 2007, 13, 5745–5755. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, Y.; Kong, W.; Fu, L.; Liu, Y.; Yao, Q.; Yuan, Y. PVT1-derived miR-1207-5p promotes breast cancer cell growth by targeting STAT6. Cancer Sci. 2017, 108, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Gu, X.; Yang, X.; Meng, Y. MiR-1204 promotes ovarian squamous cell carcinoma growth by increasing glucose uptake. Biosci. Biotechnol. Biochem. 2019, 83, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Liu, W.; Li, B.; Chen, D.; Hu, F.; Wang, L.; Liu, X.M.; Cui, R.; Liu, R. MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene 2019, 38, 4820–4834. [Google Scholar] [CrossRef]
- You, L.; Wang, H.; Yang, G.; Zhao, F.; Zhang, J.; Liu, Z.; Zhang, T.; Liang, Z.; Liu, C.; Zhao, Y. Gemcitabine exhibits a suppressive effect on pancreatic cancer cell growth by regulating processing of PVT 1 to miR1207. Mol. Oncol. 2018, 12, 2147–2164. [Google Scholar] [CrossRef]
- Das, D.K.; Ogunwobi, O.O. A novel microRNA-1207-3p/FNDC1/FN1/AR regulatory pathway in prostate cancer. RNA Dis. 2017, 4, e1503. [Google Scholar]
- Wang, W.; Zhou, R.; Wu, Y.; Liu, Y.; Su, W.; Xiong, W.; Zeng, Z. PVT1 Promotes Cancer Progression via MicroRNAs. Front. Oncol. 2019, 9, 609. [Google Scholar] [CrossRef]
- Wu, X.-Z.; Cui, H.-P.; Lv, H.-J.; Feng, L. Knockdown of lncRNA PVT1 inhibits retinoblastoma progression by sponging miR-488-3p. Biomed. Pharmacother. 2019, 112, 108627. [Google Scholar] [CrossRef]
- Hua, X.; Xiao, Y.; Pan, W.; Li, M.; Huang, X.; Liao, Z.; Xian, Q.; Yu, L. miR-186 inhibits cell proliferation of prostate cancer by targeting GOLPH3. Am. J. Cancer Res. 2016, 6, 1650–1660. [Google Scholar] [PubMed]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Carramusa, L.; Contino, F.; Ferro, A.; Minafra, L.; Perconti, G.; Giallongo, A.; Feo, S. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J. Cell. Physiol. 2007, 213, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef]
- Oh, S.; Shao, J.; Mitra, J.; Xiong, F.; D’Antonio, M.; Wang, R.; Garcia-Bassets, I.; Ma, Q.; Zhu, X.; Lee, J.-H.; et al. Enhancer release and retargeting activates disease-susceptibility genes. Nature 2021, 595, 735–740. [Google Scholar] [CrossRef]
- Olivero, C.E.; Martínez-Terroba, E.; Zimmer, J.; Liao, C.; Tesfaye, E.; Hooshdaran, N.; Schofield, J.; Bendor, J.; Fang, D.; Simon, M.D.; et al. p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cell 2020, 77, 761–774.e8. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).