Inositol in Disease and Development: Roles of Catabolism via myo-Inositol Oxygenase in Drosophila melanogaster
Abstract
1. Introduction
2. Results
2.1. MIOX Homolog Identified in D. melanogaster (CG6910) Is Regulated in Response to Dietary myo-Inositol
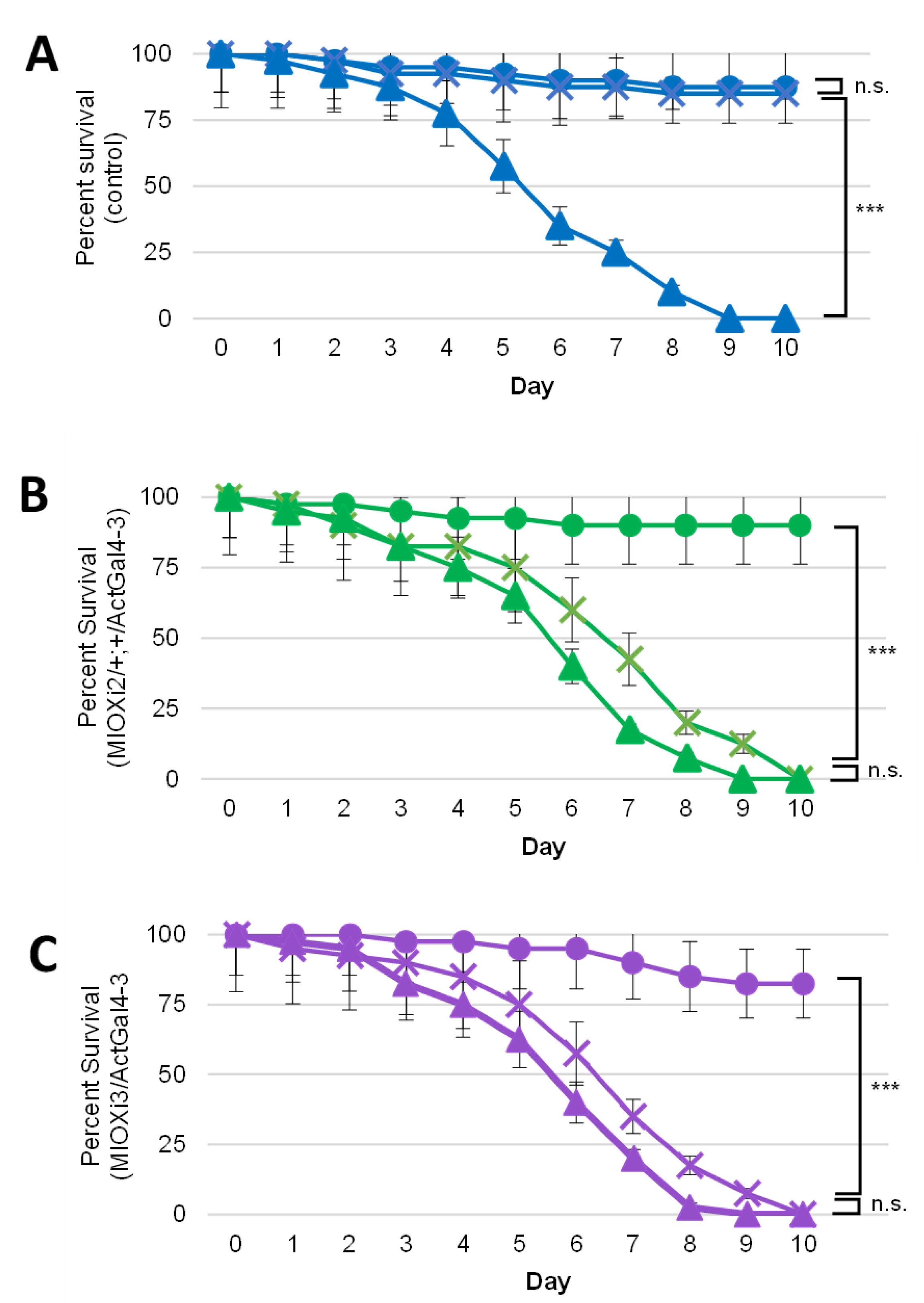
2.2. Dietary myo-Inositol Supports Survival of Wild-Type (CS) D. melanogaster Adults but Not of MIOX Knockdown Strains
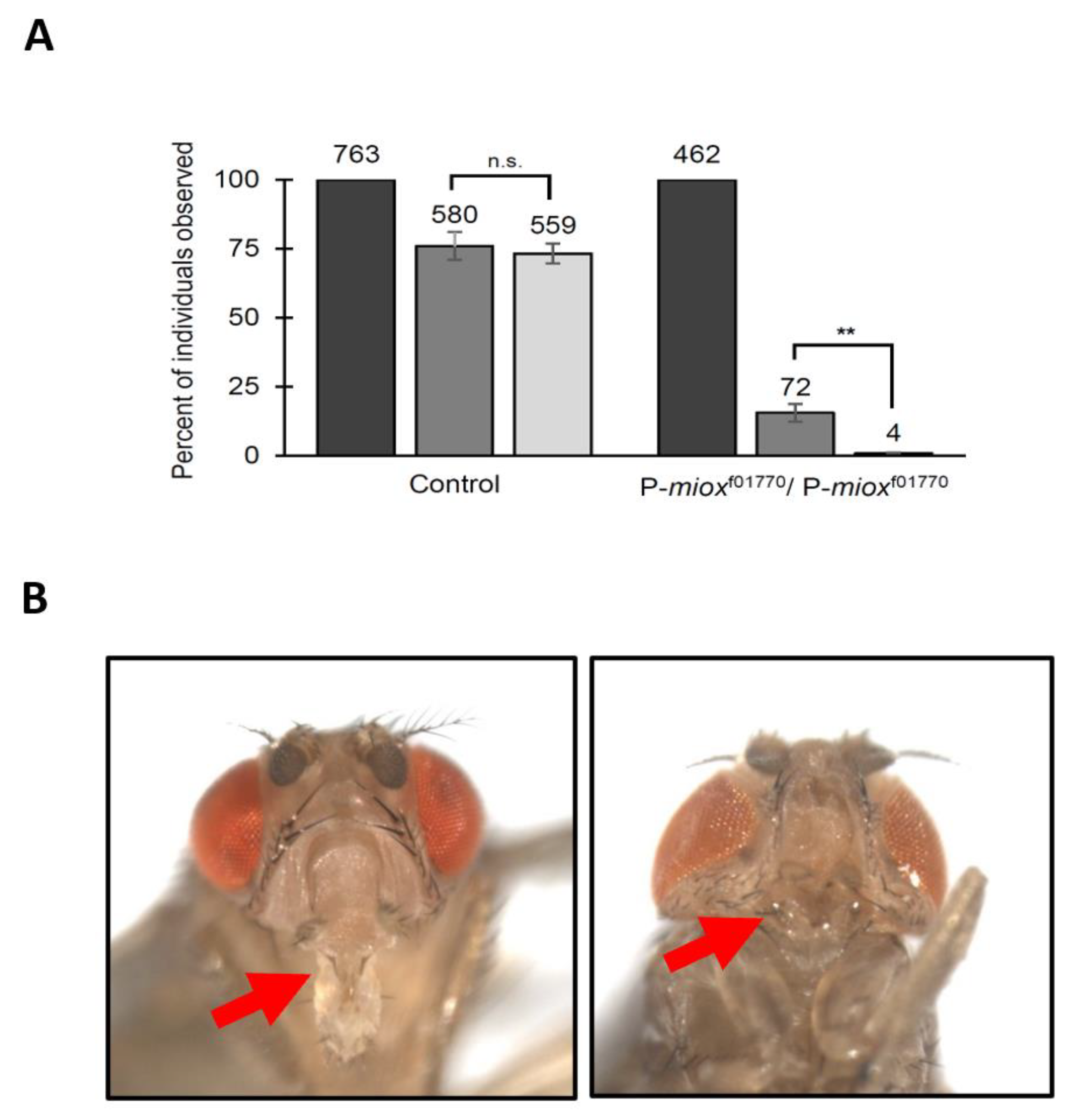
2.3. Disruption of myo-Inositol Catabolism via Piggybac WH-Element Insertion in MIOX Results in Developmental Defects
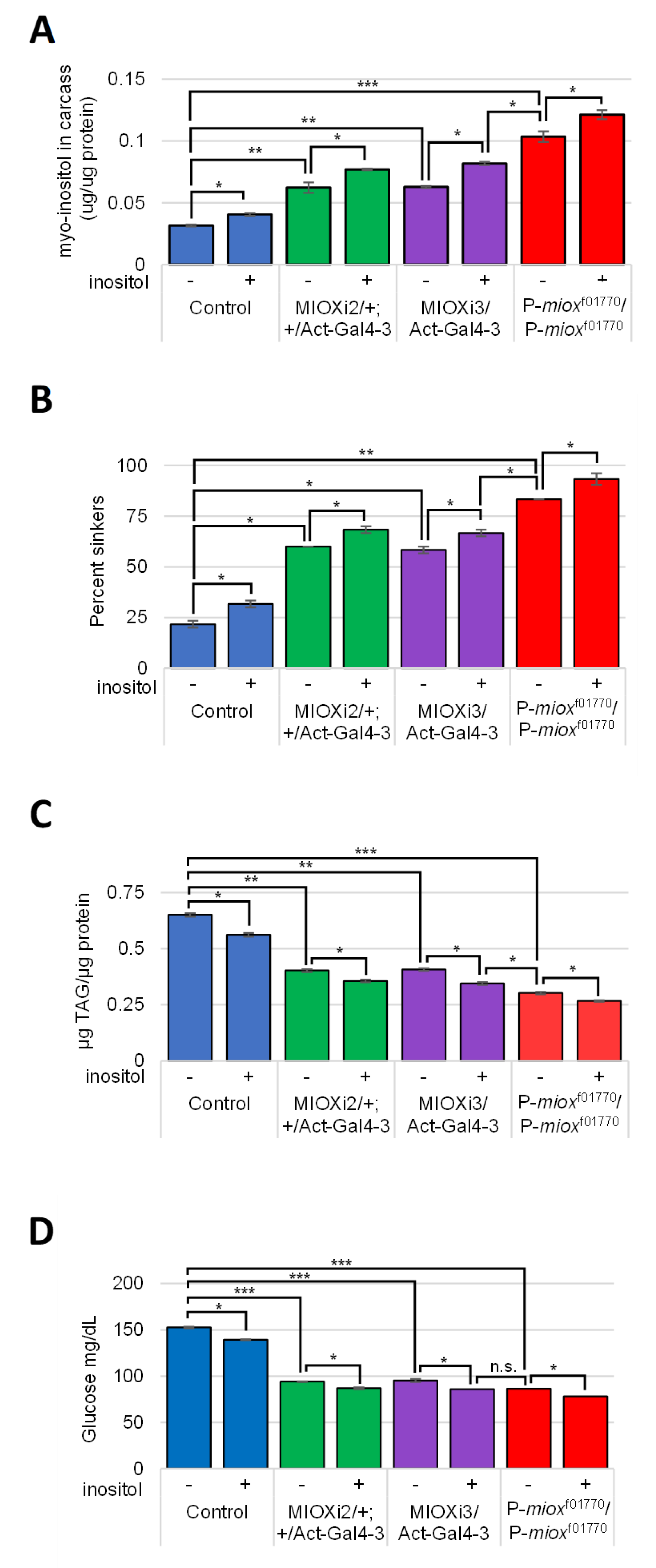
2.4. Reduced myo-Inositol Catabolism Increases myo-Inositol Levels in Larvae
2.5. Increased myo-Inositol Decreases Larval Obesity and Hemolymph Glucose
3. Discussion
4. Materials and Methods
4.1. Fly Stocks and Maintenance
4.2. RNA Extraction and qRT-PCR
4.3. Protein Extraction and the MIOX Activity Assay
4.4. Survival Studies
4.5. Pupariation and Eclosion
4.6. Light Microscopy
4.7. myo-Inositol Assay
4.8. Buoyancy Assay
4.9. Triacylglyceride (TAG) Assay
4.10. Hemolymph Glucose Assay
4.11. Computational Analyses
4.12. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bevilacqua, A.; Bizzarri, M. Inositols in insulin signaling and glucose metabolism. Int. J. Endocrinol. 2018, 2018, 1968450. [Google Scholar] [CrossRef]
- Chhetri, D.R. Myo-inositol and its derivatives: Their emerging role in the treatment of human diseases. Front. Pharmacol. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Guo, T.; Nan, Z.; Miao, C.; Jin, X.; Yang, W.; Wang, Z.; Tu, Y.; Bao, H.; Lyu, J.; Zheng, H.; et al. The autophagy-related gene Atg101 in Drosophila regulates both neuron and midgut homeostasis. J. Biol. Chem. 2019, 294, 5666–5676. [Google Scholar] [CrossRef] [PubMed]
- D’Anna, R.; Corrado, F.; Loddo, S.; Gullo, G.; Giunta, L.; Di Benedetto, A. Myoinositol plus α-lactalbumin supplementation, insulin resistance and birth outcomes in women with gestational diabetes mellitus: A randomized, controlled study. Sci. Rep. 2021, 11, 8866. [Google Scholar] [CrossRef]
- Kiani, A.K.; Paolacci, S.; Calogero, A.E.; Cannarella, R.; Di Renzo, G.C.; Gerli, S.; Della Morte, C.; Busetto, G.M.; De Berardinis, E.; Del Giudice, F.; et al. From Myo-inositol to D-chiro-inositol molecular pathways. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 2390–2402. [Google Scholar] [CrossRef]
- Patkee, P.A.; Baburamani, A.A.; Long, K.R.; Dimitrova, R.; Ciarrusta, J.; Allsop, J.; Hughes, E.; Kangas, J.; McAlonan, G.M.; Rutherford, M.A.; et al. Neurometabolite mapping highlights elevated myo-inositol profiles within the developing brain in down syndrome. Neurobiol. Dis. 2021, 153, 105316–105325. [Google Scholar] [CrossRef]
- Dinicola, S.; Unfer, V.; Facchinetti, F.; Soulage, C.O.; Greene, N.D.; Bizzarri, M.; Laganà, A.S.; Chan, S.-Y.; Bevilacqua, A.; Pkhaladze, L.; et al. Inositols: From established knowledge to novel approaches. Int. J. Mol. Sci. 2021, 22, 10575. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Sun, L.Y.; Yang, A.L.; Liao, J.; Yang, G.Y. Overview of inositol and inositol phosphates on chemoprevention of colitis-induced carcinogenesis. Molecules 2020, 26, 31. [Google Scholar] [CrossRef] [PubMed]
- Dyck, P.J.; Zimmerman, B.R.; Vilen, T.H.; Minnerath, S.R.; Karnes, J.L.; Yao, J.K.; Poduslo, J.F. Nerve Glucose, Fructose, Sorbitol, Myoinositol and Fiber Degeneration and Regeneration in Diabetic Neuropathy. New Engl. J. Med. 1988, 319, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-R.; Reddy, V.; Giblin, F.; Kador, P.; Kinoshita, J. Polyol accumulation in cultured human lens epithelial cells. Exp. Eye Res. 1991, 52, 93–100. [Google Scholar] [CrossRef]
- Henry, D.N.; Del Monte, M.; A Greene, D.; Killen, P.D. Altered aldose reductase gene regulation in cultured human retinal pigment epithelial cells. J. Clin. Investig. 1993, 92, 617–623. [Google Scholar] [CrossRef]
- Cohen, A.M.; Wald, H.; Popovtzer, M.; Rosenmann, E. Effect of myo-inositol supplementation on the development of renal pathological changes in the Cohen diabetic (type 2) rat. Diabetologia 1995, 38, 899–905. [Google Scholar] [CrossRef]
- Arner, R.J.; Prabhu, K.S.; Krishnan, V.; Johnson, M.C.; Reddy, C.C. Expression of myo-inositol oxygenase in tissues susceptible to diabetic complications. Biochem. Biophys. Res. Commun. 2006, 339, 816–820. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, C.; Miao, X.; Xu, K.; Wu, X.; Liu, C. Increased expression of myo-inositol oxygenase is involved in the tubulointerstitial injury of diabetic nephropathy. Exp. Clin. Endocrinol. Diabetes 2009, 117, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Nayak, B.; Kondeti, V.K.; Xie, P.; Lin, S.; Viswakarma, N.; Raparia, K.; Kanwar, Y.S. Transcriptional and post-translational modulation of myo-inositol oxygenase by high glucose and related pathobiological stresses. J. Biol. Chem. 2011, 286, 27594–27611. [Google Scholar] [CrossRef] [PubMed]
- Jackson, N.A.; Flores, A.M.; Eldon, E.D.; Klig, L.S. Disruption of INOS, a gene encoding myo-inositol phosphate synthase, causes male sterility in Drosophila melanogaster. G3 Genes Genomes Genet. 2018, 8, 2913–2922. [Google Scholar] [CrossRef]
- Rivera, M.J.; Contreras, A.; Nguyen, L.T.; Eldon, E.D.; Klig, L.S. Regulated inositol synthesis is critical for balanced metabolism and development in Drosophila melanogaster. Biol. Open 2021, 10, bio058833. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A.; Loewus, M.W. Myo-Inositol: Its biosynthesis and metabolism. Annu. Rev. Plant Physiol. 1983, 34, 137–161. [Google Scholar] [CrossRef]
- Holub, B.J. Metabolism and function of myo-inositol and inositol phospholipids. Annu. Rev. Nutr. 1986, 6, 563–597. [Google Scholar] [CrossRef]
- Holub, B.J. The nutritional importance of inositol and the phosphoinositides. New Engl. J. Med. 1992, 326, 1285–1287. [Google Scholar] [CrossRef]
- Henry, S.A.; Gaspar, M.L.; Jesch, S.A. The response to inositol: Regulation of glycerolipid metabolism and stress response signaling in yeast. Chem. Phys. Lipids 2014, 180, 23–43. [Google Scholar] [CrossRef]
- Basak, P.; Sangma, S.; Mukherjee, A.; Agarwal, T.; Sengupta, S.; Ray, S.; Majumder, A.L. Functional characterization of two myo-inositol-1-phosphate synthase (MIPS) gene promoters from the halophytic wild rice (Porteresia coarctata). Planta 2018, 248, 1121–1141. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Ma, A.; Wang, X.; Huang, Z. Myo-inositol enhances the low-salinity tolerance of turbot (Scophathalmus maximus) by modulating cortisol synthesis. Biochem. Biophys. Res. Commun. 2020, 526, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.F.; Anderson, L. Metabolism of myo-inositol in animals. II. Complete catabolism of myo-inositol-14C by rat kidney slices. Arch. Biochem. Biophys. 1967, 118, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Hankes, L.V.; Politzer, W.M.; Touster, O.; Anderson, L. Myo-inositol catabolism in human pentosurics: The predominant role of the glucuronate-xylulose-pentose phosphate pathway. Ann. N. Y. Acad. Sci. 1969, 165, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Arner, R.J.; Prabhu, K.S.; Thompson, J.T.; Hildenbrandt, G.R.; Liken, A.D.; Reddy, C.C. Myo-Inositol oxygenase: Molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and d-chiro-inositol. Biochem. J. 2001, 360, 313–320. [Google Scholar] [CrossRef]
- Chang, H.-H.; Chao, H.-N.; Walker, C.S.; Choong, S.-Y.; Phillips, A.; Loomes, K.M. Renal depletion of myo-inositol is associated with its increased degradation in animal models of metabolic disease. Am. J. Physiol. Physiol. 2015, 309, F755–F763. [Google Scholar] [CrossRef]
- Charalampous, F.C.; Lyras, C. Biochemical studies on inositol. IV. Conversion of inositol to glucuronic acid by rat kidney extracts. J. Biol. Chem. 1957, 228, 1–13. [Google Scholar] [CrossRef]
- Charalampous, F.C. Biochemical studies on inositol. V. Purification and properties of the enzyme that cleaves inositol to D-glucuronic acid. J. Biol. Chem. 1959, 234, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Koller, E.; Koller, F.; Hoffmann-Ostenhof, O. Myo-inositol oxygenase from oat seedlings. Mol. Cell. Biochem. 1976, 10, 33–39. [Google Scholar] [CrossRef]
- Reddy, C.C.; Swan, J.S.; Hamilton, G.A. Myo-inositol oxygenase from hog kidney. I. Purification and characterization of the oxygenase and of an enzyme complex containing the oxygenase and D-glucuronate reductase. J. Biol. Chem. 1981, 256, 8510–8518. [Google Scholar] [CrossRef] [PubMed]
- Molina, Y.; Ramos, S.E.; Douglass, T.; Klig, L.S. Inositol synthesis and catabolism in Cryptococcus neoformans. Yeast 1999, 15, 1657–1667. [Google Scholar] [CrossRef]
- Mackenzie, E.A.; Klig, L.S. Computational modeling and in silico analysis of differential regulation of myo-inositol catabolic enzymes in Cryptococcus neoformans. BMC Mol. Biol. 2008, 9, 88. [Google Scholar] [CrossRef]
- Endres, S.; Tenhaken, R. Down-regulation of the myo-inositol oxygenase gene family has no effect on cell wall composition in Arabidopsis. Planta 2011, 234, 157–169. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Wei, Y.; Liu, Y.; Xing, L.; Liu, M.; Li, P.; Lu, Q.; Peng, R. Genome-wide identification of the MIOX gene family and their expression profile in cotton development and response to abiotic stress. PLoS ONE 2021, 16, e0254111. [Google Scholar] [CrossRef]
- Brown, P.M.; Caradoc-Davies, T.T.; Dickson JM, J.; Cooper GJ, S.; Loomes, K.M.; Baker, E.N. Crystal structure of a substrate complex of myo-inositol oxygenase, A di-iron oxygenase with a key role in inositol metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 15032–15037. [Google Scholar] [CrossRef]
- Thorsell, A.-G.; Persson, C.; Voevodskaya, N.; Busam, R.D.; Hammarström, M.; Graslund, S.; Gräslund, A.; Hallberg, B.M. Structural and biophysical characterization of human myo-inositol oxygenase. J. Biol. Chem. 2008, 283, 15209–15216. [Google Scholar] [CrossRef] [PubMed]
- Musselman, L.P.; Kühnlein, R.P. Drosophilaas a model to study obesity and metabolic disease. J. Exp. Biol. 2018, 221, jeb163881. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.M.; Barry, W.E.; Cox, J.; Thummel, C.S. Methods for studying metabolism in Drosophila. Methods 2014, 68, 105–115. [Google Scholar] [CrossRef]
- Ugrankar, R.; Berglund, E.; Akdemir, F.; Tran, C.; Kim, M.S.; Noh, J.; Schneider, R.; Ebert, B.; Graff, J.M. Drosophila glucome screening identifies Ck1alpha as a regulator of mammalian glucose metabolism. Nat. Commun. 2015, 6, 7102. [Google Scholar] [CrossRef]
- Williams, M.J.; Klockars, A.; Eriksson, A.; Voisin, S.; Dnyansagar, R.; Wiemerslage, L.; Kasagiannis, A.; Akram, M.; Kheder, S.; Ambrosi, V.; et al. The drosophila ETV5 homologue Ets96B: Molecular link between obesity and bipolar disorder. PLoS Genet. 2016, 12, e1006104. [Google Scholar] [CrossRef] [PubMed]
- Graham, P.; Pick, L. Drosophila as a model for diabetes and diseases of insulin resistance. Curr. Top. Dev. Biol. 2017, 121, 397–419. [Google Scholar] [CrossRef] [PubMed]
- Mattila, J.; Hietakangas, V. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 2017, 207, 1231–1253. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wan, Z.; Wang, Z.; Zhou, B. Insulin signaling in Drosophila melanogaster mediates Aβ toxicity. Commun. Biol. 2019, 2, 13. [Google Scholar] [CrossRef]
- Gillette, C.M.; Tennessen, J.M.; Reis, T. Balancing energy expenditure and storage with growth and biosynthesis during Drosophila development. Dev. Biol. 2021, 475, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Perrimon, N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7, eabg4336. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.D.; Thummel, C.S. Diabetic larvae and obese flies—Emerging studies of metabolism in Drosophila. Cell Metab. 2007, 6, 257–266. [Google Scholar] [CrossRef]
- Reis, T.; Van Gilst, M.R.; Hariharan, I.K. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 2010, 6, e1001206. [Google Scholar] [CrossRef]
- Musselman, L.P.; Fink, J.L.; Narzinski, K.; Ramachandran, P.V.; Sukumar Hathiramani, S.; Cagan, R.L.; Baranski, T.J. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 2011, 4, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S. Controlling animal growth and body size–Does fruit fly physiology point the way? F1000 Biol. Rep. 2012, 4, 12. [Google Scholar] [CrossRef]
- Rovenko, B.M.; Kubrak, O.I.; Gospodaryov, D.V.; Perkhulyn, N.V.; Yurkevych, I.S.; Sanz, A.; Lushchak, O.V.; Lushchak, V.I. High sucrose consumption promotes obesity whereas its low consumption induces oxidative stress in Drosoph melanogaster. J. Insect Physiol. 2015, 79, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Hazegh, K.E.; Reis, T. A Buoyancy-based method of determining fat levels in Drosophila. J. Vis. Exp. 2016, 117, 54744. [Google Scholar] [CrossRef]
- Reis, T. Effects of synthetic diets enriched in specific nutrients on Drosophila development, body fat, and lifespan. PLoS ONE 2016, 11, e0146758. [Google Scholar] [CrossRef]
- Alfa, R.W.; Kim, S.K. Using Drosophila to discover mechanisms underlying type 2 diabetes. Dis. Model. Mech. 2016, 9, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.T.; Brock, K.; Musselman, L.P. Meep, a novel regulator of insulin signaling, supports development and insulin sensitivity via maintenance of protein homeostasis in Drosophila melanogaster. G3 Genes Genomes Genet. 2020, 10, 4399–4410. [Google Scholar] [CrossRef]
- Campos-Ortega, J.A.; Hartenstein, V. The Embryonic Development of Drosophila melanogaster; Springer: Berlin/Heidelberg, Germany, 1985. [Google Scholar]
- Younossi-Hartenstein, A.; Tepass, U.; Hartenstein, V. Embryonic origin of the imaginal discs of the head of Drosophila melanogaster. Roux’s Arch. Dev. Biol. 1993, 203, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Gramates, L.S.; Agapite, J.; Attrill, H.; Calvi, B.R.; Crosby, M.A.; Dos Santos, G.; Goodman, J.L.; Goutte-Gattat, D.; Jenkins, V.K.; Kaufman, T.; et al. FlyBase: A guided tour of highlighted features. Genetics 2022, 220, iyac035. [Google Scholar] [CrossRef] [PubMed]
- Casas-Vila, N.; Bluhm, A.; Sayols, S.; Dinges, N.; Dejung, M.; Altenhein, T.; Kappei, D.; Altenhein, B.; Roignant, J.Y.; Butter, F. The developmental proteome of Drosophila melanogaster. Genome Res. 2017, 27, 1273–1285. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021, 49, D899–D907. [Google Scholar] [CrossRef]
- Sharma, I.; Deng, F.; Liao, Y.; Kanwar, Y.S. Myo-inositol Oxygenase (MIOX) Overexpression Drives the Progression of Renal Tubulointerstitial Injury in Diabetes. Diabetes 2020, 69, 1248–1263. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, T.; Sharma, I.; Fujita, Y.; Doi, T.; Wallner, A.K.; Kanwar, Y.S. Myo-inositol oxygenase accentuates renal tubular injury initiated by endoplasmic reticulum stress. Am. J. Physiology. Ren. Physiol. 2019, 316, F301–F315. [Google Scholar] [CrossRef]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Green, E.W.; Fedele, G.; Giorgini, F.; Kyriacou, C.P. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nature Methods 2014, 11, 222–223. [Google Scholar] [CrossRef] [PubMed]
- Vissers, J.H.; Manning, S.A.; Kulkarni, A.; Harvey, K.F. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat. Commun. 2016, 7, 10368. [Google Scholar] [CrossRef]
- Evangelou, A.; Ignatiou, A.; Antoniou, C.; Kalanidou, S.; Chatzimatthaiou, S.; Shianiou, G.; Ellina, S.; Athanasiou, R.; Panagi, M.; Apidianakis, Y.; et al. Unpredictable effects of the genetic background of transgenic lines in physiological quantitative traits. G3 2019, 9, 3877–3890. [Google Scholar] [CrossRef]
- Seeds, A.M.; Tsui, M.M.; Sunu, C.; Spana, E.P.; York, J.D. Inositol phosphate kinase 2 is required for imaginal disc development in Drosophila. Proc. Natl. Acad. Sci. USA 2015, 112, 15660–15665. [Google Scholar] [CrossRef]
- Janardan, V.; Sharma, S.; Basu, U.; Raghu, P. A genetic screen in Drosophila to identify novel regulation of cell growth by phosphoinositide signaling. G3 2020, 10, 57–67. [Google Scholar] [CrossRef]
- Abbott, M.K.; Lengyel, J.A. Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics 1991, 129, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, A.; Agapite, J.; McCall, K.; Steller, H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 1998, 95, 331–341. [Google Scholar] [CrossRef]
- Hursh, D.A.; Stultz, B.G. Odd-Paired: The Drosophila Zic Gene. Adv. Exp. Med. Biol. 2018, 1046, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, L.; Pan, X.; Woods, A.L.; O’Connor, M.B.; Hariharan, I.K. The BMP2/4 ortholog DPP can function as an inter-organ signal that regulates developmental timing. Life Sci. Alliance 2018, 1, e201800216. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.; de la Puebla, S.F.; Guerrero, I. Drosophila Zic family member odd-paired is needed for adult post-ecdysis maturation. Open Biol. 2019, 9, 190245. [Google Scholar] [CrossRef]
- Lepore, E.; Lauretta, R.; Bianchini, M.; Mormando, M.; Di Lorenzo, C.; Unfer, V. Inositols depletion and resistance: Principal mechanisms and therapeutic strategies. Int. J. Mol. Sci. 2021, 22, 6796. [Google Scholar] [CrossRef] [PubMed]
- Hagen-Lillevik, S.; Johnson, J.; Siddiqi, A.; Persinger, J.; Hale, G.; Lai, K. Harnessing the power of purple sweet potato color and myo-inositol to treat classic galactosemia. Int. J. Mol. Sci. 2022, 23, 8654. [Google Scholar] [CrossRef]
- Lazcano, P.; Schmidtke, M.W.; Onu, C.; Greenberg, M.L. Phosphatidic acid inhibits inositol synthesis by inducing nuclear translocation of kinase IP6K1 and repression of myo-inositol-3-P synthase. J. Biol. Chem. Adv. Online Publ. 2022, 298, 102363. [Google Scholar] [CrossRef]
- Croze, M.L.; Soulage, C.O. Potential role and therapeutic interests of myo-inositol in metabolic diseases. Biochimie 2013, 95, 1811–1827. [Google Scholar] [CrossRef]
- Subramanian, M.; Metya, S.K.; Sadaf, S.; Kumar, S.; Schwudke, D.; Hasan, G. Altered lipid homeostasis in Drosophila InsP3 receptor mutants leads to obesity and hyperphagia. Dis. Model. Mech. 2013, 6, 734–744. [Google Scholar] [CrossRef]
- Bellen, H.J.; Levis, R.W.; He, Y.; Carlson, J.W.; Evans-Holm, M.; Bae, E.; Kim, J.; Metaxakis, A.; Savakis, C.; Schulze, K.L.; et al. The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics 2011, 188, 731–743. [Google Scholar] [CrossRef]
- Thibault, S.T.; Singer, M.A.; Miyazaki, W.Y.; Milash, B.; Dompe, N.A.; Singh, C.M.; Buchholz, R.; Demsky, M.; Fawcett, R.; Francis-Lang, H.L.; et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004, 36, 283–287. [Google Scholar] [CrossRef]
- Sang, J.H. Quantitative nutritional requirements of Drosophila melanogaster. J. Exp. Biology 1955, 33, 45–72. [Google Scholar] [CrossRef]
- Falk, D.R.; Nash, D. Sex-linked auxotrophic and putative auxotrophic mutants of Drosophila melanogaster. Genetics 1974, 76, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R. Total RNA Extraction from Drosophila melanogaster. In Molecular Cloning; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2012; p. 2028. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Tsuda-Sakurai, K.; Seong, K.H.; Horiuchi, J.; Aigaki, T.; Tsuda, M. Identification of a novel role for Drosophila MESR4 in lipid metabolism. Genes Cells Devoted Mol. Cell. Mech. 2015, 20, 358–365. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras, A.; Jones, M.K.; Eldon, E.D.; Klig, L.S. Inositol in Disease and Development: Roles of Catabolism via myo-Inositol Oxygenase in Drosophila melanogaster. Int. J. Mol. Sci. 2023, 24, 4185. https://doi.org/10.3390/ijms24044185
Contreras A, Jones MK, Eldon ED, Klig LS. Inositol in Disease and Development: Roles of Catabolism via myo-Inositol Oxygenase in Drosophila melanogaster. International Journal of Molecular Sciences. 2023; 24(4):4185. https://doi.org/10.3390/ijms24044185
Chicago/Turabian StyleContreras, Altagracia, Melissa K. Jones, Elizabeth D. Eldon, and Lisa S. Klig. 2023. "Inositol in Disease and Development: Roles of Catabolism via myo-Inositol Oxygenase in Drosophila melanogaster" International Journal of Molecular Sciences 24, no. 4: 4185. https://doi.org/10.3390/ijms24044185
APA StyleContreras, A., Jones, M. K., Eldon, E. D., & Klig, L. S. (2023). Inositol in Disease and Development: Roles of Catabolism via myo-Inositol Oxygenase in Drosophila melanogaster. International Journal of Molecular Sciences, 24(4), 4185. https://doi.org/10.3390/ijms24044185




