Association between Microorganisms and Microplastics: How Does It Change the Host–Pathogen Interaction and Subsequent Immune Response?
Abstract
1. Introduction
2. Interactions between Microorganisms and Microplastics
3. Immune Reactions to Microorganisms Complexed with Microplastics
4. Conclusions and Perspectives
- 1.
- Do MP/microorganism complexes cause more harm than microorganisms or MPs alone?
- 2.
- Are MP/microorganism complexes eliminated more easily by phagocytes than isolated microorganisms?
- 3.
- Is MP shape and size important in determining immune reactions to MP/microorganism complexes?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Guzzetti, E.; Sureda, A.; Tejada, S.; Faggio, C. Microplastic in marine organism: environmental and toxicological effects. Environ. Toxicol. Pharmacol. 2018, 64, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Human consumption of microplastics. Environ. Sci. Technol. 2019, 53, 7068–7074. [Google Scholar] [CrossRef] [PubMed]
- Enyoh, C.E.; Verla, A.W.; Verla, E.N.; Ibe, F.C.; Amaobi, C.E. Airborne microplastics: a review study on method for analysis, occurrence, movement and risks. Environ. Monit. Assess. 2019, 191, 668. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.W.Y.; Shiran, Y.; Bailey, R.M.; Cook, E.; Stuchtey, M.R.; Koskella, J.; Velis, C.A.; Godfrey, L.; Boucher, J.; Murphy, M.B.; et al. Evaluating scenarios toward zero plastic pollution. Science 2020, 369, 1455–1461. [Google Scholar] [CrossRef]
- Mohamed Nor, N.H.; Kooi, M.; Diepens, N.J.; Koelmans, A.A. Lifetime accumulation of microplastic in children and adults. Environ. Sci. Technol. 2021, 55, 5084–5096. [Google Scholar] [CrossRef]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: a review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef]
- Ateia, M.; Zheng, T.; Calace, S.; Tharayil, N.; Pilla, S.; Karanfil, T. Sorption behavior of real microplastics (MPs): insights for organic micropollutants adsorption on a large set of well-characterized MPs. Sci. Total Environ. 2020, 720, 137634. [Google Scholar] [CrossRef]
- Oelschlägel, K.; Pfeiffer, J.; Potthoff, A. Imitating the weathering of microplastics in the marine environment. In Proceedings of the International Conference on Microplastic Pollution in the Mediterranean Sea; Springer Water: Cham, Switzerland, 2018; pp. 171–179. [Google Scholar] [CrossRef]
- Hu, Y.L.; Gong, M.Y.; Wang, J.Y.; Bassi, A. Current research trends on microplastic pollution from wastewater systems: a critical review. Rev. Environ. Sci. Bio. 2019, 18, 207–230. [Google Scholar] [CrossRef]
- Godoy, V.; Blazquez, G.; Calero, M.; Quesada, L.; Martin-Lara, M.A. The potential of microplastics as carriers of metals. Environ. Pollut. 2019, 255, 113363. [Google Scholar] [CrossRef]
- Koelmans, A.A.; Mohamed Nor, N.H.; Hermsen, E.; Kooi, M.; Mintenig, S.M.; De France, J. Microplastics in freshwaters and drinking water: critical review and assessment of data quality. Water Res. 2019, 155, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.L.; Kelly, F.J. Plastic and human health: a micro issue? Environ. Sci. Technol. 2017, 51, 6634–6647. [Google Scholar] [CrossRef] [PubMed]
- Revel, M.; Châtel, A.; Mouneyrac, C. Micro(nano)plastics: a threat to human health? Curr. Opin. Environ. Sci. Health 2018, 1, 17–23. [Google Scholar] [CrossRef]
- Vethaak, A.D.; Legler, J. Microplastics and human health. Science 2021, 371, 672–674. [Google Scholar] [CrossRef]
- Domenech, J.; Marcos, R. Pathways of human exposure to microplastics, and estimation of the total burden. Curr. Opin. Food. Sci. 2021, 39, 144–151. [Google Scholar] [CrossRef]
- Ageel, H.K.; Harrad, S.; Abdallah, M.A. Occurrence, human exposure, and risk of microplastics in the indoor environment. Environ. Sci. Process. Impacts 2022, 24, 17–31. [Google Scholar] [CrossRef]
- Puckowski, A.; Cwiek, W.; Mioduszewska, K.; Stepnowski, P.; Bialk-Bielinska, A. Sorption of pharmaceuticals on the surface of microplastics. Chemosphere 2021, 263, 127976. [Google Scholar] [CrossRef]
- Tuson, H.H.; Weibel, D.B. Bacteria-surface interactions. Soft Matter 2013, 9, 4368–4380. [Google Scholar] [CrossRef]
- Ter Halle, A.; Ladirat, L.; Gendre, X.; Goudouneche, D.; Pusineri, C.; Routaboul, C.; Tenailleau, C.; Duployer, B.; Perez, E. Understanding the fragmentation pattern of marine plastic debris. Environ. Sci. Technol. 2016, 50, 5668–5675. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Kirstein, I.V.; Kirmizi, S.; Wichels, A.; Garin-Fernandez, A.; Erler, R.; Löder, M.; Gerdts, G. Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 2016, 120, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, P.; Pinto, M.; Proietti, M.; Reisser, J.; Ferrari, F.F.; Abbas, B.; van Loosdrecht, M.C.M.; Slat, B.; Herndl, G.J. Extracting DNA from ocean microplastics: a method comparison study. Anal. Methods 2017, 9, 1521–1526. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Loeder, M.G.; Gerdts, G.; Osborn, A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014, 90, 478–492. [Google Scholar] [CrossRef]
- Frere, L.; Maignien, L.; Chalopin, M.; Huvet, A.; Rinnert, E.; Morrison, H.; Kerninon, S.; Cassone, A.-L.; Lambert, C.; Reveillau, J.; et al. Microplastic bacterial communities in the Bay of Brest: influence of polymer type and size. Environ. Pollut. 2018, 242, 614–625. [Google Scholar] [CrossRef]
- Fernandez-Juarez, V.; Lopez-Alforja, X.; Frank-Comas, A.; Echeveste, P.; Bennasar-Figueras, A.; Ramis-Munar, G.; Gomila, R.M.; Agawin, N.S.R. “The good, the bad and the double-sword” effects of microplastics and their organic additives in marine bacteria. Front. Microbiol. 2020, 11, 581118. [Google Scholar] [CrossRef] [PubMed]
- Amato-Lourenço, L.F.; de Souza Xavier Costa, N.; Dantas, K.C.; Dos Santos Galvão, L.; Moralles, F.N.; Lombardi, S.; Mendroni Junior, A.; Lauletta Linoso, J.A.; Ando, R.A.; Gallego Lima, F.; et al. Airborne microplastics and SARS-CoV-2 in total suspended particles in the area surrounding the largest medical centre in Latin America. Environ. Pollut. 2022, 292, 118299. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornberg, N.J.; Garber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Gassilloud, B.; Huguet, L.; Maul, A.; Gantzer, C. Development of a viral concentration method for bottled water stored in hydrophobic support. J. Virol. Methods 2007, 142, 98–104. [Google Scholar] [CrossRef]
- Zhang, G.F.; Cao, G.L.; Luo, R.H.; Song, Q.L.; Zeng, Y.Q.; Liu, K.; Qu, J.; Lin, X.; Liu, F.-L.; Wang, G.; et al. Microplastics interact with SARS-CoV-2 and facilitate host cell infection. Environ. Sci. Nano 2022, 9, 2653–2664. [Google Scholar] [CrossRef]
- Gassilloud, B.; Gantzer, C. Adhesion-aggregation and inactivation of poliovirus 1 in groundwater stored in a hydrophobic container. Appl. Environ. Microbiol. 2005, 71, 912–920. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Grainger, D.C.; Busby, S.J. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010, 13, 773–780. [Google Scholar] [CrossRef]
- Devaraj, A.; Justice, S.S.; Bakaletz, L.O.; Goodman, S.D. DNABII proteins play a central role in UPEC biofilm structure. Mol. Microbiol. 2015, 96, 1119–1135. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.; Arora, K.; Gupta, A.; Guptasarma, P. The DNA-binding protein HU is a molecular glue that attaches bacteria to extracellular DNA in biofilms. J. Biol. Chem. 2021, 296, 100532. [Google Scholar] [CrossRef] [PubMed]
- Holloczki, O.; Gehrke, S. Nanoplastics can change the secondary structure of proteins. Sci. Rep. 2019, 9, 16013. [Google Scholar] [CrossRef] [PubMed]
- Arias-Andres, M.; Klumper, U.; Rojas-Jimenez, K.; Grossart, H.P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018, 237, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, W.; Yang, X.; Wang, J.; Lin, H.; Yang, Y. Microplastics are a hotspot for antibiotic resistance genes: progress and perspective. Sci. Total Environ. 2021, 773, 145643. [Google Scholar] [CrossRef]
- Liu, J.; Lv, M.; Sun, A.; Ding, J.; Wang, Y.; Chang, X.; Chen, L. Exposure to microplastics reduces the bioaccumulation of sulfamethoxazole but enhances its effects on gut microbiota and the antibiotic resistome of mice. Chemosphere 2022, 294, 133810. [Google Scholar] [CrossRef]
- Lu, K.; Zhan, D.; Fang, Y.; Li, L.; Chen, G.; Chen, S.; Wang, L. Micropalstics, potential threat to patients with lung diseases. Front. Toxicol. 2022, 4, 958414. [Google Scholar] [CrossRef]
- Molina, E.; Benede, S. Is there evidence of health risks from exposure to micro- and nanoplastics in foods? Front. Nutr. 2022, 9, 910094. [Google Scholar] [CrossRef] [PubMed]
- Anagnosti, L.; Varvaresou, A.; Pavlou, P.; Protopapa, E.; Carayanni, V. Worldwide actions against plastic pollution from microbeads and microplastics in cosmetics focusing on European policies. Has the issue been handled effectively? Mar. Pollut. Bull. 2021, 162, 111883. [Google Scholar] [CrossRef]
- Napper, I.E.; Bakir, A.; Rowland, S.J.; Thompson, R.C. Characterisation, quantity and sorptive properties of microplastics extracted from cosmetics. Mar. Pollut. Bull. 2015, 99, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Gouin, T.; Roche, N.; Lohmann, R.; Hodges, G. A thermodynamic approach for assessing the environmental exposure of chemicals absorbed to microplastic. Environ. Sci. Technol. 2011, 45, 1466–1472. [Google Scholar] [CrossRef]
- Eriksson, K.; Wiklund, L. Dermal exposure to styrene in the fibreglass reinforced plastics industry. Ann. Occup. Hyg. 2004, 48, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Stracke, F.; Hansen, S.; Schaefer, U.F. Nanoparticles and their interactions with the dermal barrier. Dermato-Endocrinology 2009, 1, 197–206. [Google Scholar] [CrossRef]
- Li, L.; Xu, Y.; Li, S.; Zhang, X.; Feng, H.; Dai, Y.; Zhao, J.; Yue, T. Molecular modeling of nanoplastic transformations in alveolar fluid and impacts on the lung surfactant film. J. Hazard. Mater. 2022, 427, 127872. [Google Scholar] [CrossRef]
- Shi, W.; Cao, Y.; Chai, X.; Zhao, Q.; Geng, Y.; Liu, D.; Tian, S. Potential health risks of the interaction of microplastics and lung surfactant. J. Hazard. Mater. 2022, 429, 128109. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, Y.; Deng, Y.; Jiang, W.; Zhao, Y.; Geng, J.; Ding, L.; Ren, H. Uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver. Environ. Sci. Technol. 2016, 50, 4054–4060. [Google Scholar] [CrossRef]
- Smith, M.; Love, D.C.; Rochman, C.M.; Neff, R.A. Microplastics in seafood and the implications for human health. Curr. Environ. Health Rep. 2018, 5, 375–386. [Google Scholar] [CrossRef]
- Yang, Y.F.; Chen, C.Y.; Lu, T.H.; Liao, C.M. Toxicity-based toxicokinetic/toxicodynamic assessment for bioaccumulation of polystyrene microplastics in mice. J. Hazard. Mater. 2019, 366, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Yang, K.; Cheng, X.; Guo, C.; Xing, X.; Sun, H.; Liu, D.; Liu, X.; Wang, D. Accumulation of polystyrene microplastics induces liver fibrosis by activating cGAS/STING pathway. Environ. Pollut. 2022, 300, 118986. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, Y.; Lemos, B.; Ren, H. Tissue accumulation of microplastics in mice and biomarker responses suggest widespread health risks of exposure. Sci. Rep. 2017, 7, 46687. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bai, J.; Ning, B.; Fan, L.; Sun, T.; Fang, Y.; Wu, J.; Li, S.; Duan, C.; Zhang, Y.; et al. Effects of bisphenol A and nanoscale and microscale polystyrene plastic exposure on particle uptake and toxicity in human Caco-2 cells. Chemosphere 2020, 254, 126788. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.; Lindeque, P.; Fileman, E.; Halsband, C.; Goodhead, R.; Moger, J.; Galloway, T.S. Microplastic ingestion by zooplankton. Environ. Sci. Technol. 2013, 47, 6646–6655. [Google Scholar] [CrossRef]
- Murphy, K.; Weaver, C.; Berg, L.J. Janeway’s Immunobiology, 10th ed.; W.W. Norton & Co.: New York, NY, USA, 2022. [Google Scholar]
- Tong, X.; Li, B.; Li, J.; Li, L.; Zhang, R.; Du, Y.; Zhang, Y. Polyethylene microplastics cooperate with Helicobacter pylori to promote gastric injury and inflammation in mice. Chemosphere 2022, 288, 132579. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Bio. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Ramsperger, A.; Narayana, V.K.B.; Gross, W.; Mohanraj, J.; Thelakkat, M.; Greiner, A.; Schmalz, H.; Kress, H.; Laforsch, C. Environmental exposure enhances the internalization of microplastic particles into cells. Sci. Adv. 2020, 6, eabd1211. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Zhang, Z.; Penn, R.; Barclay, W.S.; Giotis, E.S. Naive human macrophages are refractory to SARS-CoV-2 infection and exhibit a modest inflammatory response early in infection. Viruses 2022, 14, 441. [Google Scholar] [CrossRef]
- Labzin, L.I.; Chew, K.Y.; Wang, X.; Esposito, T.; Stocks, C.J.; Rae, J.; Yordanov, T.; Holley, C.L.; Emming, S.; Fritzlar, S.; et al. ACE2 is necessary for SARS-CoV-2 infection and sensing by macrophages but not sufficient for productive viral replication. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhang, G.; Cong, Y.; Liu, F.L.; Sun, J.; Zhang, J.; Cao, G.; Zhou, L.; Yang, W.; Song, Q.; Wang, F.; et al. A nanomaterial targeting the spike protein captures SARS-CoV-2 variants and promotes viral elimination. Nat. Nanotechnol. 2022, 17, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Italiani, P.; Palomba, R.; Decuzzi, P.; Duschl, A.; Fadeel, B.; Moghimi, S.M. Nanoparticles and innate immunity: new perspectives on host defence. Semin. Immunol. 2017, 34, 33–51. [Google Scholar] [CrossRef]
- Grainger, J.R.; Konkel, J.E.; Zangerle-Murray, T.; Shaw, T.N. Macrophages in gastrointestinal homeostasis and inflammation. Pflugers Arch. 2017, 469, 527–539. [Google Scholar] [CrossRef]
- Cannon, G.J.; Swanson, J.A. The macrophage capacity for phagocytosis. J. Cell Sci. 1992, 101, 907–913. [Google Scholar] [CrossRef]
- Champion, J.A.; Mitragotri, S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA 2006, 103, 4930–4934. [Google Scholar] [CrossRef]
- Adlerz, K.M.; Aranda-Espinoza, H.; Hayenga, H.N. Substrate elasticity regulates the behavior of human monocyte-derived macrophages. Eur. Biophys. J. 2016, 45, 301–309. [Google Scholar] [CrossRef]
- Baranska, A.; Shawket, A.; Jouve, M.; Baratin, M.; Malosse, C.; Voluzan, O.; Vu Manh, T.P.; Fiore, F.; Bajénoff, M.; Benaroch, O.; et al. Unveiling skin macrophage dynamics explains both tattoo persistence and strenuous removal. J. Exp. Med. 2018, 215, 1115–1133. [Google Scholar] [CrossRef]
- Prietl, B.; Meindl, C.; Roblegg, E.; Pieber, T.R.; Lanzer, G.; Frohlich, E. Nano-sized and micro-sized polystyrene particles affect phagocyte function. Cell Biol. Toxicol. 2014, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Mollerherm, H.; von Kockritz-Blickwede, M.; Branitzki-Heinemann, K. Antimicrobial activity of mast cells: role and relevance of extracellular DNA traps. Front. Immunol. 2016, 7, 265. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Gold, J.A.; Andina, N.; Lee, J.J.; Kelly, A.M.; Kozlowski, E.; Schmid, I.; Straumann, A.; Reichenbach, J.; Gleich, G.J.; et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 2008, 14, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Schorn, C.; Janko, C.; Latzko, M.; Chaurio, R.; Schett, G.; Herrmann, M. Monosodium urate crystals induce extracellular DNA traps in neutrophils, eosinophils, and basophils but not in mononuclear cells. Front. Immunol. 2012, 3, 277. [Google Scholar] [CrossRef]
- Morshed, M.; Hlushchuk, R.; Simon, D.; Walls, A.F.; Obata-Ninomiya, K.; Karasuyama, H.; Djonov, V.; Eggel, A.; Kaufmann, T.; Simon, H.-U.; et al. NADPH oxidase-independent formation of extracellular DNA traps by basophils. J. Immunol. 2014, 192, 5314–5323. [Google Scholar] [CrossRef]
- von Kockritz-Blickwede, M.; Goldmann, O.; Thulin, P.; Heinemann, K.; Norrby-Teglund, A.; Rohde, M.; Medina, E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008, 111, 3070–3080. [Google Scholar] [CrossRef]
- Delgado-Rizo, V.; Martinez-Guzman, M.A.; Iniguez-Gutierrez, L.; Garcia-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil extracellular traps and its implications in inflammation: an overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef]
- Goldmann, O.; Medina, E. The expanding world of extracellular traps: not only neutrophils but much more. Front. Immunol. 2012, 3, 420. [Google Scholar] [CrossRef]
- Doster, R.S.; Rogers, L.M.; Gaddy, J.A.; Aronoff, D.M. Macrophage extracellular traps: a scoping review. J. Innate Immun. 2018, 10, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Farrera, C.; Bhattacharya, K.; Lazzaretto, B.; Andon, F.T.; Hultenby, K.; Kotchey, G.P.; Star, A.; Fadeel, B. Extracellular entrapment and degradation of single-walled carbon nanotubes. Nanoscale 2014, 6, 6974–6983. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Konduru, N.V.; Feng, W.; Allen, B.L.; Conroy, J.; Volkov, Y.; Vlasova, I.I.; Belikova, N.A.; Yanamala, N.; Kapralov, A.; et al. Carbon nanotubes degraded by neutrophil myeloperoxidase induce less pulmonary inflammation. Nat. Nanotechnol. 2010, 5, 354–359. [Google Scholar] [CrossRef]
- Shvedova, A.A.; Pietroiusti, A.; Fadeel, B.; Kagan, V.E. Mechanisms of carbon nanotube-induced toxicity: focus on oxidative stress. Toxicol. Appl. Pharmacol. 2012, 261, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Peng, L.; Song, E.; Song, Y. Polystyrene nanoplastics induce neutrophil extracellular traps in mice neutrophils. Chem. Res. Toxicol. 2022, 35, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 2018, 18, 134–147. [Google Scholar] [CrossRef]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, S.M.; Farhangrazi, Z.S. Nanomedicine and the complement paradigm. Nanomedicine 2013, 9, 458–460. [Google Scholar] [CrossRef]
- Szebeni, J. The interaction of liposomes with the complement system. Crit. Rev. Ther. Drug 1998, 15, 57–88. [Google Scholar] [CrossRef] [PubMed]
- Escamilla-Rivera, V.; Solorio-Rodríguez, A.; Uribe-Ramirez, M.; Lozano, O.; Lucas, S.; Chagolla-López, A.; Winkler, R.; De Vizcaya-Ruiz, A. Plasma protein adsorption on Fe3O4-PEG nanoparticles activates the complement system and induces an inflammatory response. Int. J. Nanomedicine 2019, 14, 2055–2067. [Google Scholar] [CrossRef]
- La-Beck, N.M.; Islam, M.R.; Markiewski, M.M. Nanoparticle-induced complement activation: implications for cancer nanomedicine. Front. Immunol. 2020, 11, 603039. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, G.; Griffin, J.I.; Brenneman, B.; Banda, N.K.; Holers, V.M.; Backos, D.S.; Wu, L.P.; Moghimi, S.M.; Simberg, D. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 2017, 12, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Domenech, M.; Ramos-Sevillano, E.; Garcia, E.; Moscoso, M.; Yuste, J. Biofilm formation avoids complement immunity and phagocytosis of Streptococcus pneumoniae. Infect. Immun. 2013, 81, 2606–2615. [Google Scholar] [CrossRef] [PubMed]
- Pier, G.B.; Coleman, F.; Grout, M.; Franklin, M.; Ohman, D.E. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 2001, 69, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- Kristian, S.A.; Birkenstock, T.A.; Sauder, U.; Mack, D.; Gotz, F.; Landmann, R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 2008, 197, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- De Smet, K.; Contreras, R. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol. Lett. 2005, 27, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Kristensen, K.; Henriksen, J.R.; Andresen, T.L. Adsorption of cationic peptides to solid surfaces of glass and plastic. PLoS ONE 2015, 10, e0122419. [Google Scholar] [CrossRef]
- Kurtz, J. Specific memory within innate immune systems. Trends Immunol. 2005, 26, 186–192. [Google Scholar] [CrossRef]
- Melillo, D.; Marino, R.; Italiani, P.; Boraschi, D. Innate immune memory in invertebrate metazoans: a critical appraisal. Front. Immunol. 2018, 9, 1915. [Google Scholar] [CrossRef]
- Seeley, J.J.; Ghosh, S. Molecular mechanisms of innate memory and tolerance to LPS. J. Leukoc. Biol. 2017, 101, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Quintin, J.; van der Meer, J.W. Trained immunity: a memory for innate host defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Lebre, F.; Boland, J.B.; Gouveia, P.; Gorman, A.L.; Lundahl, M.L.E.; Lynch, R.I.; O’Brien, F.J.; Coleman, J.; Lavelle, E.C. Pristine graphene induces innate immune training. Nanoscale 2020, 12, 11192–11200. [Google Scholar] [CrossRef] [PubMed]
- Swartzwelter, B.J.; Fux, A.C.; Johnson, L.; Swart, E.; Hofer, S.; Hofstatter, N.; Geppert, M.; Italiani, P.; Boraschi, D.; Duschl, A.; et al. The impact of nanoparticles on innate immune activation by live bacteria. Int. J. Mol. Sci. 2020, 21, 9695. [Google Scholar] [CrossRef]
- Rohde, K.; Yates, R.M.; Purdy, G.E.; Russell, D.G. Mycobacterium tuberculosis and the environment within the phagosome. Immunol. Rev. 2007, 219, 37–54. [Google Scholar] [CrossRef]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The immune escape mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Eisenbarth, S.C.; Colegio, O.R.; O’Connor, W.; Sutterwala, F.S.; Flavell, R.A. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 2008, 453, 1122–1126. [Google Scholar] [CrossRef]
- Hornung, V.; Bauernfeind, F.; Halle, A.; Samstad, E.O.; Kono, H.; Rock, K.L.; Fitzgerald, K.A.; Latz, E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 2008, 9, 847–856. [Google Scholar] [CrossRef]
- Martinon, F.; Petrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Ziglari, T.; Wang, Z.; Holian, A. Contribution of particle-induced lysosomal membrane hyperpolarization to lysosomal membrane permeabilization. Int. J. Mol. Sci. 2021, 22, 2277. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Ackerman, A.L.; Cody, V.; Giodini, A.; Hinson, E.R.; Cresswell, P.; Edelson, R.L.; Saltzman, W.M.; Hanlon, D.J. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology 2006, 117, 78–88. [Google Scholar] [CrossRef] [PubMed]
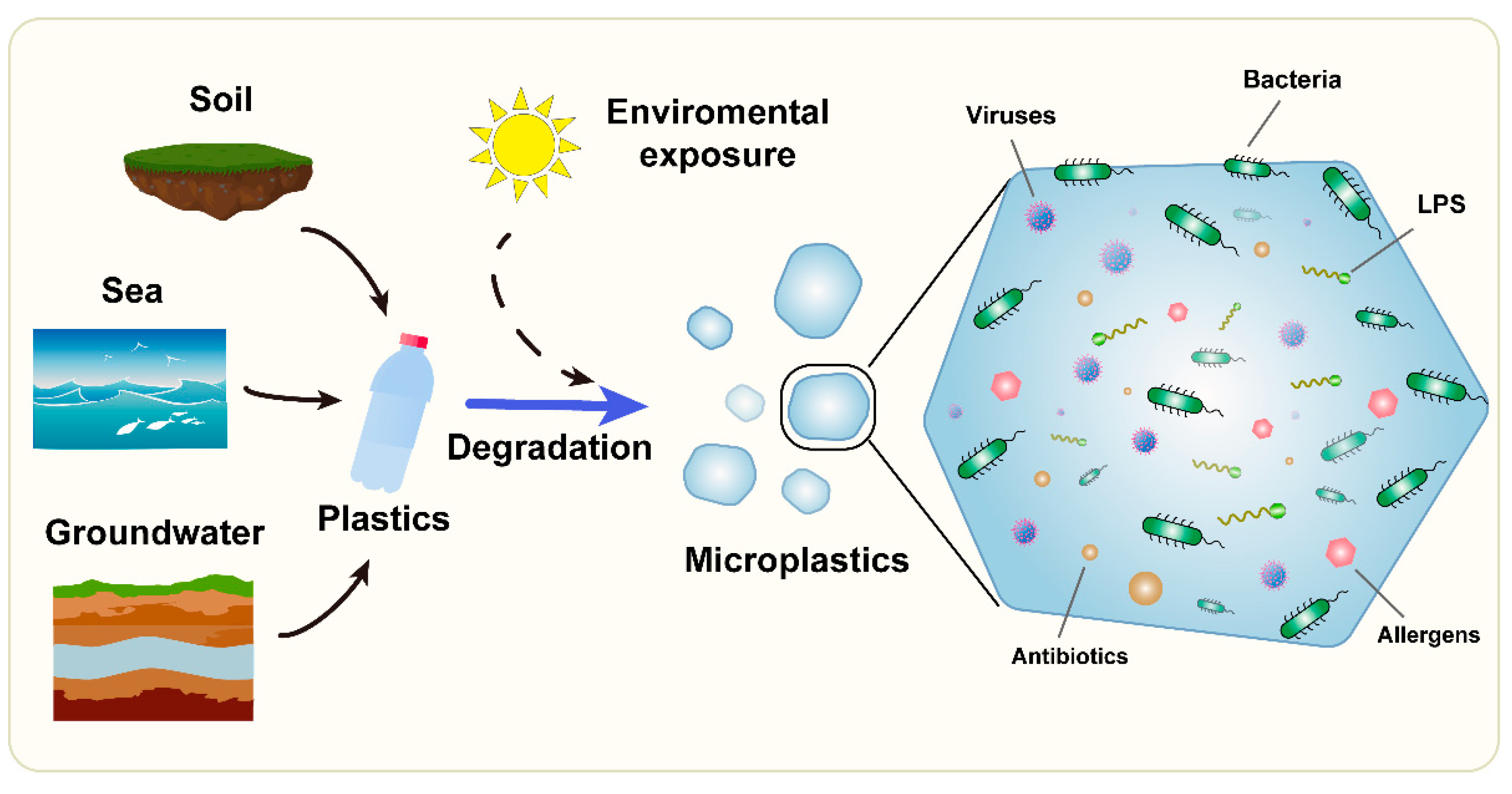
| Microplastic origin | seawater, freshwater, soil, airborne dusts, cosmetics, textiles, food packaging |
| Exposure route | dermal, inhalation, ingestion |
| Microorganisms on MPs | viruses, bacteria; selection/concentration of specific strains, biofilm formation |
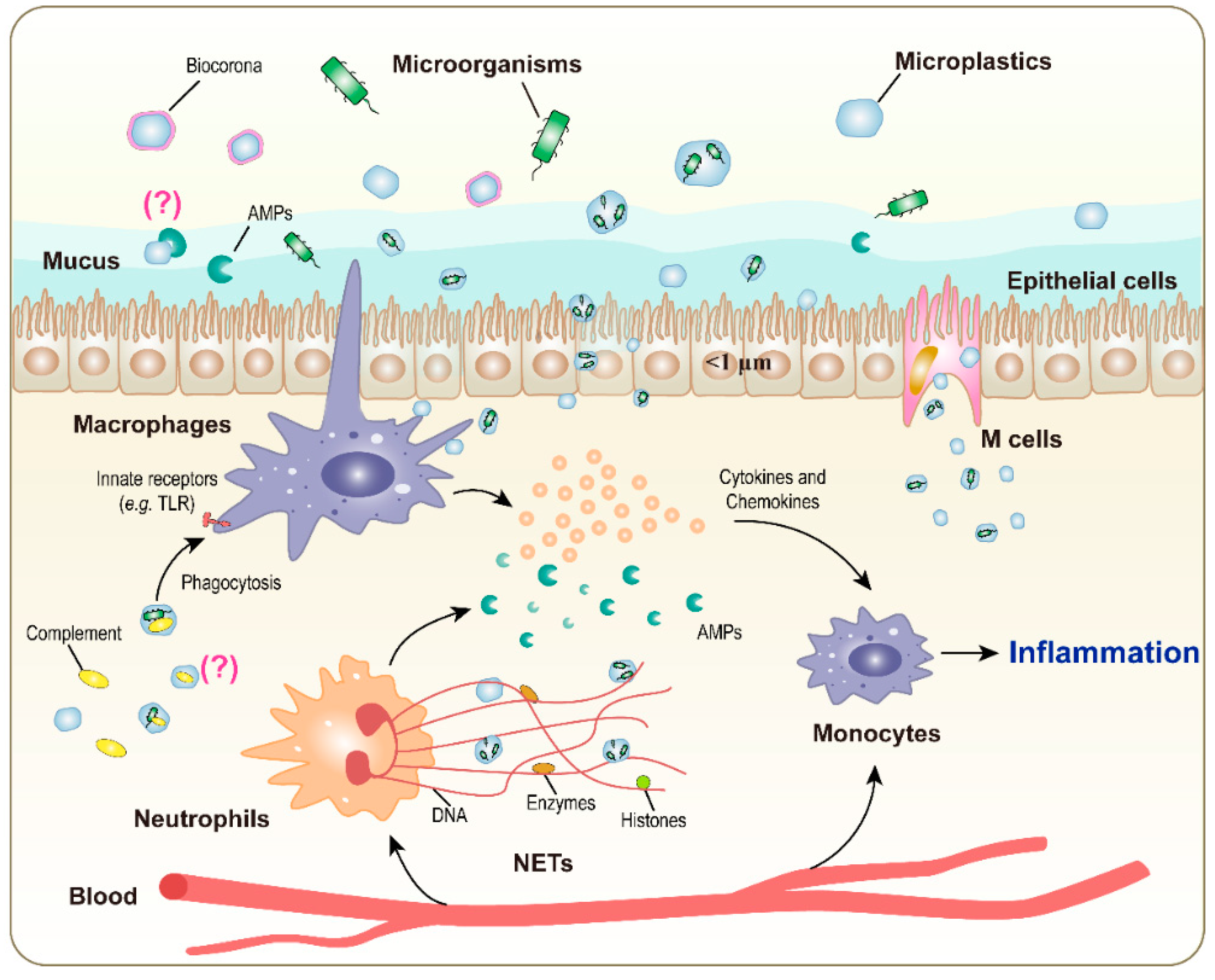
| Main immune cells involved | macrophages, neutrophils, mast cells (innate immune cells) |
| Main soluble immune factors involved | antimicrobial peptides, complement components (innate immune factors) |
| Effects | changes in the modality of infection (facilitation of entry via phagocytosis) |
| Detrimental | increased entry into target cells |
| Beneficial | increased microorganism intracellular killing, antigen processing and presentation, and establishment of adaptive immunity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Li, Y.; Boraschi, D. Association between Microorganisms and Microplastics: How Does It Change the Host–Pathogen Interaction and Subsequent Immune Response? Int. J. Mol. Sci. 2023, 24, 4065. https://doi.org/10.3390/ijms24044065
Yang W, Li Y, Boraschi D. Association between Microorganisms and Microplastics: How Does It Change the Host–Pathogen Interaction and Subsequent Immune Response? International Journal of Molecular Sciences. 2023; 24(4):4065. https://doi.org/10.3390/ijms24044065
Chicago/Turabian StyleYang, Wenjie, Yang Li, and Diana Boraschi. 2023. "Association between Microorganisms and Microplastics: How Does It Change the Host–Pathogen Interaction and Subsequent Immune Response?" International Journal of Molecular Sciences 24, no. 4: 4065. https://doi.org/10.3390/ijms24044065
APA StyleYang, W., Li, Y., & Boraschi, D. (2023). Association between Microorganisms and Microplastics: How Does It Change the Host–Pathogen Interaction and Subsequent Immune Response? International Journal of Molecular Sciences, 24(4), 4065. https://doi.org/10.3390/ijms24044065







