A Satellite-Free Centromere in Equus przewalskii Chromosome 10
Abstract
1. Introduction
2. Results
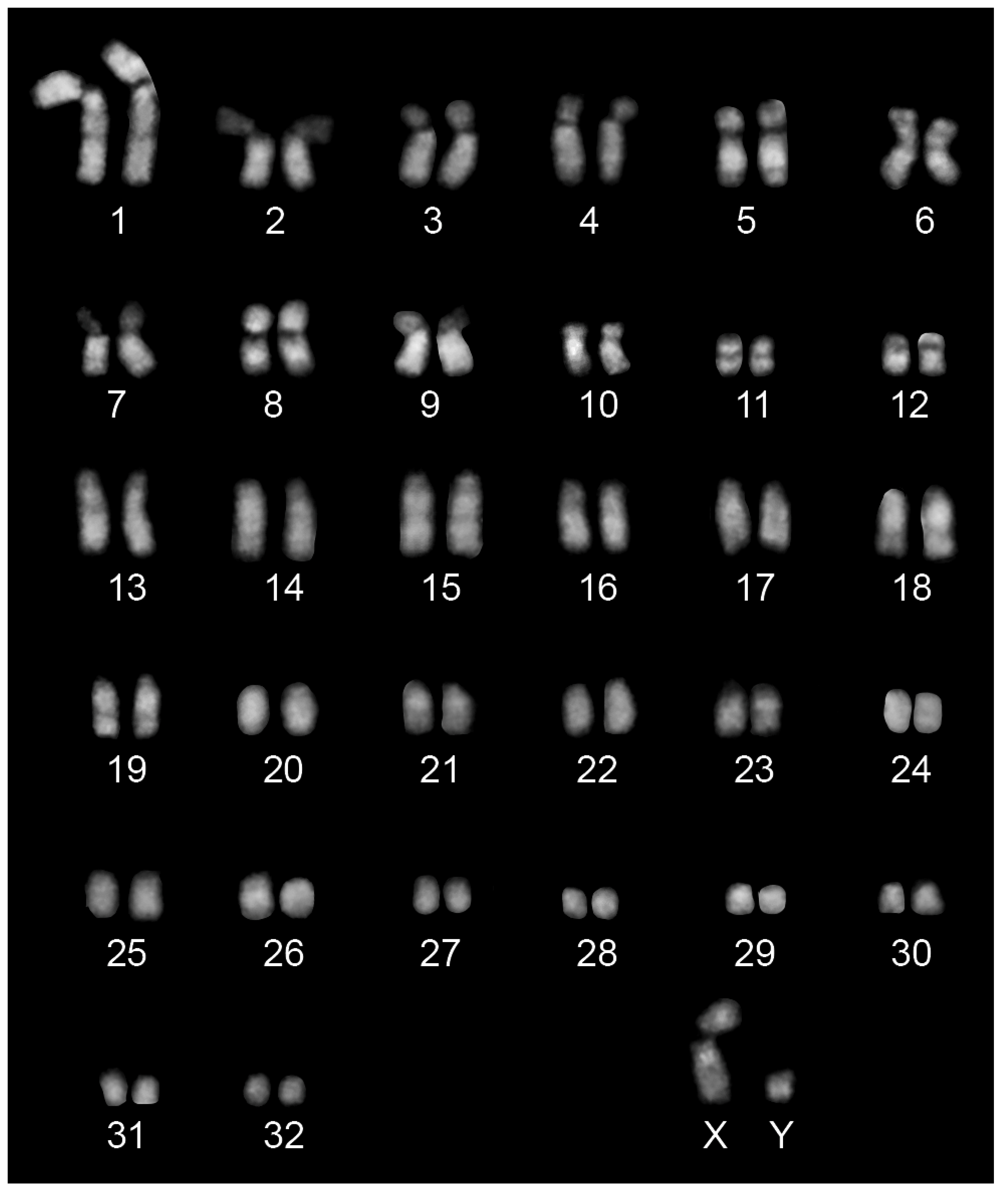
2.1. Characterization of a Fibroblasts Cell Line from Equus przewalskii
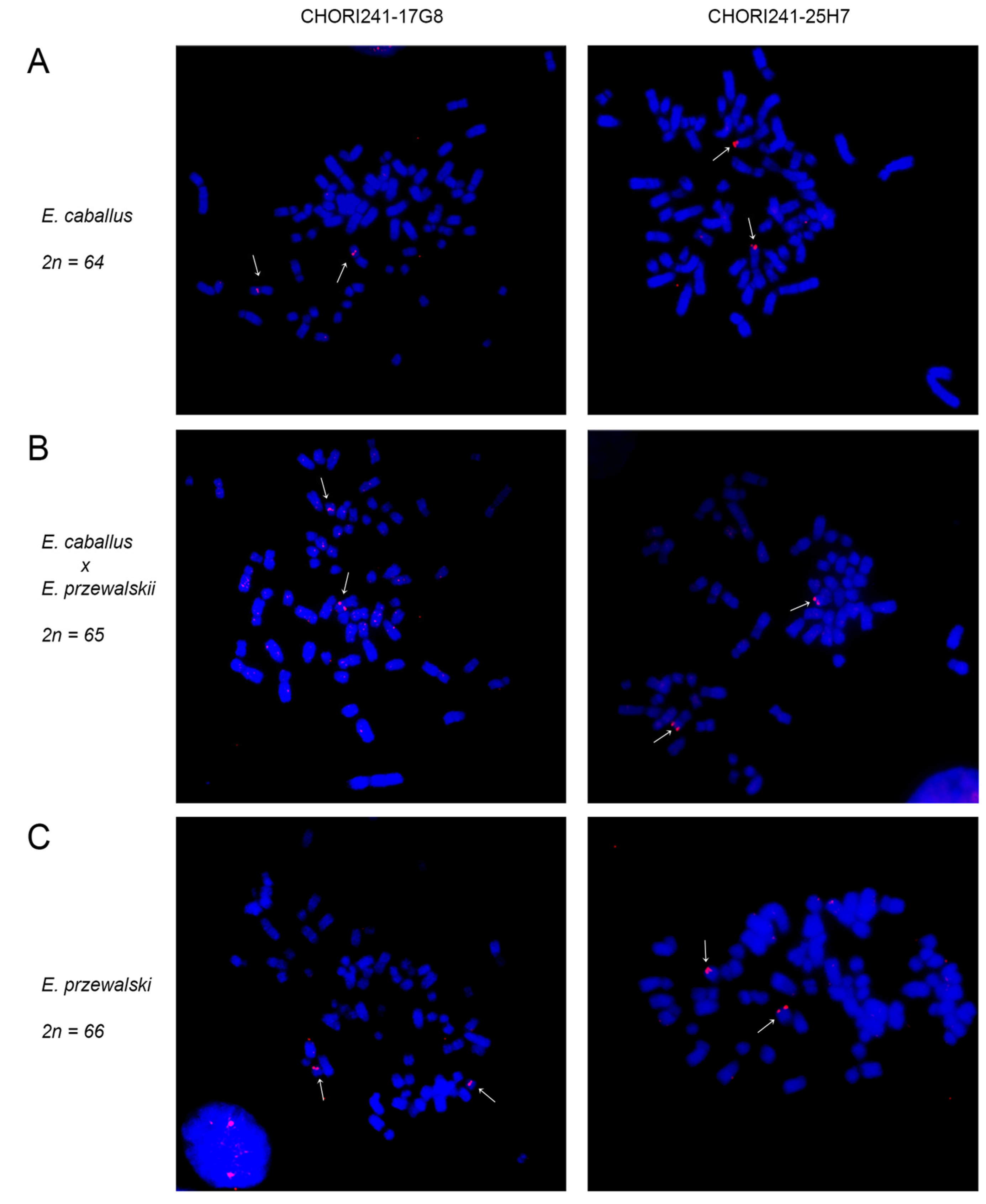
2.2. Chromosomal Distribution of Satellite DNA Families in Equus przewalskii
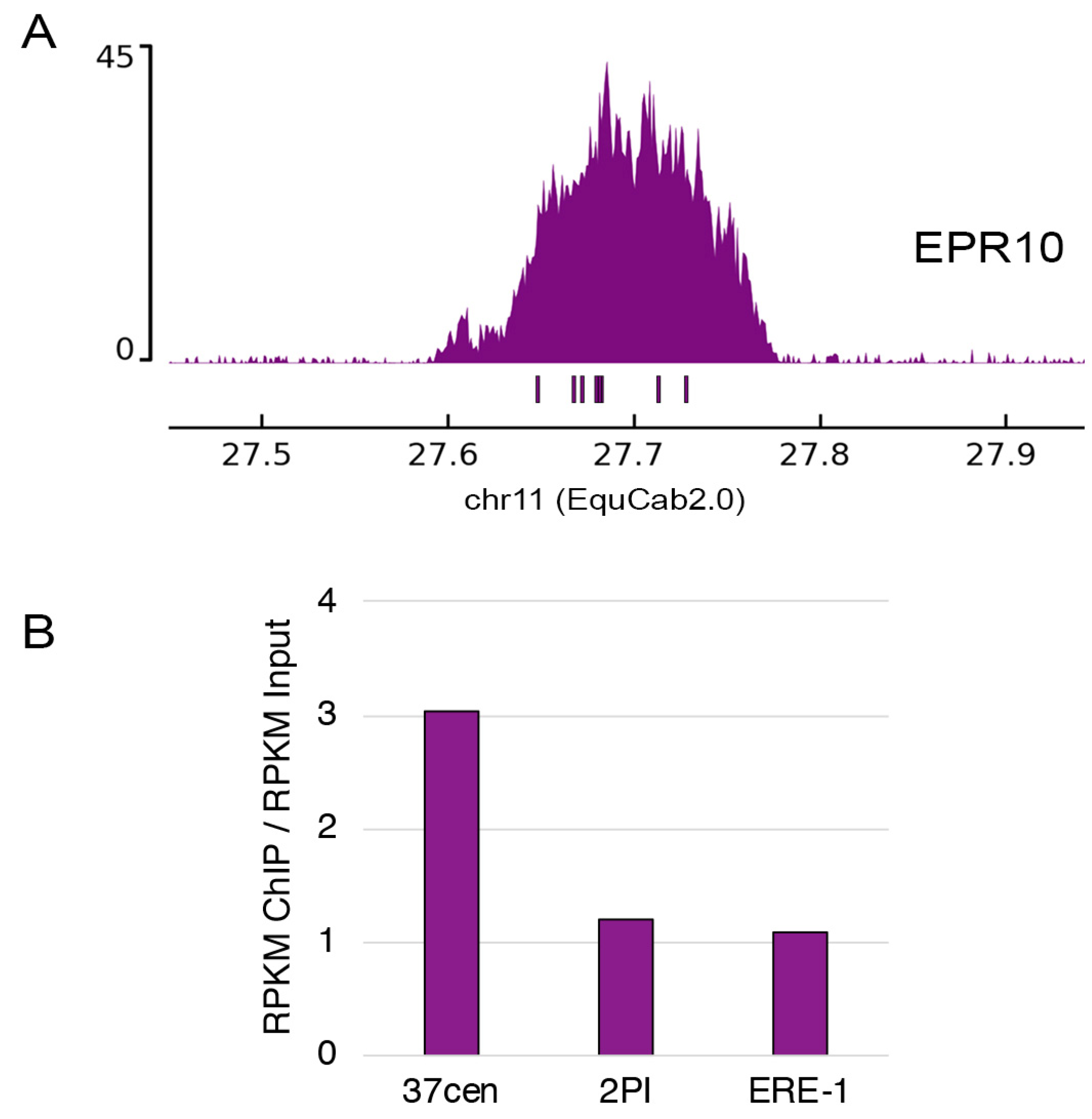
2.3. ChIP-Seq Analysis of Centromeric Domains
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Metaphase Spread Preparation and FISH
4.3. ChIP-Seq
4.4. Bioinformatic Analysis of ChIP-Seq Data
4.5. SNV Analysis
4.6. Mitogenomic Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allshire, R.C.; Karpen, G.H. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 2008, 9, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Choo, K.H. Centromerization. Trends Cell Biol. 2000, 10, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Marshall, O.J.; Chueh, A.C.; Wong, L.H.; Choo, K.H. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008, 82, 261–282. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Migeon, B.R. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 1985, 92, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Voullaire, L.E.; Slater, H.R.; Petrovic, V.; Choo, K.H. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am. J. Hum. Genet. 1993, 52, 1153–1163. [Google Scholar]
- Plohl, M.; Luchetti, A.; Mestrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Piras, F.M.; Gamba, R.; Corbo, M.; Cerutti, F.; McCarter, J.G.W.; Cappelletti, E.; Gozzo, F.; Harman, R.M.; Antczak, D.F.; et al. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 2018, 28, 789–799. [Google Scholar] [CrossRef]
- Cappelletti, E.; Piras, F.M.; Badiale, C.; Bambi, M.; Santagostino, M.; Vara, C.; Masterson, T.A.; Sullivan, K.F.; Nergadze, S.G.; Ruiz-Herrera, A.; et al. CENP-A binding domains and recombination patterns in horse spermatocytes. Sci. Rep. 2019, 9, 15800. [Google Scholar] [CrossRef]
- Purgato, S.; Belloni, E.; Piras, F.M.; Zoli, M.; Badiale, C.; Cerutti, F.; Mazzagatti, A.; Perini, G.; Della Valle, G.; Nergadze, S.G.; et al. Centromere sliding on a mammalian chromosome. Chromosoma 2015, 124, 277–287. [Google Scholar] [CrossRef]
- Peng, S.; Petersen, J.L.; Bellone, R.R.; Kalbfleisch, T.; Kingsley, N.B.; Barber, A.M.; Cappelletti, E.; Giulotto, E.; Finno, C.J. Decoding the Equine Genome: Lessons from ENCODE. Genes 2021, 12, 1707. [Google Scholar] [CrossRef]
- Carbone, L.; Nergadze, S.G.; Magnani, E.; Misceo, D.; Francesca Cardone, M.; Roberto, R.; Bertoni, L.; Attolini, C.; Francesca Piras, M.; de Jong, P.; et al. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 2006, 87, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Piras, F.M.; Cappelletti, E.; Santagostino, M.; Nergadze, S.G.; Giulotto, E.; Raimondi, E. Molecular Dynamics and Evolution of Centromeres in the Genus Equus. Int. J. Mol. Sci. 2022, 23, 4183. [Google Scholar] [CrossRef] [PubMed]
- Cappelletti, E.; Piras, F.M.; Sola, L.; Santagostino, M.; Abdelgadir, W.A.; Raimondi, E.; Lescai, F.; Nergadze, S.G.; Giulotto, E. Robertsonian Fusion and Centromere Repositioning Contributed to the Formation of Satellite-free Centromeres During the Evolution of Zebras. Mol. Biol. Evol. 2022, 39, msac162. [Google Scholar] [CrossRef]
- Roberti, A.; Bensi, M.; Mazzagatti, A.; Piras, F.M.; Nergadze, S.G.; Giulotto, E.; Raimondi, E. Satellite DNA at the Centromere is Dispensable for Segregation Fidelity. Genes 2019, 10, 469. [Google Scholar] [CrossRef] [PubMed]
- Piras, F.M.; Nergadze, S.G.; Magnani, E.; Bertoni, L.; Attolini, C.; Khoriauli, L.; Raimondi, E.; Giulotto, E. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010, 6, e1000845. [Google Scholar] [CrossRef]
- Giulotto, E.; Raimondi, E.; Sullivan, K.F. The Unique DNA Sequences Underlying Equine Centromeres. Prog. Mol. Subcell Biol. 2017, 56, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.A.; Musilova, P.; Kulemsina, A.I. Chromosome evolution in Perissodactyla. Cytogenet. Genome Res. 2012, 137, 208–217. [Google Scholar] [CrossRef]
- Trifonov, V.A.; Stanyon, R.; Nesterenko, A.I.; Fu, B.; Perelman, P.L.; O’Brien, P.C.; Stone, G.; Rubtsova, N.V.; Houck, M.L.; Robinson, T.J.; et al. Multidirectional cross-species painting illuminates the history of karyotypic evolution in Perissodactyla. Chromosome Res. 2008, 16, 89–107. [Google Scholar] [CrossRef]
- Musilova, P.; Kubickova, S.; Vahala, J.; Rubes, J. Subchromosomal karyotype evolution in Equidae. Chromosome Res. 2013, 21, 175–187. [Google Scholar] [CrossRef]
- Montefalcone, G.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Centromere repositioning. Genome Res. 1999, 9, 1184–1188. [Google Scholar] [CrossRef]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Piras, F.M.; Nergadze, S.G.; Poletto, V.; Cerutti, F.; Ryder, O.A.; Leeb, T.; Raimondi, E.; Giulotto, E. Phylogeny of horse chromosome 5q in the genus Equus and centromere repositioning. Cytogenet. Genome Res. 2009, 126, 165–172. [Google Scholar] [CrossRef]
- Wijers, E.R.; Zijlstra, C.; Lenstra, J.A. Rapid evolution of horse satellite DNA. Genomics 1993, 18, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Sakagami, M.; Hirota, K.; Awata, T.; Yasue, H. Molecular cloning of an equine satellite-type DNA sequence and its chromosomal localization. Cytogenet. Cell Genet. 1994, 66, 27–30. [Google Scholar] [CrossRef]
- Broad, T.E.; Forrest, J.W.; Lewis, P.E.; Pearce, P.D.; Phua, S.H.; Pugh, P.A.; Stewart-Scott, I.A. Cloning of a DNA repeat element from horse: DNA sequence and chromosomal localization. Genome 1995, 38, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Broad, T.E.; Ede, A.J.; Forrest, J.W.; Lewis, P.E.; Phua, S.H.; Pugh, P.A. Families of tandemly repeated DNA elements from horse: Cloning, nucleotide sequence, and organization. Genome 1995, 38, 1285–1289. [Google Scholar] [CrossRef]
- Cerutti, F.; Gamba, R.; Mazzagatti, A.; Piras, F.M.; Cappelletti, E.; Belloni, E.; Nergadze, S.G.; Raimondi, E.; Giulotto, E. The major horse satellite DNA family is associated with centromere competence. Mol. Cytogenet. 2016, 9, 35. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Belloni, E.; Piras, F.M.; Khoriauli, L.; Mazzagatti, A.; Vella, F.; Bensi, M.; Vitelli, V.; Giulotto, E.; Raimondi, E. Discovery and comparative analysis of a novel satellite, EC137, in horses and other equids. Cytogenet. Genome Res. 2014, 144, 114–123. [Google Scholar] [CrossRef]
- Ryder, O. Genetic studies of Przewalski’s horses and their impact on conservation. In Przewalski’s Horse: The History and Biology of an Endangered Species; Boyd, L., Houpt, K.A., Eds.; The State University of New York Press: Albany, NY, USA, 1994; pp. 75–92. [Google Scholar]
- Ransom, J.; Kaczensky, P. Wild Equids: Ecology, Management, and Conservation; JHU Press: Baltimore, MD, USA, 2016; pp. 1–229. [Google Scholar]
- Goto, H.; Ryder, O.A.; Fisher, A.R.; Schultz, B.; Kosakovsky Pond, S.L.; Nekrutenko, A.; Makova, K.D. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski’s horses. Genome Biol. Evol. 2011, 3, 1096–1106. [Google Scholar] [CrossRef]
- Wallner, B.; Brem, G.; Müller, M.; Achmann, R. Fixed nucleotide differences on the Y chromosome indicate clear divergence between Equus przewalskii and Equus caballus. Anim. Genet. 2003, 34, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Ginolhac, A.; Zhang, G.; Froese, D.; Albrechtsen, A.; Stiller, M.; Schubert, M.; Cappellini, E.; Petersen, B.; Moltke, I.; et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 2013, 499, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Der Sarkissian, C.; Ermini, L.; Schubert, M.; Yang, M.A.; Librado, P.; Fumagalli, M.; Jónsson, H.; Bar-Gal, G.K.; Albrechtsen, A.; Vieira, F.G.; et al. Evolutionary Genomics and Conservation of the Endangered Przewalski’s Horse. Curr. Biol. 2015, 25, 2577–2583. [Google Scholar] [CrossRef] [PubMed]
- Gaunitz, C.; Fages, A.; Hanghøj, K.; Albrechtsen, A.; Khan, N.; Schubert, M.; Seguin-Orlando, A.; Owens, I.J.; Felkel, S.; Bignon-Lau, O.; et al. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 2018, 360, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L. Ancient Genomes Reveal Unexpected Horse Domestication and Management Dynamics. Bioessays 2020, 42, e1900164. [Google Scholar] [CrossRef] [PubMed]
- Bouman, D.T.; Bouman, J.G. The history of Przewalski’s Horse. In Przewalski’s Horse: The History and Biology of an Endangered Species; Boyd, L., Houpt, K.A., Eds.; State University of New York Press: Albany, NY, USA, 1994; pp. 5–38. [Google Scholar]
- Jiang, Z.; Zong, H. Reintroduction of the Przewalski’s Horse in China: Status Quo and Outlook. Nat. Conserv. Res. 2019, 4, 15–22. [Google Scholar] [CrossRef]
- Turghan, M.A.; Jiang, Z.; Niu, Z. An Update on Status and Conservation of the Przewalski’s Horse (Equus ferus przewalskii): Captive Breeding and Reintroduction Projects. Animals 2022, 12, 3158. [Google Scholar] [CrossRef]
- Oakenfull, E.A.; Ryder, O.A. Mitochondrial control region and 12S rRNA variation in Przewalski’s horse (Equus przewalskii). Anim. Genet. 1998, 29, 456–459. [Google Scholar] [CrossRef]
- Kerekes, V.; Sándor, I.; Nagy, D.; Ozogány, K.; Göczi, L.; Ibler, B.; Széles, L.; Barta, Z. Trends in demography, genetics, and social structure of Przewalski’s horses in the Hortobagy National Park, Hungary over the last 22 years. Glob. Ecol. Conserv. 2021, 25, e01407. [Google Scholar] [CrossRef]
- Bakirova, R.T.; Zharkikh, T.L. Programme on Establishing a Semi-Free Population of Przewalski’s Horse in Orenburg State Nature Reserve: The First Successful Project on the Reintroduction of the Species in Russia. Nat. Conserv. Res. 2019, 4, 57–64. [Google Scholar] [CrossRef]
- Bernatkova, A.; Oyunsaikhan, G.; Šimek, J.; Komárková, M.; Bobek, M.; Ceacero, F. Influence of weather on the behaviour of reintroduced Przewalski’s horses in the Great Gobi B Strictly Protected Area (Mongolia): Implications for conservation. BMC Zool. 2022, 7. [Google Scholar] [CrossRef]
- Oakenfull, E.A.; Lim, H.N.; Ryder, O.A. A survey of equid mitochondrial DNA: Implications for the evolution, genetic diversity and conservation of Equus. Conserv. Genet. 2000, 1, 341–355. [Google Scholar] [CrossRef]
- Jansen, T.; Forster, P.; Levine, M.A.; Oelke, H.; Hurles, M.; Renfrew, C.; Weber, J.; Olek, K. Mitochondrial DNA and the origins of the domestic horse. Proc. Natl. Acad. Sci. USA 2002, 99, 10905–10910. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Hooshiar Kashani, B.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Ryder, O.A.; Hansen, S.K. Molecular cytogenetics of the Equidae. I. Purification and cytological localization of a (G + C)-rich satellite DNA from Equus przewalskii. Chromosoma 1979, 72, 115–129. [Google Scholar] [CrossRef]
- Myka, J.L.; Lear, T.L.; Houck, M.L.; Ryder, O.A.; Bailey, E. FISH analysis comparing genome organization in the domestic horse (Equus caballus) to that of the Mongolian wild horse (E. przewalskii). Cytogenet. Genome Res. 2003, 102, 222–225. [Google Scholar] [CrossRef]
- Yang, F.; Fu, B.; O’Brien, P.C.; Robinson, T.J.; Ryder, O.A.; Ferguson-Smith, M.A. Karyotypic relationships of horses and zebras: Results of cross-species chromosome painting. Cytogenet. Genome Res. 2003, 102, 235–243. [Google Scholar] [CrossRef]
- Ahrens, E.; Stranzinger, G. Comparative chromosomal studies of E. caballus (ECA) and E. przewalskii (EPR) in a female F1 hybrid. J. Anim. Breed Genet. 2005, 122 (Suppl. S1), 97–102. [Google Scholar] [CrossRef] [PubMed]
- Kvist, L.; Niskanen, M. Modern Northern Domestic Horses Carry Mitochondrial DNA Similar to Przewalski’s Horse. J. Mamm. Evol. 2021, 28, 371–376. [Google Scholar] [CrossRef]
- Anglana, M.; Bertoni, L.; Giulotto, E. Cloning of a polymorphic sequence from the nontranscribed spacer of horse rDNA. Mamm. Genome 1996, 7, 539–541. [Google Scholar] [CrossRef]
- Benirschke, K.; Malouf, N.; Low, R.J.; Heck, H. Chromosome complement: Differences between Equus caballus and Equus przewalskii, Poliakoff. Science 1965, 148, 382–383. [Google Scholar] [CrossRef] [PubMed]
- Fry, K.; Salser, W. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Salser, W.; Bowen, S.; Browne, D.; el-Adli, F.; Fedoroff, N.; Fry, K.; Heindell, H.; Paddock, G.; Poon, R.; Wallace, B.; et al. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed. Proc. 1976, 35, 23–35. [Google Scholar] [PubMed]
- Ryder, O.A.; Epel, N.C.; Benirschke, K. Chromosome banding studies of the Equidae. Cytogenet Cell Genet. 1978, 20, 332–350. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xu, C.Q.; Cao, Q.; Zimmermann, W.; Songer, M.; Zhao, S.S.; Li, K.; Hu, D.F. Mitochondrial and pedigree analysis in Przewalski’s horse populations: Implications for genetic management and reintroductions. Mitochondrial DNA 2014, 25, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.; Downs, P.; Irvin, Z.; Bell, K. Intrageneric amplification of horse microsatellite markers with emphasis on the Przewalski’s horse (E. przewalskii). Anim. Genet. 1994, 25, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Shafer, A.B.A.; Zimmermann, W.; Hu, D.; Wang, W.; Chu, H.; Cao, J.; Zhao, C. Evaluating the reintroduction project of Przewalski’s horse in China using genetic and pedigree data. Biol. Conserv. 2014, 171, 288–298. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, G.; Zhao, S.; Li, K.; Zhang, D.; Liu, S.; Hu, D. Major Histocompatibility Complex (MHC) Diversity of the Reintroduction Populations of Endangered Przewalski’s Horse. Genes 2022, 13, 928. [Google Scholar] [CrossRef]
- Jónsson, H.; Schubert, M.; Seguin-Orlando, A.; Ginolhac, A.; Petersen, L.; Fumagalli, M.; Albrechtsen, A.; Petersen, B.; Korneliussen, T.S.; Vilstrup, J.T.; et al. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc. Natl. Acad. Sci. USA 2014, 111, 18655–18660. [Google Scholar] [CrossRef]
- Vidale, P.; Magnani, E.; Nergadze, S.G.; Santagostino, M.; Cristofari, G.; Smirnova, A.; Mondello, C.; Giulotto, E. The catalytic and the RNA subunits of human telomerase are required to immortalize equid primary fibroblasts. Chromosoma 2012, 121, 475–488. [Google Scholar] [CrossRef]
- Vidale, P.; Piras, F.M.; Nergadze, S.G.; Bertoni, L.; Verini-Supplizi, A.; Adelson, D.; Guérin, G.; Giulotto, E. Chromosomal assignment of six genes (EIF4G3, HSP90, RBBP6, IL8, TERT, and TERC) in four species of the genus Equus. Anim. Biotechnol. 2011, 22, 119–123. [Google Scholar] [CrossRef]
- Santagostino, M.; Khoriauli, L.; Gamba, R.; Bonuglia, M.; Klipstein, O.; Piras, F.M.; Vella, F.; Russo, A.; Badiale, C.; Mazzagatti, A.; et al. Genome-wide evolutionary and functional analysis of the Equine Repetitive Element 1: An insertion in the myostatin promoter affects gene expression. BMC Genet. 2015, 16, 126. [Google Scholar] [CrossRef]
- Santagostino, M.; Piras, F.M.; Cappelletti, E.; Del Giudice, S.; Semino, O.; Nergadze, S.G.; Giulotto, E. Insertion of Telomeric Repeats in the Human and Horse Genomes: An Evolutionary Perspective. Int. J. Mol. Sci. 2020, 21, 2838. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Lupotto, M.; Pellanda, P.; Santagostino, M.; Vitelli, V.; Giulotto, E. Mitochondrial DNA insertions in the nuclear horse genome. Anim. Genet. 2010, 41 (Suppl. S2), 176–185. [Google Scholar] [CrossRef]
- Raimondi, E.; Piras, F.M.; Nergadze, S.G.; Di Meo, G.P.; Ruiz-Herrera, A.; Ponsà, M.; Ianuzzi, L.; Giulotto, E. Polymorphic organization of constitutive heterochromatin in Equus asinus (2n = 62) chromosome 1. Hereditas 2011, 148, 110–113. [Google Scholar] [CrossRef]
- Vangipuram, M.; Ting, D.; Kim, S.; Diaz, R.; Schüle, B. Skin punch biopsy explant culture for derivation of primary human fibroblasts. J. Vis. Exp. 2013, 77, e3779. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Delisle, L.; Rabbani, L.; Wolff, J.; Bhardwaj, V.; Backofen, R.; Grüning, B.; Ramírez, F.; Manke, T. pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 2021, 37, 422–423. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
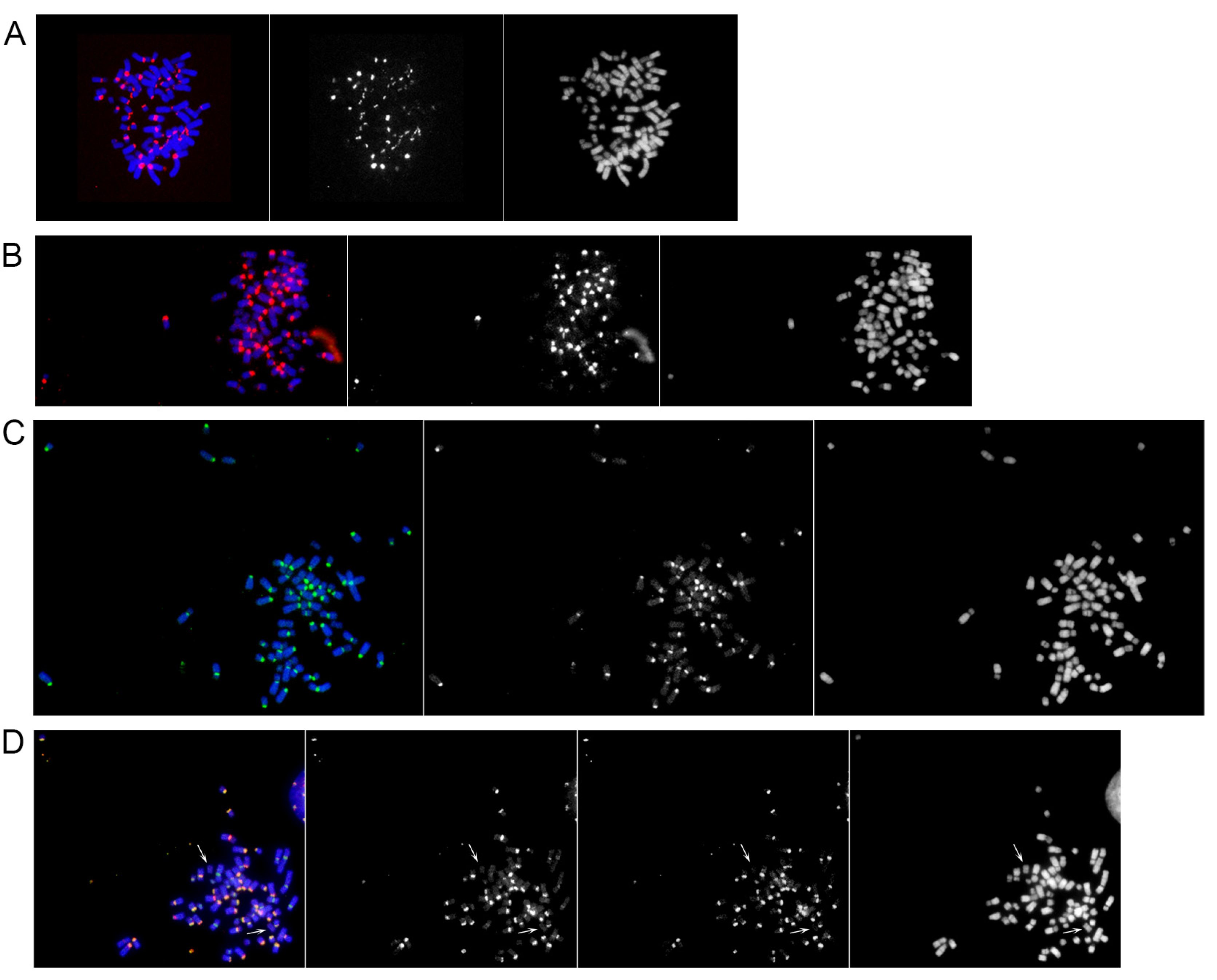
| EPR Chromosome | 37cen | 2PI | Genomic DNA | |||
|---|---|---|---|---|---|---|
| Number of Labeled Chromosomes (%) | Number of Analyzed Chromosomes | Number of Labeled Chromosomes (%) | Number of Analyzed Chromosomes | Number of Labeled Chromosomes (%) | Number of Analyzed Chromosomes | |
| 1cen | 60 (100) | 60 | 35 (85.4) | 41 | 10 (100) | 10 |
| 2cen | 3 (5.1) | 59 | 37 (100) | 37 | 10 (100) | 10 |
| 3cen | 53 (100) | 53 | 34 (100) | 34 | 10 (100) | 10 |
| 4cen | 57 (100) | 57 | 27 (71.1) | 38 | 10 (100) | 10 |
| 5cen | 51 (100) | 51 | 36 (100) | 36 | 10 (100) | 10 |
| 6cen | 12 (20.3) | 59 | 36 (100) | 36 | 1 (10) | 10 |
| 7cen | 52 (100) | 52 | 38 (100) | 38 | 10 (100) | 10 |
| 8cen | 56 (100) | 56 | 38 (100) | 38 | 10 (100) | 10 |
| 9cen | 49 (100) | 49 | 39 (100) | 39 | 9 (100) | 9 |
| 10cen | 0 (0) | 58 | 0 (0) | 34 | 0 (0) | 10 |
| 11cen | 48 (100) | 48 | 11 (40.7) | 27 | 13 (100) | 13 |
| 12cen | 48 (100) | 48 | 29 (100) | 29 | 9 (100) | 9 |
| 13cen | 47 (100) | 47 | 34 (100) | 34 | 9 (100) | 9 |
| 14cen | 41 (100) | 41 | 33 (100) | 33 | 8 (100) | 8 |
| 15cen | 45 (100) | 45 | 32 (100) | 32 | 9 (100) | 9 |
| 16cen | 33 (100) | 33 | 28 (100) | 28 | 8 (100) | 8 |
| 17cen | 30 (100) | 30 | 27 (100) | 27 | 8 (100) | 8 |
| 18cen | 17 (44.7) | 38 | 28 (100) | 28 | 8 (100) | 8 |
| 19cen | 45 (100) | 45 | 25 (100) | 25 | 10 (100) | 10 |
| 20cen | 24 (100) | 24 | 23 (100) | 23 | 8 (100) | 8 |
| 21cen | 24 (100) | 24 | 25 (100) | 25 | 8 (100) | 8 |
| 22cen | 20 (100) | 20 | 24 (100) | 24 | 8 (100) | 8 |
| 23cen | 20 (100) | 20 | 24 (100) | 24 | 8 (100) | 8 |
| 24cen | 20 (100) | 20 | 24 (100) | 24 | 8 (100) | 8 |
| 25cen | 20 (100) | 20 | 24 (100) | 24 | 8 (100) | 8 |
| 26cen | 18 (100) | 18 | 24 (100) | 24 | 8 (100) | 8 |
| 27cen | 18 (100) | 18 | 24 (100) | 24 | 8 (100) | 8 |
| 28cen | 18 (100) | 18 | 23 (100) | 23 | 7 (100) | 7 |
| 29cen | 4 (10.5) | 38 | 8 (36.4) | 22 | 11 (100) | 11 |
| 30cen | 18 (100) | 18 | 24 (100) | 24 | 8 (100) | 8 |
| 31cen | 17 (100) | 17 | 24 (100) | 24 | 8 (100) | 8 |
| 32cen | 17 (100) | 17 | 24 (100) | 24 | 8 (100) | 8 |
| Xcen | 29 (100) | 29 | 15 (71.4) | 21 | 5 (100) | 5 |
| Xqinter | 0 (0) | 29 | 0 (0) | 21 | 5 (100) | 5 |
| Ycen | 17 (100) | 17 | 3 (25) | 12 | 6 (100) | 6 |
| Position (chr11, EquCab2.0 Reference) | Reference Allele | Alternative Allele | Number of Reads Reference Allele | Number of Reads Alternative Allele | Total Number of Reads | Reference Allele Frequency | Alternative Allele Frequency |
|---|---|---|---|---|---|---|---|
| 27648739 | T | C | 8 | 15 | 23 | 0.3 | 0.7 |
| 27667251 | T | C | 20 | 15 | 35 | 0.6 | 0.4 |
| 27671967 | C | G | 14 | 18 | 32 | 0.4 | 0.6 |
| 27680741 | G | C | 7 | 9 | 16 | 0.4 | 0.6 |
| 27680744 | C | T | 8 | 8 | 16 | 0.5 | 0.5 |
| 27680746 | C | T | 9 | 9 | 18 | 0.5 | 0.5 |
| 27712271 | C | T | 11 | 19 | 30 | 0.4 | 0.6 |
| 27728102 | T | A | 8 | 16 | 24 | 0.3 | 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piras, F.M.; Cappelletti, E.; Abdelgadir, W.A.; Salamon, G.; Vignati, S.; Santagostino, M.; Sola, L.; Nergadze, S.G.; Giulotto, E. A Satellite-Free Centromere in Equus przewalskii Chromosome 10. Int. J. Mol. Sci. 2023, 24, 4134. https://doi.org/10.3390/ijms24044134
Piras FM, Cappelletti E, Abdelgadir WA, Salamon G, Vignati S, Santagostino M, Sola L, Nergadze SG, Giulotto E. A Satellite-Free Centromere in Equus przewalskii Chromosome 10. International Journal of Molecular Sciences. 2023; 24(4):4134. https://doi.org/10.3390/ijms24044134
Chicago/Turabian StylePiras, Francesca M., Eleonora Cappelletti, Wasma A. Abdelgadir, Giulio Salamon, Simone Vignati, Marco Santagostino, Lorenzo Sola, Solomon G. Nergadze, and Elena Giulotto. 2023. "A Satellite-Free Centromere in Equus przewalskii Chromosome 10" International Journal of Molecular Sciences 24, no. 4: 4134. https://doi.org/10.3390/ijms24044134
APA StylePiras, F. M., Cappelletti, E., Abdelgadir, W. A., Salamon, G., Vignati, S., Santagostino, M., Sola, L., Nergadze, S. G., & Giulotto, E. (2023). A Satellite-Free Centromere in Equus przewalskii Chromosome 10. International Journal of Molecular Sciences, 24(4), 4134. https://doi.org/10.3390/ijms24044134









