Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants
Abstract
1. Introduction
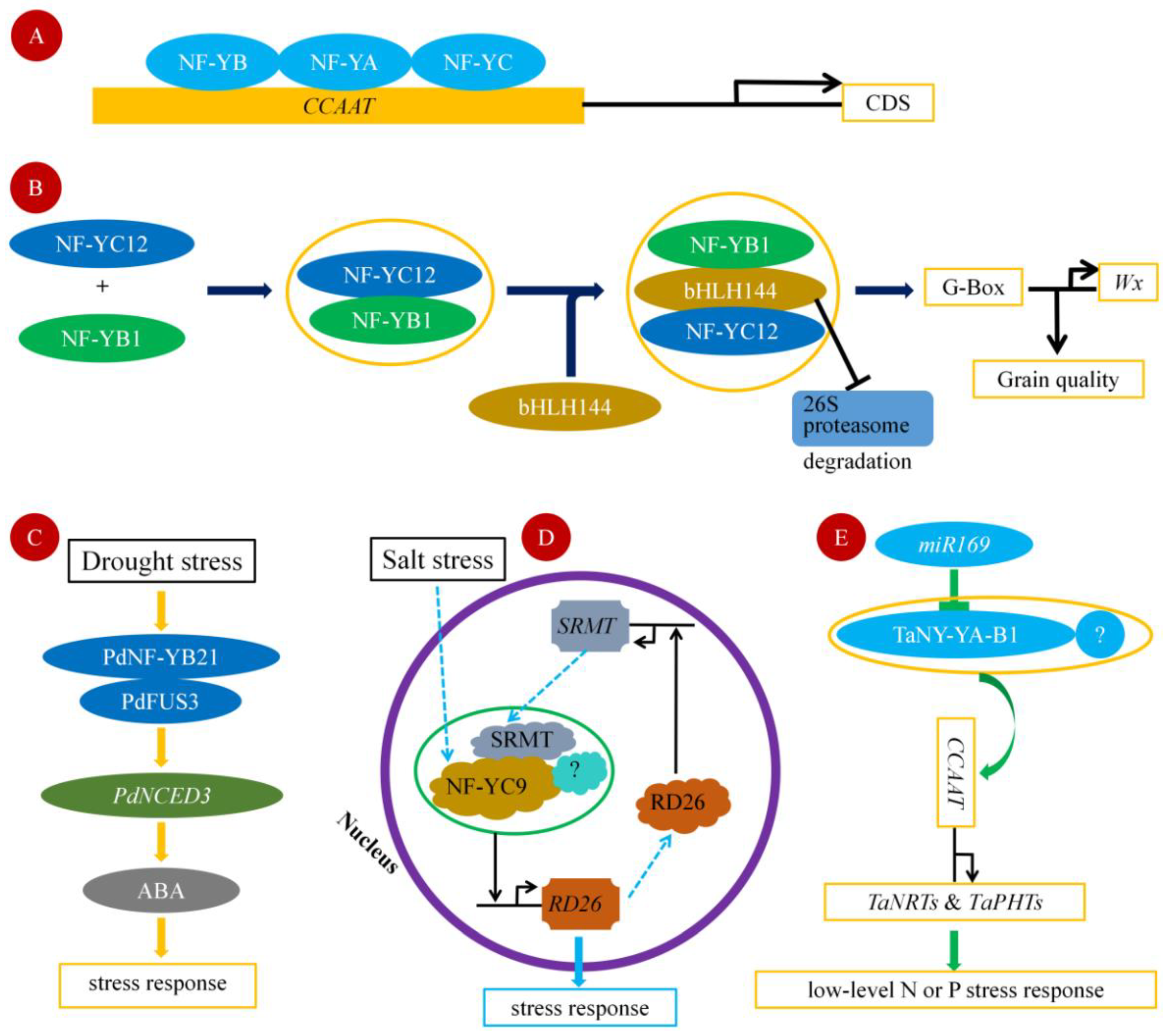
2. Functional Mechanism of NF-Y Transcription Factor
3. Drought Stress
4. Salt Stress
5. Nutrient Stress
6. Osmotic Stress
7. Other Stress
8. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sinha, S.; Kim, I.S.; Sohn, K.Y.; De Crombrugghe, B.; Maity, S.N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996, 16, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Gusmaroli, G.; Tonelli, C.; Mantovani, R.J.G. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 2002, 283, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Bucher, P.; Trifonov, E.N. CCAAT box revisited: Bidirectionality, location and context. J. Biomol. Struct. Dyn. 1988, 5, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Baudin, M.; Laloum, T.; Lepage, A.; Ripodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar]
- Guntram, S. NF-Y and SP transcription factors-New insights in a long-standing liaison. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 590–597. [Google Scholar]
- Albani, D.; Robert, L.S. Cloning and characterization of a Brassica napus gene encoding a homologue of the B sub-unit of a heteromeric CCAAT-binding factor. Gene 1995, 167, 209–213. [Google Scholar] [CrossRef]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Yan, H.L.; Wu, F.W.; Jiang, G.X.; Xiao, L.; Li, Z.W.; Duan, X.W.; Jiang, Y.M. Genome-wide identification, characterization and expression analysis of NF-Y gene family in relation to fruit ripening in banana. Postharvest Biol. Technol. 2019, 151, 98–110. [Google Scholar] [CrossRef]
- Panahi, B.; Mohammadi, S.A.; Ruzicka, K.; Abbasi Holaso, H.; Mehrjerdi, M.Z. Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol. Mol. Biol. Plants 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Liu, R.; Wu, M.; Liu, H.L.; Gao, Y.M.; Chen, J.; Yan, H.W.; Xiang, Y. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Physiol. Plant 2021, 3, 309–327. [Google Scholar] [CrossRef]
- Su, H.H.; Cao, Y.Y.; Ku, L.X.; Yao, W.; Cao, Y.Y.; Ren, Z.Z.; Dou, D.D.; Wang, H.T.; Ren, Z.B.; Liu, H.F.; et al. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 2018, 69, 5177–5189. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, K.; Ju, Z.; Cao, D.Y.; Fu, D.Q.; Zhu, H.L.; Zhu, B.Z.; Luo, Y.B. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.L.; Li, Q.; Yao, K.; Zhang, Y.; Meng, S.; Yin, W.L.; Xia, X.L. Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Front. Plant Sci. 2018, 9, 954. [Google Scholar] [CrossRef]
- Huang, M.; Hu, Y.; Liu, X.; Li, Y.; Hou, X. Arabidopsis Leafy Cotyledon1 mediates postembryonic development via interacting with Phytochrome-Interacting Factor4. Plant Cell 2015, 27, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Zhang, Y.; Wang, X.W.; Han, X.; An, Y.; Lin, S.W.; Shen, C.; Wen, J.L.; Liu, C.; Yin, W.L.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Wu, X.L.; Shi, H.F.; Guo, Z.F. Overexpression of a NF-YC gene results in enhanced drought and salt tolerance in transgenic Seashore Paspalum. Front. Plant Sci. 2018, 9, 1355. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ji, H.A. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Mantovani, R. The molecular biology of the CCAAT-binding factor NF-Y. Gene 1999, 239, 15–27. [Google Scholar] [CrossRef]
- Myers, Z.A.; Holt, B.F., III. Nuclear Factor-Y: Still complex after all these years? Curr. Opin. Plant Biol. 2018, 45, 96–102. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar] [CrossRef]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.; Horner, D.S.; Holt, B.F., III; Mantovani, R. Constans imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. Constans and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Mach, J. Constans companion: CO binds the NF-YB/NF-YC dimer and confers sequence-specific DNA binding. Plant Cell 2017, 29, 1183. [Google Scholar] [CrossRef]
- Chaves-Sanjuan, A.; Gnesutta, N.; Gobbini, A.; Martignago, D.; Bernardini, A.; Fornara, F.; Mantovani, R.; Nardini, M. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J. 2020, 105, 49–61. [Google Scholar] [CrossRef]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef]
- Tang, N.; Ma, S.; Zong, W.; Yang, N.; Lv, Y.; Yan, C.; Guo, Z.; Li, J.; Li, X.; Xiang, Y.; et al. MODD Mediates Deactivation and Degradation of OsbZIP46 to Negatively Regulate ABA Signaling and Drought Resistance in Rice. Plant Cell 2016, 28, 2161–2177. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback Regulation of ABA Signaling and Biosynthesis by a bZIP Transcription Factor Targets Drought-Resistance-Related Genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef]
- Chong, L.; Xu, R.; Huang, P.; Guo, P.; Zhu, M.; Du, H.; Sun, X.; Ku, L.; Zhu, J.K.; Zhu, Y. The tomato OST1-VOZ1 module regulates drought-mediated flowering. Plant Cell 2022, 34, 2001–2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Bu, T.T.; Cheng, Q.; Dong, L.D.; Su, T.; Chen, Z.M.; Kong, F.J.; Gong, Z.Z.; Liu, B.H.; Li, M.N. Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 2021, 229, 2660–2675. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.D.; Jian, C.; Cheng, X.X.; Chen, B.; Mei, F.M.; Li, F.F.; Zhang, Y.F.; Li, S.M.; Du, L.Y.; Li, T.; et al. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol. J. 2022, 20, 846–861. [Google Scholar] [CrossRef]
- Xuanyuan, G.; Lu, C.; Zhang, R.; Jiang, J. Overexpression of StNFYB3.1 reduces photosynthetic capacity and tuber production, and promotes ABA-mediated stomatal closure in potato (Solanum tuberosum L.). Plant Sci. 2017, 261, 50–59. [Google Scholar] [CrossRef]
- Bi, C.; Ma, Y.; Wang, X.F.; Zhang, D.P. Overexpression of the transcription factor NF-YC9 confers abscisic acid hypersensitivity in Arabidopsis. Plant Mol. Biol. 2017, 955, 425–439. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.W.; Huang, M.K.; Tang, Y.; Li, Y.G.; Li, L.; Hou, X.L. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Han, X.; Tang, S.; An, Y.; Zheng, D.C.; Xia, X.L.; Yin, W.L. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef]
- Chu, H.D.; Nguyen, K.H.; Watanabe, Y.; Le, D.T.; Pham, T.L.T.; Mochida, K.; Tran, L.P. Identification, structural characterization and gene expression analysis of members of the nuclear factor-Y family in Chickpea (Cicer arietinum L.) under dehydration and abscisic acid treatments. Int. J. Mol. Sci. 2018, 19, 3290. [Google Scholar] [CrossRef]
- Yu, T.F.; Liu, Y.; Fu, J.D.; Ma, J.; Fang, Z.W.; Chen, J.; Zheng, L.; Lu, Z.W.; Zhou, Y.B.; Chen, M.; et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 2021, 19, 2589–2605. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Li, Y.J.; Fang, Y.; Fu, Y.R.; Huang, J.G.; Wu, C.A.; Zheng, C.C. NFYA1 is involved in regulation of post germination growth arrest under salt stress in Arabidopsis. PLoS ONE 2013, 8, e61289. [Google Scholar]
- Mu, J.Y.; Tan, H.L.; Hong, S.L.; Liang, Y.; Zuo, J.R. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant 2013, 6, 188–201. [Google Scholar] [CrossRef]
- Ma, X.Y.; Zhu, X.L.; Li, C.L.; Song, Y.L.; Zhang, W.; Xia, G.M.; Wang, M. Overexpression of wheat NF-YA10 gene regulates the salinity stress response in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 86, 34–43. [Google Scholar] [CrossRef]
- Miyoshi, K.; Ito, Y.; Serizawa, A.; Kurata, N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003, 36, 532–540. [Google Scholar] [CrossRef]
- Li, S.G.; Zhang, N.; Zhu, X.; Ma, R.; Liu, S.Y.; Wang, X.; Yang, J.W.; Si, H.J. Genome-wide analysis of NF-Y genes in potato and functional identification of StNF-YC9 in drought tolerance. Front. Plant Sci. 2021, 12, 749688. [Google Scholar] [CrossRef]
- Pereira, S.L.S.; Martins, C.P.S.; Sousa, A.O.; Camillo, L.R.; Araújo, C.P.; Alcantara, G.M.; Camargo, D.S.; Cidade, L.C.; de Almeida, A.F.; Costa, M.G.C. Genome-wide characterization and expression analysis of citrus NUCLEAR FACTOR-Y (NF-Y) transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance. PLoS ONE 2018, 13, e0199187. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Zhang, Y.Y.; Li, T.J.; Ni, C.Y.; Bai, X.Y.; Lin, R.Z.; Xiao, K. TaNF-YA7-5B, a gene encoding nuclear factor Y (NF-Y) subunit A in Triticum aestivum, confers plant tolerance to PEG-inducing dehydration simulating drought through modulating osmotic stress-associated physiological processes. Plant Physiol. Biochem. 2022, 188, 81–96. [Google Scholar] [CrossRef]
- Qu, B.Y.; He, X.; Wang, J.; Zhao, Y.Y.; Teng, W.; Shao, A.; Zhao, X.Q.; Ma, W.Y.; Wang, J.Y.; Li, B.; et al. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 2015, 167, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, Y.Y.; Sun, Y.T.; Peng, X.; Zhang, X.; Zhou, X.; Jiao, J.; Zhai, Z.F.; Xiao, Y.Q.; Wang, W.L.; et al. Expression of the Malus sieversii NF-YB21 encoded gene confers tolerance to osmotic stresses in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 9777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, D.; Liu, Y.J.; Luo, C.B.; Zhou, Y.N.; Zhang, L.Y. Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 94, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor (NF-Y) YB subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A role for Gigantea. Plant Signal. Behav. 2013, 8, e24820. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Zhao, S.S.; Zhang, Q.K.; Liu, M.Y.; Zhou, H.P.; Ma, C.L.; Wang, P.P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Liu, M.M.; Pan, Z.X.; Yu, J.; Zhu, L.; Zhao, M.Z.; Wang, Y.F.; Chen, P.; Liu, C.; Hu, J.; Liu, T.; et al. Transcriptome-wide characterization, evolutionary analysis, and expression pattern analysis of the NF-Y transcription factor gene family and salt stress response in Panax ginseng. BMC Plant Biol. 2022, 22, 320. [Google Scholar] [CrossRef]
- An, Y.X.; Suo, X.; Niu, Q.C.; Yin, S.X.; Chen, L. Genome-wide identification and analysis of the NF-Y transcription factor family reveal its potential roles in salt stress in Alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 2022, 23, 6426. [Google Scholar] [CrossRef]
- Wei, Q.; Wen, S.Y.; Lan, C.Y.; Yu, Y.; Chen, G.J. Genome-wide identification and expression profile analysis of the NF-Y transcription factor gene family in Petunia hybrida. Plants 2020, 9, 336. [Google Scholar] [CrossRef]
- Tong, S.F.; Wang, Y.B.; Chen, N.N.; Wang, D.Y.; Liu, B.; Wang, W.W.; Chen, Y.; Liu, J.Q.; Ma, T.; Jiang, Y.Z. PtoNF-YC9-SRMT-PtoRD26 module regulates the high saline tolerance of a triploid poplar. Genome Biol. 2022, 23, 148. [Google Scholar] [CrossRef]
- Yang, W.L.; Wang, B.M.; Wang, J.M.; He, C.M.; Zhang, D.F.; Li, P.; Zhang, J.R.; Li, Z.X. Transcription factors ZmNF-YA1 and ZmNF-YB16 regulate plant growth and drought tolerance in maize. Plant Physiol. 2022, 190, 1506–1525. [Google Scholar] [CrossRef]
- Lu, L.; Wei, W.; Tao, J.J.; Lu, X.; Bian, X.H.; Hu, Y.; Cheng, T.; Yin, C.C.; Zhang, W.K.; Chen, S.Y.; et al. Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 2362–2379. [Google Scholar] [CrossRef]
- Ma, X.Y.; Li, C.L.; Wang, M. Wheat NF-YA10 functions independently in salinity and drought stress. Bioengineered 2015, 4, 245–247. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. Wrky transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Upreti, K.K.; Murti, G.S.R. Response of grape rootstocks to salinity: Changes in root growth, polyamines and abscisic acid. Biol. Plant 2010, 54, 730–734. [Google Scholar] [CrossRef]
- Xie, K.; Ren, Y.H.; Chen, A.Q.; Yang, C.F.; Zheng, Q.S.; Chen, J.; Wang, D.S.; Li, Y.T.; Hu, S.J.; Xu, G.H. Plant nitrogen nutrition: The roles of arbuscular mycorrhizal fungi. J. Plant Physiol. 2022, 269, 153591. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Takahashi, H. Improving nitrogen use efficiency: From cells to plant systems. J. Exp. Bot. 2020, 71, 4359–4364. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Gao, S.P.; Chu, C.C. Improvement of nutrient use efficiency in rice: Current toolbox and future perspectives. Theor. Appl. Genet. 2020, 133, 1365–1384. [Google Scholar] [CrossRef]
- Zheng, X.R.; Zhang, H.; Zhang, L.M.; Xu, F.S.; Shi, L.; Wang, S.L.; Hong, J.; Ding, G.D. Identification and comprehensive analysis of the Nuclear Factor-Y family genes reveal their multiple roles in response to nutrient deficiencies in Brassica napus. Int. J. Mol. Sci. 2021, 22, 10354. [Google Scholar] [CrossRef]
- Bu, F.; Rutten, L.; Roswanjaya, Y.P.; Kulikova, O.; Rodriguez-Franco, M.; Ott, T.; Bisseling, T.; van Zeijl, A.; Geurts, R. Mutant analysis in the nonlegume Parasponia andersonii identifies NIN and NF-YA1 transcription factors as a core genetic network in nitrogen-fixing nodule symbioses. New Phytol. 2020, 226, 541–554. [Google Scholar] [CrossRef]
- Li, L.; Zheng, W.G.; Zhu, Y.B.; Ye, H.X.; Tang, B.Y.; Arendsee, Z.W.; Jones, D.; Li, R.; Ortiz, D.; Zhao, X.F.; et al. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 14734–14739. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, L.Y.; Chen, X.; Zhou, M.Y.; Shi, W.G.; Deng, S.R.; Luo, Z.B. Genome-wide identification and characterization of the NF-YA gene family and its expression in response to different nitrogen forms in Populus × canescens. Int. J. Mol. Sci. 2022, 23, 11217. [Google Scholar] [CrossRef]
- Leyva-González, M.A.; Ibarra-Laclette, E.; Cruz-Ramírez, A.; Herrera-Estrella, L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 2012, 7, e48138. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef]
- Zhao, M.; Ding, H.; Zhu, J.-K.; Zhang, F.; Li, W.-X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef]
- Ruan, W.F.; Cai, H.B.; Xu, X.M.; Man, Y.; Wang, R.; Tai, Y.P.; Chen, Z.B.; Vymazal, J.; Chen, J.X.; Yang, Y.; et al. Efficiency and plant indication of nitrogen and phosphorus removal in constructed wetlands: A field-scale study in a frost-free area. Sci. Total Environ. 2021, 799, 149301. [Google Scholar] [CrossRef]
- Bremer, E.; Krämer, R. Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 2019, 73, 313–334. [Google Scholar] [CrossRef]
- Rodas-Junco, B.A.; Racagni-Di-Palma, G.E.; Canul-Chan, M.; Usorach, J.; Hernández-Sotomayor, S.M.T. Link between lipid second messengers and osmotic stress in plants. Int. J. Mol. Sci. 2021, 22, 2658. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Luo, L.; Zhang, X.R.; Lv, Y.Y.; Zhu, S.Q.; Kong, L.R.; Wan, Y.S.; Liu, F.Z.; Zhang, K. Genome-wide identification and abiotic stress response pattern analysis of NF-Y gene family in Peanut (Arachis Hypogaea, L.). Trop. Plant Biol. 2021, 14, 329–344. [Google Scholar] [CrossRef]
- Yang, J.; Wan, X.L.; Guo, C.; Zhang, J.W.; Bao, M.Z. Identification and expression analysis of nuclear factor Y families in Prunus mume under different abiotic stresses. Biol. Plant 2016, 60, 419–426. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, D.; Keetman, U.; Grimm, B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 2012, 13, 3458–3477. [Google Scholar] [CrossRef]
- Rao, S.; Gupta, A.; Bansal, C.; Sorin, C.; Crespi, M.; Mathur, S. A conserved HSF: miR169:NF-YA loop involved in tomato and Arabidopsis heat stress tolerance. Plant J. 2022, 112, 7–26. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef]
- Maheshwari, P.; Kummari, D.; Palakolanu, S.R.; Tejaswi, U.N.; Nagaraju, M.; Rajasheker, G.; Jawahar, G.; Jalaja, N.; Rathnagiri, P.; Kishor, P.B.K. Genome-wide identification and expression profile analysis of nuclear factor Y family genes in Sorghum bicolor L. (Moench). PLoS ONE 2019, 14, e0222203. [Google Scholar] [CrossRef]
- Gyula, P.; Ivett, B.; Tamas, T.; Irina, M.; Tamas, D.; György, S. Ambient temperature regulates the expression of a small set of sRNAs infuencing plant development through NFYA2 and YUC2. Plant Cell Environ. 2018, 41, 2404–2417. [Google Scholar] [CrossRef]
- Zheng, T.; Lv, J.; Sadeghnezhad, E.; Cheng, J.; Jia, H. Transcriptomic and metabolomic profiling of strawberry during postharvest cooling and heat storage. Front. Plant Sci. 2022, 12, 1009747. [Google Scholar] [CrossRef]
| Gene | Species | Phenotypic Manifestation | References |
|---|---|---|---|
| PdNF-YB21 | Populus | Enhanced drought tolerance and promoted root development. | [16] |
| PdNF-YB7 | Populus | Enhanced sensitivity to ABA and reduced water loss. | [39] |
| GmNF-YA16/YB2/YC14 | Glycine max L. | Enhanced tolerance to drought stress and increased sensitivity to ABA. | [41] |
| AtNF-YC3/YC4/YC9 | Arabidopsis thaliana | Enhanced drought tolerance and promoted flowering. | [19] |
| StNF-YC9 | Solanum tuberosum L. | Better growth under drought. | [47] |
| CsNF-YA5 | Citrus sinensis | Enhanced leaf water retention capacity. | [48] |
| TaNF-YA7/YB2/YC7 | Triticum aestivum L. | Regulated stomatal closure, enhanced leaf water retention capacity and maintained cell ROS homeostasis. | [49] |
| PgNF-YB9/C2/C7 | Panax ginseng | Response to salt stress. | [60] |
| MsNF-YB2 | Medicago L. | Regulation of Ca2+ signaling pathway under salt stress. | [61] |
| NF-YC9 | Populus | Interaction with MYB transcription factor enhances salt tolerance of poplar. | [63] |
| ZmNF-YA1 | Zea mays L. | Activate the salt tolerance gene. | [64] |
| TaNF-YA10 | Triticum aestivum L. | Enhanced plant sensitivity to salinity. | [66] |
| AtNF-YA1 | Arabidopsis thaliana | Negative regulation of plant development under salt stress. | [43] |
| GmNF-YA1/YB1 | Glycine max L. | Provide carbohydrates. | [50] |
| GmNF-YC4 | Glycine max L. | Increased protein level and reduced starch content. | [74] |
| AtNF-YA4/YB3/YC2 | Arabidopsis thaliana | Up-regulated the expression of ER stress-induced genes. | [84] |
| AtNF-YA2/YB3 | Arabidopsis thaliana | Regulation of plant heat tolerance. | [87] |
| SbNF-YA7/A8 | Sorghum bicolor L. | Response temperature change. | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Liu, S.; Ren, T.; Niu, M.; Liu, X.; Liu, C.; Wang, H.; Yin, W.; Xia, X. Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. Int. J. Mol. Sci. 2023, 24, 4426. https://doi.org/10.3390/ijms24054426
Zhang H, Liu S, Ren T, Niu M, Liu X, Liu C, Wang H, Yin W, Xia X. Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. International Journal of Molecular Sciences. 2023; 24(5):4426. https://doi.org/10.3390/ijms24054426
Chicago/Turabian StyleZhang, Han, Shujing Liu, Tianmeng Ren, Mengxue Niu, Xiao Liu, Chao Liu, Houling Wang, Weilun Yin, and Xinli Xia. 2023. "Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants" International Journal of Molecular Sciences 24, no. 5: 4426. https://doi.org/10.3390/ijms24054426
APA StyleZhang, H., Liu, S., Ren, T., Niu, M., Liu, X., Liu, C., Wang, H., Yin, W., & Xia, X. (2023). Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. International Journal of Molecular Sciences, 24(5), 4426. https://doi.org/10.3390/ijms24054426






