Abstract
Until a few years ago, many studies focused on the transcriptomic response to single stresses. However, tomato cultivations are often constrained by a wide range of biotic and abiotic stress that can occur singularly or in combination, and several genes can be involved in the defensive mechanism response. Therefore, we analyzed and compared the transcriptomic responses of resistant and susceptible genotypes to seven biotic stresses (Cladosporium fulvum, Phytophthora infestans, Pseudomonas syringae, Ralstonia solanacearum, Sclerotinia sclerotiorum, Tomato spotted wilt virus (TSWV) and Tuta absoluta) and five abiotic stresses (drought, salinity, low temperatures, and oxidative stress) to identify genes involved in response to multiple stressors. With this approach, we found genes encoding for TFs, phytohormones, or participating in signaling and cell wall metabolic processes, participating in defense against various biotic and abiotic stress. Moreover, a total of 1474 DEGs were commonly found between biotic and abiotic stress. Among these, 67 DEGs were involved in response to at least four different stresses. In particular, we found RLKs, MAPKs, Fasciclin-like arabinogalactans (FLAs), glycosyltransferases, genes involved in the auxin, ET, and JA pathways, MYBs, bZIPs, WRKYs and ERFs genes. Detected genes responsive to multiple stress might be further investigated with biotechnological approaches to effectively improve plant tolerance in the field.
1. Introduction
Plants are sessile living organisms that developed many strategies for quickly adapting to environmental changes. Despite this, adverse environmental factors (abiotic stress), such as drought, salinity, low temperatures, oxidative stress and plant pathogen or pest attacks (biotic stress), can negatively affect plant growth and production []. Simultaneous exposition to biotic and abiotic stress can induce tremendous crop yield losses. Therefore, the resistance mechanisms to various tomato stresses have been under investigation for a long time. Until a few years ago, most stress-related studies focused on the single stress response mechanism. Recently, more emphasis has been given to studies investigating the plant response to combinations of multiple stresses []. An increasing number of studies have been conducted to identify new forms of resistance to multiple stressors, and several genes that recognize both biotic and abiotic stress have been found [,,,,]. In tomato, different genes for perception, signaling, hormone balancing, and transcription modulation are involved in various biotic and abiotic stress responses [,,,,]. The modulation of these pathways leads to the activation or repression of several responsive proteins. In this context, genetic engineering became a fundamental tool for creating new plants that quickly adapt to biotic and abiotic stress without compromising plants’ phenotypical traits and yields [,]. For this purpose, studying transcriptomic alterations in plants subjected to various stress could aid the identification of principal genes that participate in resistance or susceptibility processes []. High-throughput sequencing RNA-seq technology quantifies gene expression levels with high accuracy []. To date, RNA-seq has been extensively used to investigate plant stress interactions, and raw reads of each experiment are deposited in sequence databases. Therefore, published sequencing data could represent a valuable resource to further explore plant stress responses by analyzing and comparing different studies to identify genes responsive to various stresses.
In tomato, a comparative analysis using the microarray gene expression technique identified 1862 and 835 genes responding to biotic and abiotic stress, respectively []. However, RNA-seq technology showed a higher sensitivity for gene expression than microarray technology and better genome coverage []. So far, Illumina technology is the most used sequencing platform for RNA samples due to its accuracy, rapidity, and moderate price []. In our work, RNA-seq raw datasets of tomato-stressed samples were assessed to identify genes involved in both biotic and abiotic stress. Raw data included tomato transcriptional response to 12 different stressors, eight biotic collected by [], and four abiotic stress collected from other works [,,]. In particular, genes involved in cell wall metabolism, membrane receptors, transcription factors (TFs), and phytohormones modulation were deeply investigated. The final goal of our analysis was to prioritize a list of responsive genes to multiple stress as potential candidates to be employed in genetic engineering programs. Indeed, genes resulting from our comparative analysis could be further characterized through biotechnological approaches to investigate their role in tomato response to multiple stresses.
2. Results
This work aimed to explore the transcriptomic alterations of tomato plants exposed to pathogens and environmental stresses to identify genes involved in response to multiple stressors. To examine and characterize genes responding to different stresses, we exhaustively re-analyzed twelve publicly available RNA-seq studies of resistant (R) and susceptible (S) tomatoes challenged by different biotic and abiotic stresses (Table 1, Supplementary Table S1).

Table 1.
Transcriptomic responses to biotic and abiotic stress analyzed in this study. The number of up and downregulated genes and total differentially expressed genes (DEGs) has been reported for each experiment. Si = susceptible stressed; Ri = resistant stressed; Sni = susceptible not stressed; Rni = resistant not stressed; Ti = tolerant stressed; Tni = tolerant not stressed; Dpi = days post-induced stress.
The experiments were singularly analyzed, and the lists of resulting DEGs were compared to identify genes involved in response to different stresses (Supplementary Tables S2 and S3). A high variable number of DEGs was found among studies depending on the induced stress (Table 1). P. infestans, P. syringae, S. sclerotiorum, and T. absoluta (T genotype) caused the differential expression (DE) of a considerable number of tomato genes (higher than 10,000). TSWV-stressed plants showed a DEGs peak at twenty-one dpi (1490 DEGs), while R. solanacearum induced an increased number of DEGs at two dpi. The R and S genotypes to C. fulvum showed a decrease in DEGs from seven to twenty dpi. Among abiotic stress analyzed, DEGs ranged from 2000 to 6000, except for the low temperatures experiment (12836 DEGs).
To have an overview of DEGs responsive to different stresses, we first analyzed biotic and abiotic stress separately, seeking DEGs in more than a single experiment. Then, we extended the comparison of the results for detecting genes involved in both biotic and abiotic responses.
2.1. Exploration of the Datasets of DEGs under Various Biotic Stress
Transcriptomic datasets of plants subjected to biotic stress were gathered in four broad groups: (1) fungi, including C. fulvum, P. infestans (R and S response) and S. sclerotiorum (S response); (2) bacteria, containing R. solanacearum (R and S response) and P. Syringae (S response); (3) TSWV (R response), and (4) T. absoluta (T and Sresponse). To better analyze the data within each biotic stress group, we considered the more similar time points. Therefore, tomato response to fungi, TSWV and T. absoluta was investigated at more than 20 dpi, whereas transcriptomic changes inplants infected with bacteria were investigated at 2 dpi. Supplementary Figure S1 shows the DEGs induced or repressed by each tomato interaction studied.
The DEGs found in each group (fungi, bacteria, TSWV, and T. absoluta) were intersected to identify R and S common genes. The S genotypes to fungi showed 913 down and 560 common-upregulated DEGs (Figure 1A), while R plants showed an overlap of 1841 and 1770 up and downregulated genes, respectively (Figure 1B). The S tomatoes to bacteria shared 755 up and 732 downregulated genes (Figure 1C) while the R genotype showed 1247 down and 1135 upregulated genes (Figure 1D). Considering that in this study TSWV and T. absoluta could not be compared with other viruses or pests, we used their whole datasets of DEGs for further analysis.
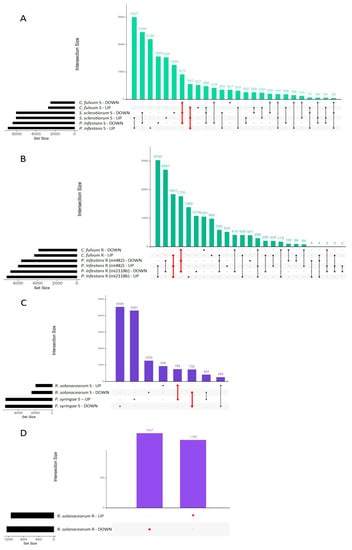
Figure 1.
The intersection of up and down DEGs induced by tomato genotypes infected with fungi and bacteria (first 30 intersections). (A) Susceptible genotypes infected with fungi. (B) Resistant genotypes infected with fungi. (C) Susceptible genotypes infected with bacteria. (D) Resistant genotype infected with bacteria. In red are DEGs with the same expression trend in all the analyzed datasets.
Contrasting expression patterns among interactions were also found. For example, in R genotypes to fungi, 298 genes were downregulated in C. fulvum and upregulated in P. infestans. On the contrary, 592 genes were upregulated in C. fulvum and downregulated in P. infestans (Figure 1). To identify genes associated with resistance or susceptibility within each pathogen group, lists of common DEGs identified in R and S genotypes were compared (Figure 2). The R genotypes to fungi specifically induced and repressed 1457 and 1090 genes, respectively (Figure 2), while S genotypes showed 176 and 233 privately up and downregulated genes. Similarly, the R genotypes to bacteria showed the private activation of 826 genes, while 989 were downregulated. The S genotypes displayed 446 and 474 specific up and downregulated genes (Figure 2). The R genotype to T. absoluta showed the private differential regulation of 2432 induced and 2773 repressed genes, while the S genotype showed 536 upregulated and 419 downregulated private genes (Figure 2). Lists of private DEGs were used to analyze processes related to biotic stress response in R and S genotypes.
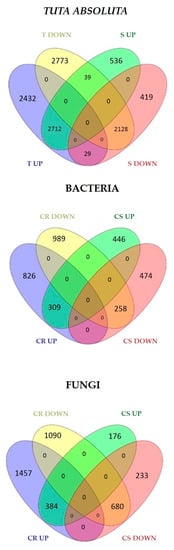
Figure 2.
Private DEGs identified in R and S genotypes to fungi, bacteria, and Tuta absoluta. CR = common DEGs among R genotypes; CS = common DEGs among S genotypes; T = tolerant; S = susceptible genotypes.
2.1.1. Analysis of Processes Putatively Involved in Biotic Stress Response, Privately Activated by Resistant and Susceptible Genotypes
The R genotypes to fungi showed the activation of genes involved in ethylene (ET) and jasmonic acid (JA) pathways, while several genes for the synthesis of abscisic acid (ABA) were repressed. By contrast, S plants showed the repression of the ET pathway, while ABA metabolism was generally activated. In addition, R and S genotypes differed in regulating genes participating in cell wall metabolism, in signaling or encoding for transcription factors (TFs), such as WRKYs and MYBs, and pathogen-related proteins (PR-proteins) which were mainly upregulated in R genotypes (Figure 3).
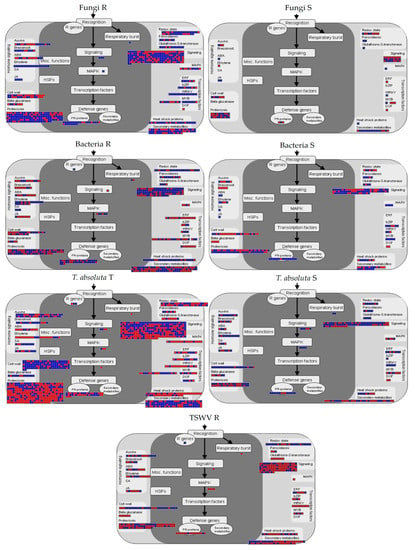
Figure 3.
Genes putatively involved in biotic stress response under fungi, bacteria, TSWV, and T. absoluta infections. Blue: upregulated, red: downregulated genes. R = resistant; S = susceptible; T = tolerant genotype.
The R genotype to bacteria showed the induction of genes for salicylic acid (SA) synthesis, while ABA and JA synthesis were predominantly repressed. In addition, the R genotype to bacteria showed the downregulation of different TFs, especially ERFs, DOFs, and MYBs. On the other hand, different MYBs were upregulated in S genotypes (Figure 3).
The T genotype to T. absoluta upregulated genes involved in lignin, AGPs, and hemicellulose biosynthesis, simultaneously activating SA synthesis. By contrast, we observed the repression of genes involved in ET and ABA biosynthesis. Moreover, several genes participating in signaling and proteolysis processes were DE. Contrarily, the S genotype mainly upregulated the ET pathway (Figure 3).
The R genotype to TSWV showed general repression of the defense signaling and the activation of cell wall metabolism (Figure 3).
2.1.2. Differentially Regulation of Cell Wall Precursors under Various Biotic Stress
Due to the cell wall’s role in different stress responses, we focused on genes participating in the biosynthesis of the cell wall precursors, with specific reference to genes participating in cellulose and pectin metabolism such as Fasciclin-like arabinogalactans (FLAs), expansins (EXPs), pectate lyases, UDP-glucose-4-epimerases (UGEs) and polygalacturonases (PGs) (Figure 4).
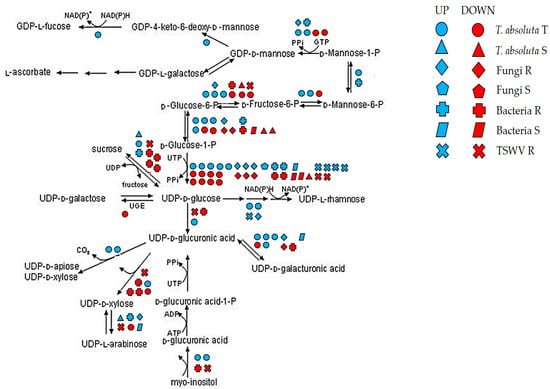
Figure 4.
DEGs under various biotic stress involved in the biosynthesis of cell wall precursors. Blue: upregulated; red: downregulated. R = resistant; S = susceptible; T = tolerant genotype.
Cellulose is one of the main components of the cell wall. In this study, two FLAs (Solyc01g091530 and Solyc10g005960) were downregulated among R genotypes to fungi, while Solyc07g045440 and Solyc07g053540 were also repressed by bacteria S and induced by TSWV R. The Solyc06g075220 (FLA8) was downregulated by S genotypes to bacteria and upregulated by the T genotype to T. absoluta. By contrast, Solyc12g015690 (FLA11) was upregulated by both T. absoluta T and TSWV R genotypes. Cellulose disassembly could also be promoted by EXP genes. Interestingly, an Expansin-like protein, Solyc08g077900 (EXLB1), was upregulated by R genotypes to fungi.
Among genes involved in pectin degradation, we found that pectate lyases were involved in response to different pathogens. For example, the R genotypes to fungi and T. absoluta activated Solyc03g111690, while R genotypes to TSWV and T. absoluta commonly induced Solyc05g014000 and Solyc06g083580. Similarly, two pectate lyases (Solyc09g008380 and Solyc09g091430) were activated by the R genotype to TSWV and the T genotype to T. absoluta and repressed by S genotypes to bacteria.
Other important enzymes involved in pectin metabolism were found DE under various biotic stress. In particular, a PG (Solyc08g060970) and a UGE (Solyc02g030230) were induced by R genotypes to fungi and bacteria. Similarly, the R genotypes to bacteria showed the downregulation of Solyc12g010540. By contrast, a different UGE (Solyc07g043550) was activated by bacteria S genotypes and repressed by the T. absoluta T genotype.
Galacturonic acid (GalA) is utilized to synthesize hemicellulose and pectin. Our results showed that T plants to T. absoluta activated four genes participating in the GalA epimerization (GAE) (Solyc01g091200, Solyc09g092330, Solyc05g050990, and Solyc10g018260), while Solyc08g079440 and Solyc08g082440, involved in UDP-galactose production, were repressed (Figure 4).
In general, we observed that R and T genotypes to fungi and T. absoluta showed a high number of common DEGs. Among these, we found the activation of an endo-1,4 beta-glucanase (Solyc05g005080), an endo-1,4-mannosidase (Solyc10g074920), and a 3,5-epimerase/4-reductase (Solyc08g080140).
2.1.3. Identification of Genes Differentially Expressed under Different Biotic Stress and Involved in the Signaling Process
The investigation of DEGS involved in the plant signaling process allowed the identification of 30 genes encoding for RLPs (Figure 5). In particular, the analysis of the seven expression profiles pointed out a clear divergent regulation. Interestingly, an LRR-RLK (Solyc08g061560) was suppressed by R genotypes to fungi and bacteria and induced by T. absoluta T and TSWV R genotypes. Similarly, Solyc03g093460 was activated by the T genotype to T. absoluta (Figure 5) and repressed by R genotypes to fungi and bacteria. We also found two common RLKs (Solyc02g068830 and Solyc06g048740) differentially regulated among R genotypes to fungi, bacteria, and TSWV, while Solyc11g006040 was induced in R genotypes to fungi, T. absoluta and TSWV (Figure 5).

Figure 5.
Heat map of DEGs participants in the signaling process. The expression pattern of DEGs identified and their function have been reported. Blue = upregulated, and red = downregulated genes. R = resistant; S = susceptible; T = tolerant genotype.
2.2. Exploration of the Datasets of Differentially Expressed Genes under Various Abiotic Stress
Lists of DEGs resulting from tomatoes stressed with drought, low temperature, salinity, and oxidative stress were compared to identify genes involved in response to various abiotic stress. The two studies on drought stress showed similar expression profiles except for a few genes (Figure 6A). Therefore, to select common DEGs in the susceptible responses to drought, we compared DEGs of M82 and Jinlingmeiyu (S cultivars), identifying 176 up and 347 downregulated genes (Figure 6B). Moreover, we compared DEGs of S genotypes with those of the IL9-1 (T plants), discovering 28 up and 32 downregulated genes privately DE by the T genotype (Figure 6B).
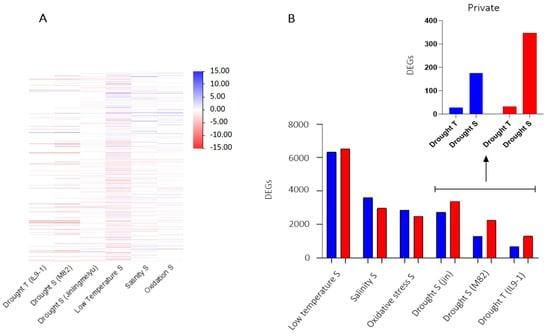
Figure 6.
(A) Heatmap of DEGs induced during different abiotic stress; (B) Number of DEGs induced by different abiotic stress with a focus on private DEGs induced by tolerant and susceptible genotypes to drought. Blue = Upregulated and Red = Downregulated. R = resistant; S = susceptible; T = tolerant genotype.
2.2.1. Investigation of Hormones and TFs Differentially Regulated under Various Abiotic Stress
To identify genes responsive to multiple abiotic stress, we focused on four primary defensive processes: signaling, hormone modulation, TFs regulation and biosynthesis of cell wall precursors. Our results showed that the salinity induced DEGs implicated in ABA, ET, and SA pathways. By contrast, low temperatures caused the downregulation of genes involved in SA, auxins, and brassinosteroids (BRs) pathways (Figure 7).
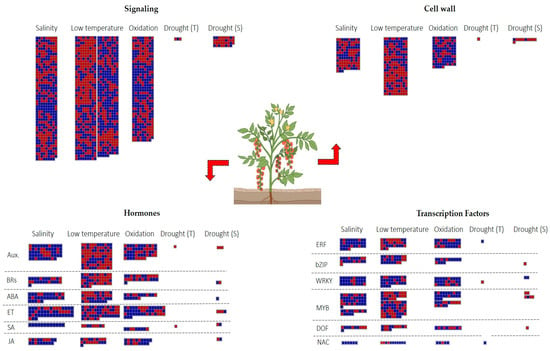
Figure 7.
DEGs involved in signaling, cell wall, hormones, and TFs. Blue = upregulated; red = downregulated genes.
Among the 32 private DEGs found in the drought T genotype, Solyc01g110680 and Solyc12g096820 were, respectively, involved in the auxin and SA biosynthesis and were suppressed. Contrarily, the S genotypes revealed the activation of a gene involved in ABA signaling (HVA22d) and the repression of genes involved in ET pathways (Solyc02g064950, Solyc07g049550, Solyc08g079750) (Figure 7).
Differences in TFs regulation included ERFs, which were mainly induced during salinity and oxidative stresses, while Solyc07g054220 (ERF2a) was also upregulated by the drought T genotype. In addition, the drought T genotype showed the activation of various NAC genes, including Solyc01g009860. Different WRKYs were upregulated during salinity, low temperature, oxidative stress, and in S genotypes to drought. On the other hand, the T genotype to drought showed the downregulation of Solyc07g055280 (WRKY78).
2.2.2. Investigating Genes Involved in Different Abiotic Stress Responses and Participating in Signaling and Cell Wall Processe
Comparison of genes involved in the signaling process led to the identification of genes encoding for Calnexins, RLKs, and Glutamate receptors (GLRs) (Table 2). Interestingly, Solyc03g118040 (Calnexin) was exclusively downregulated by the T genotype to drought and upregulated during all the other stress. At the same time, two RLKs (Solyc02g072310 and Solyc05g056370) were repressed during low temperature, salinity, and oxidative stress. Genes encoding for Glutamate receptors were mainly downregulated under different abiotic stress. In particular, Solyc06g063180 and Solyc07g052390 were repressed by salinity and low temperatures. The Solyc07g052400 was downregulated during drought S, oxidation, and low temperatures, while low temperatures and oxidation repressed Solyc05g045650. By contrast, Solyc02g067030 (SNF1) was induced by salinity and downregulated by low temperatures (Table 2).

Table 2.
Genes identified as responsive to different abiotic stress and involved in signaling and cell wall-related processes.
The cell wall precursors showed common DE of glucose 6-dehydrogenases (UGDs), glucuronate 4-epimerases (GAEs), glucose-4-epimerases (UGEs) and cellulose synthase-like (CSL) genes, repressed during salinity and oxidative stress and induced by low temperatures (Table 2). Two GAEs showed a divergent expression pattern during salinity, low temperature, and drought stress (Solyc07g006220 and Solyc05g050990), while the drought T genotype repressed a glycosyltransferase (Solyc12g096830). Finally, a CSL (Solyc03g097050) was induced by all the abiotic stress analyzed and was exclusively repressed by the drought T genotype.
2.3. Identification of Common Genes Differentially Expressed under Biotic and Abiotic Stress
Comparing lists of private DEGs obtained for each group of biotic (fungi, bacteria, TSWV and T. absoluta) and abiotic stress (drought, low temperature, salinity, and oxidative stress), we identified common genes related to signaling, cell wall, hormones, and TFs, activated or repressed under different stresses. Generally, the R genotypes to pathogens showed a higher number of common DEGs with abiotic stresses than their S counterparts for most of the gene classes (Figure 8). An exception was the identification in R and S plants to bacteria of a similar number of common DEGs, involved in the signaling and cell wall process, with low temperature stress. Due to the low number of private DEGs in T and S genotypes to drought, few genes were identified in common with biotic stress (Figure 8) and have been reported in Supplementary Table S4.
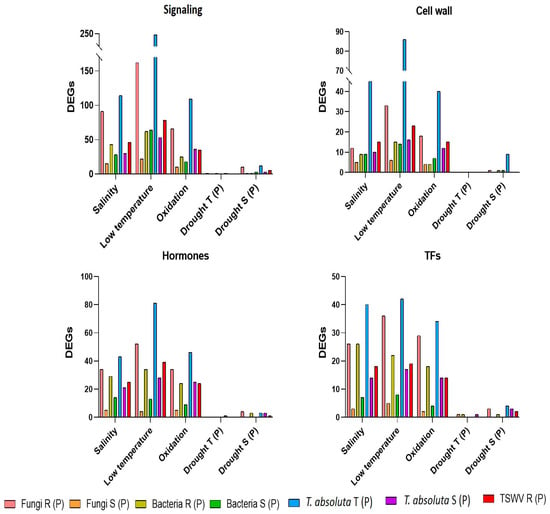
Figure 8.
Common DEGs in biotic and abiotic stress related to signaling, cell wall, hormones and transcription factors (TFs). (P) = private DEGs; R = resistant genotype; S = susceptible genotypes; T = tolerant genotype.
A total of 1474 common DEGs were found between biotic and abiotic stress (Supplementary Table S5). Therefore, we focused on those genes simultaneously DE in at least one abiotic stress and three groups of biotic stresses (Figure 9). Ten genes were involved in the biosynthesis of cell wall compounds, thirty-five DEGs resulted involved in signaling, thirteen DEGs encoded for hormones, and nine DEGs were annotated as TFs (Figure 9). DEGs involved in the synthesis of cell wall compounds included FLAs, glycosyltransferases (GTs), beta-xylosidase, GAEs, polygalacturonases, pectate lyases, and peptidoglycan-binding LysM domain-containing proteins (Figure 9).
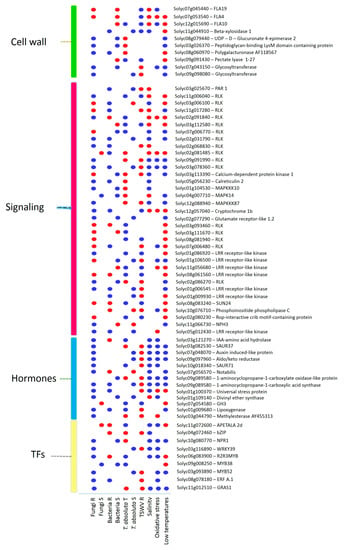
Figure 9.
DEGs responsive to biotic and abiotic stresses, encoding for cell wall precursors, signaling proteins, hormones, and TFs. Blue = upregulated; red = downregulated genes. R = resistant; S = susceptible; T = tolerant genotypes.
Genes belonging to the signaling process included RLKs, mitogen-activated protein kinases (MAPKs), a pathogenesis-related (PAR) gene involved in calcium signaling, a phosphoinositide phospholipase C, a Rop-interactive crib motif-containing protein, and genes involved in the photosynthetic pathway (Figure 9). Interestingly, we also found the upregulation of the Ve2 gene (Solyc09g005080) in the C. fulvum R genotype, S. sclerotiorum, P. syringae, oxidative stress, and its downregulation in the TSWV R genotype.
Hormone regulation showed various genes involved in the auxin and ABA pathways mainly activated by R genotypes to fungi and abiotic stress, while ET and JA synthesis showed a divergent expression profile during various biotic and abiotic stress (Figure 9). TFs included genes encoding for MYBs, WRKYs, ERFs, bZIPs, GRAS, and AP2/ERF (Figure 9).
3. Discussion
Using a common pipeline to analyze and compare publicly RNA-seq studies of tomato plants exposed to different stresses, we obtained an overview of the pathways involved in cellular reprogramming under different stresses. Moreover, we shed light on genes differentially expressed during various tomato stress interactions. The comparative analysis of tomato transcriptomic profiles provided a better understanding of the complex mechanisms underpinning the plant defense process.
3.1. Biotic Stressors Induced a Variegated Transcriptomic Response in Tomato
Comparative analysis of eight transcriptomics experiments, including resistant (R) and susceptible (S) genotypes to fungi, bacteria, T. absoluta and TSWV, allowed us to look into the intricate process of the tomato response to biotic stress. Plant-pathogen interaction leads to a deep remodeling of transcriptomic and metabolic pathways, such as phytohormones, signaling proteins, TFs, and cell wall-related processes [,,,].
Plant hormones are wellknown as critical regulators of signal defense responses in plants. In this work, we focused on the process involved in basal resistance mechanisms. Our findings suggested that the simultaneous activation of JA and the repression of ABA signaling could be a key factor in activating resistant basal response to fungi []. However, it is important to mention that different fine-tuned hormonal regulations could be observed depending on the pathogens’ lifestyle [,]. In contrast with fungi, the repression of JA and the activation of SA promoted the immune response in the R genotype to R. solanacerarum, confirming previous findings [,]. Differences in hormone regulation may also induce plants’ anti-herbivore characteristics []. In particular, we found that several genes involved in SA metabolism were induced by the T genotype to T. absoluta and could be involved in the tolerance process [].
The cell wall represents the first barrier for pathogen perception and the activation of defensive responses against multiple biotic stressors immediately downstream of the cuticle layer. Hence, changes in cell wall composition and structure may lead to specific signaling cascades and pathogen responses []. An interesting subclass of arabinogalactan proteins (AGPs) involved in cellulose metabolism, named FLAs, was highly challenged in our samples. Such proteins can have multiple roles in plant signaling, growth, development and stress responses [,,] and may regulate lignin and cellulose synthesis/deposition in response to mechanical stimuli [].
In our study, FLAs were particularly induced by fungi R genotypes. For example, Solyc07g045440 (FLA2) was upregulated by fungi R and T. absoluta T, while Solyc01g091530 (FLA13) was activated by fungi R and TSWV R genotypes. A recent study reported that in Nicotiana benthamiana different FLAs were selectively repressed after the infection with turnip mosaic virus (TuMV) and Pseudomonas syringae pv tomato strain DC3000 (Pst DC3000) []. These genes were also identified as induced during M. incognita infections and downregulated during the combination of water stress and nematodes []. Other important cell wall components, such as hemicellulose and pectin, were affected by multiple biotic stress. Interestingly, the R genotype to bacteria showed the downregulation of a GDP-mannose-4,6-dehydratase (Solyc12g010540), involved in the pectin metabolism []. Moreover, R genotypes to fungi and bacteria showed the common activation of a UGE (Solyc02g030230) and a PG (Solyc08g060970) that could have a role in the resistance against pathogens and abiotic stress [,,]. Pathogens often induce enzymes implicated in pectin degradation, such as pectate lyases, to promote plant infections []. Here, we found that two pectate lyases were induced by the T. absoluta T genotype and repressed by the R genotype to bacteria. Hence, we speculate that their downregulation may be important for maintaining proper cell wall properties during bacterial infections. In our work, several pectate lyases were responsive to T. absoluta and TSWV. For instance, Solyc06g083580 and Solyc05g014000, induced by both T and R genotypes, have been found in other studies as involved in mites and potato P. infestans susceptibility [,]. It is worth noting that pectate lyases may have a key role in biotic stress response since their silencing can provide resistance against different pathogens [].
Receptors and signaling molecules play a crucial role in the capability of plants to respond adequately to specific stresses []. Our study found RLKs with divergent expression patterns during various stress. For example, Solyc06g048740 was induced by R genotypes to fungi and bacteria and repressed by R plants to TSWV. This gene was also upregulated in a tomato C. fulvum R genotype []. Moreover, Solyc07g006480 was exclusively activated by R plants to bacteria and repressed by fungi and TSWV R plants. This gene was also activated in salinity-tolerant plant roots []. Our results suggest that RLKs may play essential roles in fine-tuning the signaling process and their expression pattern can promote specific defense responses against different stressors. The Solyc04g014400, a Pseudomonas-responsive RLP gene [], also showed a common downregulation in all R genotypes, as well as Solyc11g056680 that was induced by potato cyst nematode (PCN) in tomato roots [], and could be involved in response to multiple pathogens.
3.2. Tomato Transcriptomic Reprogramming under Different Abiotic Stresses
This work analyzed tomato response to four different abiotic stress (drought, salinity, low temperature and oxidative stress). Adaptive plant responses to specific abiotic stresses are fine-tuned by a network of hormonal signaling cascades, including ABA, ET, JA, and SA [,]. In our work, the auxins pathway was mainly repressed during drought stress, accordingly to []. We also found that the R genotype to drought downregulated the Solyc12g096830, orthologs of AT1G05680 (UGT74E2) involved in auxin distribution and drought stress response []. By contrast, the susceptible genotypes showed the induction of Solyc11g010930, which encodes for a drought-responsive protein also involved in salinity stress response []. Furthermore, S genotypes to drought showed the repression of genes involved in ET biosyntheses such as Solyc07g049550 (ACCO) and Solyc08g079750 (ACC synthase). The crosstalk between ABA and ET synthesis is important for the proper growth in drought conditions and for the closure and opening of stomata cells under drought and salinity stress []. In addition, we found several genes involved in SA biosynthesis induced under salinity to mitigate the deleterious effect of the stress, as reported in []. Despite this, the excessive production of SA could decrease plant tolerance []. On the contrary, plants subjected to low temperatures downregulated genes for SA, which instead could enhance plant growth and production [,].
TFs are fundamental tools to coordinate the transduction of various stress signals in plants during abiotic stresses. In this work, we found the common upregulation of Solyc07g054220 (ERF2a) during salt, oxidative, and drought T responses. This gene is known to exhibit different expression patterns under various abiotic stress and could be crucial for governing multiple abiotic stress interactions []. Moreover, the drought T genotype showed the private upregulation of a NAC gene (Solyc01g009860) orthologs of AT5G13180 (ANAC083) involved in leaf senescence, ABA and drought stress responses, negatively regulating the xylem vessels formation [,].
Changes in Ca2+ concentration led to the activation of plant signaling. Interestingly, a calnexin (Solyc03g118040) was induced during salinity, low temperatures and oxidative stresses and exclusively repressed by the T genotype to drought. This gene is involved in calcium homeostasis by binding the Ca2+ and participates in plant adaption to unfavorable environmental conditions [,]. This study identified four GLRs (Solyc06g063180, Solyc07g052390, Solyc07g052400, and Solyc05g045650) reported as putative PM-Localized Proteins related to Ca2+ transport that may induce a specific spectrum of downstream responses for fine-tuning adaptive responses to abiotic stress []. Among these, Solyc07g052400 was also found localized in a QTL region for tolerance to water deficit in tomato [].
Salinity and oxidative stress induced the upregulation of four genes encoding for AXS, UGD, and GAE proteins that were downregulated during low temperatures. The SNF1 gene showed an opposite regulation during salinity and low temperatures and was identified as an important factor in response to drought and cold stress by interacting with a second gene (ShCIGT), promoting plant resistance against multiple abiotic stress []. Interestingly, we also found a cellulose synthase-like (CSL) gene (Solyc03g097050) that was DE during all the abiotic stress and showed an opposite regulation between the T and the S genotypes to drought. Cellulose synthase genes might enhance plant tolerance against drought and oxidative stress and are involved in ABA modulation during abiotic stress [,,]. A recent study suggested that this gene could also be related to tomato yellow leaf curl virus (TYLCV) infections [].
3.3. Biotic and Abiotic Stress Transcriptomic Profiles Comparison
In this study, we analyzed and compared the transcriptomic response of R and S tomato plants exposed to fungi, bacteria, TSWV and T. absoluta as biotic factors and drought, salinity, low temperatures and oxidative stress as abiotic factors. A total of sixty-seven shared DEGs among different stresses, involved in cell wall metabolism, signaling TFs, and hormone pathways were found.
The cell wall is a structure with sophisticated composition and organization, which interacts dynamically with sub-localized components. Perturbations in cell wall composition may induce different plant stress responses []. FLAs, a class of genes involved in cellulose deposition, were mainly activated by R plants to TSWV and during oxidative stress. By contrast, UGEs participating in pectin metabolism were induced by R genotypes to fungi and during low temperatures conditions. The R genotypes to fungi also activated GTs, which utilize nucleotide sugars as donor substrates to generate cell wall polysaccharides, and their implication in fungi resistance has been reported in different plant species [,].
A multitude of signaling events occurring downstream of the initial stress perception requires several RLKs, MAPKs and genes involved in calcium signaling. A list of signaling-responsive genes to both biotic and abiotic stress was obtained in our study. Among these, various RLKs (Solyc03g006100, Solyc11g017280 and Solyc02g091840) were found involved in response to Phytophthora infestans and Colletotrichum coccodes [] or indicated as potentially involved in drought and pathogens recognition [,,]. Interestingly, an RLK (Solyc03g078360) was activated by the R genotype to R. solanacerarum and resulted repressed in S plants inoculated with Xanthomonas perforans []. Fluctuations in calcium concentration can lead to phosphorylation events that promote plant stress response. Here, we found Solyc03g113390, a calcium-dependent protein kinase (CDPK), induced during abiotic stress and in response against different pathogens that may lead to specific defensive responses [,]. Moreover, the Verticillium wilt disease resistance Ve gene (Solyc09g005080) was found DE during different biotic stresses and in response to oxidative stress. It is worth noting that this gene was also found to be upregulated in a resistant line to TYLCV [].
A combination of biotic and abiotic stress can trigger specific plant hormonal pathways. In this work, thirteen genes encoding for phytohormones and responsive to both biotic and abiotic stress were pointed out. Auxin-related genes, such as Small Auxin Up RNAs (SAURs), were activated during abiotic stress and by R genotypes to fungi, and four of them were repressed by the R genotype to TSWV. The induction of auxin-related genes can promote plant defense against abiotic stress and fungal pathogens by interacting with other phytohormones []. We also noted that ET (Solyc09g089580 andSolyc01g095080) was mainly activated during abiotic stress and fungi R plants and repressed by the TSWV R genotype. Furthermore, we found two genes involved in JA signaling that were downregulated during salinity (Solyc01g109140) and low temperature (Solyc07g054580) and induced by the R genotypes to TSWV and fungi. These results confirm that JA acts antagonistically with ET and auxin and its upregulation under TSWV infections could be particularly important in promoting plant resistance []. Finally, ABA (Solyc07g056570) was activated during different biotic and abiotic stress, supporting its participation in multiple stress responses [,].
Transcriptomic changes promoting defense against multiple stress are mediated by different TFs classes. The MYBs family was the most responsive family in our work since it is involved in the crosstalk response among stresses []. Interestingly, Solyc06g083900 (SlMYB13) was induced during salinity and repressed during low temperatures. This gene promotes tolerance to 2,4,6-trichlorophenol [] and is activated in response to different pathogens [,]. Among TFs responsive to multiple stress, we also found a bZIP (NPR1; Solyc10g080770) involved in the regulation of PR gene expression []. It was induced by all the abiotic stress and by the R genotypes to fungi, while it was repressed by the R genotype to T. absoluta. The Solyc03g116890 (WRKY39) was involved in response to fungi, T. absoluta, TSWV, salinity and oxidative stresses. This gene is induced under heat and H2O2 stresses, and its overexpression induces R. solanacerarum resistance in cotton [,]. ERFs regulators control the expression of ET-dependent genes, which are well known to be involved in biotic and abiotic stress responses []. In this study, Solyc08g078180 (SlERFa1) was upregulated by R genotypes to fungi and bacteria and by oxidative stress, while it was downregulated in the TSWV R genotype. The Solyc08g078180, involved in cell death and signaling pathways against different pathogens, is induced by bacterial pathogens []. Moreover, Solyc11g072600, an AP2/ERF gene (APETALA2d), showed an opposite regulation during salinity and T. absoluta (upregulated) and low temperatures and R genotypes to bacteria (downregulated). Intriguingly, overexpression of a microRNA, conferring resistance against Phytophthora infestans infection, caused the downregulation of the Solyc11g072600 [].
4. Materials and Methods
4.1. Bibliographic Research, Studys Selection and Transcriptomic Dataset Downloadg
A large-scale literature search was performed to find publicly available tomato (Solanum lycopersicum) RNA-seq experiments. Dozens of papers evaluating tomato stress response were collected. However, a further screening was made using the following criteria: (i) tomato plants were subjected to biotic or abiotic stress; (ii) sequencing was performed with Illumina technology; (iii) at least three biological replicates for treatment were used (Supplementary Table S1) except for the T. absoluta [] and drought [] experiments. Information on tomato transcriptomic studies of tomatoes exposed to biotic stress were mainly retrieved by []. In addition, three studies of tomatoes exposed to four different abiotic stresses were selected. In total, selected datasets comprised seven biotic stresses and five abiotic stresses (Table 1). Biotic stressors challenging tomatoes were: Cladosporium fulvum infecting tomato at 7 and 20 dpi (days post-infection), Phytophthora infestans at 40 dpi, Pseudomonas syringae, and Ralstonia solanacearum at 1 and 2 dpi, Sclerotinia sclerotiorum at 30 dpi, Tomato spotted wilt virus (TSWV) at 4, 7, 14, 21 and 35 dpi, and Tuta absoluta at 40 dpi. Transcriptomic studies of plants treated with C. fulvum, P. infestans, R. solanacerarum, TSWV, T. absoluta, and drought allowed the exploration of resistant (R), tolerant (T), and susceptible (S) responses. Response to abiotic stress was assessed by analyzing studies of tomato plants treated with drought (two different studies), salt, low temperature and oxidative stress (Table 1).
SRA accession number was used to access and download the corresponding raw sequencing data (fastq file) from the NCBI repository (Sequence Read Archive) (https://www.ncbi.nlm.nih.gov/sra, accessed on 9 September 2020 [,,] using SRA Toolkit (2.10.8) (https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit, accessed on 9 September 2020). After raw data collection, samples were grouped for experiments and comparative conditions.
4.2. Data Processing and Differential Expression Analysis
Collected raw data, including both pair-and and single-end reads, were analyzed with a standard pipeline through the online bioinformatics software AIR (Sequentia Biotech, Barcelona, Spain) using the RNA-seq package (https://transcriptomics.sequentiabiotech.com/, accessed on 11 January 2021). The analysis and comparison of RNA samples were conducted through algorithms described by []. After a quality check, which included the adapter removal and the trimming of low-quality reads, the clean reads were mapped against the reference tomato genome (version SL3.0). The number of reads before and after the quality check, the mean GC content, and the sequence length are reported in Supplementary Materials (Supplementary Table S1). FeatureCounts was then used to obtain gene expression values (raw reads counts) []. The identification of differentially expressed genes (DEGs) was performed using the DESeq2 package []. Genes with a false discovery rate (FDR) < 0.05, using the Benjamini–Hochberg method, were considered differentially expressed (DE) and used for further analysis. DEGs with positive or negative logFC values were classified as upregulated (logFC > 0) or downregulated (logFC < 0), respectively. This workflow allowed us to compare and analyze different tomato studies starting from transcriptomic raw data.
4.3. DEGs Filtering Criteria
After AIR analysis, DEGs from biotic and abiotic experiments were downloaded and grouped. Then, comparisons among different RNA-seq studies were made by searching for common DEGs among datasets using the Microsoft Excel ®2013 (Ver. 15.0.5519) software.
4.4. Pathway Analysis
The differentially expressed genes were mapped to pathways using MapMan software (version 3.6.0, http://mapman.gabipd.org, accessed on 19 June 2022) []. In order to assign MapMan ontology to DEGs, we first downloaded the tomato protein annotation file in fasta format (ITAG3.0_proteins.fasta) from the Solgenomics database (https://solgenomics.net/ftp/tomato_genome/annotation/ITAG3.0_release/, accessed on 18 May 2022). Then, we used the Mercator tool (version 3.6) to create an ad hoc protein annotation file []. The Mercator file was then uploaded in MapMan version 3.6 and used as a mapping file together with the list of DEG considered for each comparison analyzed in this study.
5. Conclusions
In this study, a comparative analysis of twelve RNA-sequencing experiments of tomatoes exposed to biotic and abiotic stress was carried out to identify genes involved in defensive response against different stressors. Tomato response to biotic factors, such as C. fulvum, P. infestans, and S. sclerotiorum (fungi), P. syringae, and R. solanacearum (bacteria), Tomato spotted wilt virus (TSWV), and T. absoluta, was investigated. In addition, tomato challenged by abiotic stress (drought, salinity, low temperature, and oxidative stress) was also analyzed. This study allowed the identification of commonresponsive genes encoding for signaling proteins, cell wall precursors, TFs, and hormones, involved in response to biotic and abiotic stress. In particular, we analyzed in detail sixty-seven genes associated with the response to at least four different stresses. Among these, we found ten genes involved in the biosynthesis of cell wall compounds, thirty-six DEGs involved in signaling, thirteen DEGs participants in hormone biosynthesis, and eight DEGs encoding for TFs. Above all, we found different RLKs, MAPKs, Fasciclin-like arabinogalactans (FLAs), two glycosyltransferases, ten genes involved in the auxin., ET, and JA pathways, three MYBs, two bZIP, a WRKY, and an ERF gene. Our study provides a list of genes involved in response to multiple biotic and abiotic stress that could be tested in genetic engineering programs to improve tomato multiple-stress resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24044061/s1.
Author Contributions
C.G.A. was centrally involved in manuscript writing, transcriptomic analyses and results interpretation. D.D. was involved in data interpretation and manuscript revision. R.A.C. contributed to transcriptomic analyses. M.R.E. conceived the research idea, coordinated the project, and contributed to results interpretation and manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This study was carried out within the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022). This manuscript reflects only the authors’ views and opinions; neither the European Union nor the European Commission can be considered responsible for them.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available at Sequence Read Archive of NCBI repository (https://www.ncbi.nlm.nih.gov/sra, accessed on 9 September 2020) using the SRA codes reported in Table 1.
Acknowledgments
We thank authors of articles listed in Table 1 for making their data public available.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cappetta, E.; Andolfo, G.; Di Matteo, A.; Ercolano, M.R. Empowering Crop Resilience to Environmental Multiple Stress through the Modulation of Key Response Components. J. Plant Physiol. 2020, 246–247, 153134. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-Wide Identification and Characterization of the Brassinazole-Resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Routray, S.; Mohapatra, C.; Saha, D.; Ram, C.; Siddique, K.H.M.; Kumar, A.; et al. Genome-Wide Analysis and Characterization of the Proline-Rich Extensin-like Receptor Kinases (PERKs) Gene Family Reveals Their Role in Different Developmental Stages and Stress Conditions in Wheat (Triticum aestivum L.). Plants 2022, 11, 496. [Google Scholar] [CrossRef]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-Wide Identification of Wheat WRKY Gene Family Reveals That TaWRKY75-A Is Referred to Drought and Salt Resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef]
- Shao, W.; Chen, W.; Zhu, X.; Zhou, X.; Jin, Y.; Zhan, C.; Liu, G.; Liu, X.; Ma, D.; Qiao, Y. Genome-Wide Identification and Characterization of Wheat 14-3-3 Genes Unravels the Role of Tagrf6-a in Salt Stress Tolerance by Binding Myb Transcription Factor. Int. J. Mol. Sci. 2021, 22, 1904. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.-I. Plant Immunity in Signal Integration between Biotic and Abiotic Stress Responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Krishna, R.; Karkute, S.G.; Ansari, W.A.; Jaiswal, D.K.; Verma, J.P.; Singh, M. Transgenic Tomatoes for Abiotic Stress Tolerance: Status and Way Ahead. 3 Biotech 2019, 9, 143. [Google Scholar] [CrossRef]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.F.; van der Linden, G. Plant Behaviour under Combined Stress: Tomato Responses to Combined Salinity and Pathogen Stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) Are Potential Mediators of Auxin Action in Tomato Response to Biotic and Abiotic Stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic Leucine Zipper Transcription Factor SlbZIP1 Mediates Salt and Drought Stress Tolerance in Tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Li, J.; Ouyang, B.; Wang, T.; Luo, Z.; Yang, C.; Li, H.; Sima, W.; Zhang, J.; Ye, Z. HyPRP1 Gene Suppressed by Multiple Stresses Plays a Negative Role in Abiotic Stress Tolerance in Tomato. Front. Plant Sci. 2016, 7, 967. [Google Scholar] [CrossRef]
- Mackelprang, R.; Lemaux, P.G. Genetic Engineering and Editing of Plants: An Analysis of New and Persisting Questions. Annu. Rev. Plant Biol. 2020, 71, 659–687. [Google Scholar] [CrossRef]
- Andolfo, G.; Villano, C.; Errico, A.; Frusciante, L.; Carputo, D.; Aversano, R.; Ercolano, M.R. Inferring RPW8-NLRs’s Evolution Patterns in Seed Plants: Case Study in Vitis Vinifera. Planta 2020, 251, 32. [Google Scholar] [CrossRef]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of Differential Gene Expression Results of RNA-Seq Data: Review and Integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef]
- Ashrafi-Dehkordi, E.; Alemzadeh, A.; Tanaka, N.; Razi, H. Meta-Analysis of Transcriptomic Responses to Biotic and Abiotic Stress in Tomato. PeerJ 2018, 6, e4631. [Google Scholar] [CrossRef]
- Finotello, F.; Di Camillo, B. Measuring Differential Gene Expression with RNA-Seq: Challenges and Strategies for Data Analysis. Brief. Funct. Genom. 2015, 14, 130–142. [Google Scholar] [CrossRef]
- Rasheed, M. Next Generation Sequencing as an Emerging Technology in Rare Disease Genetics. J. Islam. Med. Dent. Coll. 2020, 9, 1–3. [Google Scholar] [CrossRef]
- Calle García, J.; Guadagno, A.; Paytuvi-Gallart, A.; Saera-Vila, A.; Amoroso, C.G.; D’Esposito, D.; Andolfo, G.; Aiese Cigliano, R.; Sanseverino, W.; Ercolano, M.R.; et al. PRGdb 4.0: An Updated Database Dedicated to Genes Involved in Plant Disease Resistance Process. Nucleic Acids Res. 2022, 50, D1483–D1490. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Zhao, T.; Ottosen, C.O.; Rosenqvist, E.; Wu, Z. Physiological Analysis and Transcriptome Sequencing Reveal the Effects of Combined Cold and Drought on Tomato Leaf. BMC Plant Biol. 2019, 19, 377. [Google Scholar] [CrossRef]
- Keshishian, E.A.; Hallmark, H.T.; Ramaraj, T.; Plačková, L.; Sundararajan, A.; Schilkey, F.; Novák, O.; Rashotte, A.M. Salt and Oxidative Stresses Uniquely Regulate Tomato Cytokinin Levels and Transcriptomic Response. Plant Direct 2018, 2, e00071. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, H.; Zhao, G.; Huang, Q.; Lu, Y.; Ouyang, B. Profiling of Drought-Responsive MicroRNA and MRNA in Tomato Using High-Throughput Sequencing. BMC Genom. 2017, 18, 481. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Liu, W.; Zhao, Z.; Yang, H.; Bao, Y.; Zhang, D.; Wang, Z.; Jiang, J.; Xu, Y.; Zhang, H.; et al. Transcriptome Profiling Reveals the Response Process of Tomato Carrying Cf-19 and Cladosporium fulvum Interaction. BMC Plant Biol. 2019, 19, 572. [Google Scholar] [CrossRef] [PubMed]
- Canto-Pastor, A.; Santos, B.A.M.C.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced Resistance to Bacterial and Oomycete Pathogens by Short Tandem Target Mimic RNAs in Tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Wang, M.M.; Yin, Y.L.; Onac, E.; Zhou, G.F.; Peng, S.; Xia, X.J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. RNA-Seq Analysis Reveals the Role of Red Light in Resistance against Pseudomonas syringae Pv. Tomato DC3000 in Tomato Plants. BMC Genom. 2015, 16, 120. [Google Scholar] [CrossRef]
- French, E.; Kim, B.S.; Rivera-Zuluaga, K.; Iyer-Pascuzzi, A.S. Whole Root Transcriptomic Analysis Suggests a Role for Auxin Pathways in Resistance to Ralstonia solanacearum in Tomato. Mol. Plant-Microbe Interact. 2018, 31, 432–444. [Google Scholar] [CrossRef]
- Badet, T.; Voisin, D.; Mbengue, M.; Barascud, M.; Sucher, J.; Sadon, P.; Balagué, C.; Roby, D.; Raffaele, S. Parallel Evolution of the POQR Prolyl Oligo Peptidase Gene Conferring Plant Quantitative Disease Resistance. PLoS Genet. 2017, 13, e1007143. [Google Scholar] [CrossRef]
- Padmanabhan, C.; Ma, Q.; Shekasteband, R.; Stewart, K.S.; Hutton, S.F.; Scott, J.W.; Fei, Z.; Ling, K.S. Comprehensive Transcriptome Analysis and Functional Characterization of PR-5 for Its Involvement in Tomato Sw-7 Resistance to Tomato Spotted Wilt Tospovirus. Sci. Rep. 2019, 9, 7673. [Google Scholar] [CrossRef]
- D’Esposito, D.; Manzo, D.; Ricciardi, A.; Garonna, A.P.; De Natale, A.; Frusciante, L.; Pennacchio, F.; Ercolano, M.R. Tomato Transcriptomic Response to Tuta absoluta Infestation. BMC Plant Biol. 2021, 21, 358. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant Cell Wall-Mediated Immunity: Cell Wall Changes Trigger Disease Resistance Responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor Kinases in Plant-Pathogen Interactions: More than Pattern Recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.J.; Van Den Abeele, C.; Ortega-Salazar, I.; Papin, V.; Adaskaveg, J.A.; Wang, D.; Casteel, C.L.; Seymour, G.B.; Blanco-Ulate, B. Host Susceptibility Factors Render Ripe Tomato Fruit Vulnerable to Fungal Disease despite Active Immune Responses. J. Exp. Bot. 2021, 72, 2696–2709. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.; Rajendran, S.R.C.K.; Gaur, M.; Sajeesh, P.K.; Kumar, A. Plant Defense Signaling and Responses Against Necrotrophic Fungal Pathogens. J. Plant Growth Regul. 2016, 35, 1159–1174. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Scalschi, L.; Llorens, E.; García-Agustín, P.; Vicedo, B. Role of Jasmonic Acid Pathway in Tomato Plant-Pseudomonas syringae Interaction. Plants 2020, 9, 136. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a Bacterial Speck Resistant Tomato by CRISPR/Cas9-Mediated Editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of Plant Defense against Insect Herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Pulga, P.S.; Henschel, J.M.; de Resende, J.T.V.; Zeist, A.R.; Moreira, A.F.P.; Gabriel, A.; Silva, M.B.; Gonçalves, L.S.A. Salicylic Acid Treatments Induce Resistance to Tuta absoluta and Tetranychus urticae on Tomato Plants. Hortic. Bras. 2020, 38, 288–294. [Google Scholar] [CrossRef]
- Hamann, T. Plant Cell Wall Integrity Maintenance as an Essential Component of Biotic Stress Response Mechanisms. Front. Plant Sci. 2012, 3, 77. [Google Scholar] [CrossRef]
- Liu, E.; MacMillan, C.P.; Shafee, T.; Ma, Y.; Ratcliffe, J.; van de Meene, A.; Bacic, A.; Humphries, J.; Johnson, K.L. Fasciclin-Like Arabinogalactan-Protein 16 (FLA16) Is Required for Stem Development in Arabidopsis. Front. Plant Sci. 2020, 11, 615392. [Google Scholar] [CrossRef]
- Meng, J.; Hu, B.; Yi, G.; Li, X.; Chen, H.; Wang, Y.; Yuan, W.; Xing, Y.; Sheng, Q.; Su, Z.; et al. Genome-Wide Analyses of Banana Fasciclin-like AGP Genes and Their Differential Expression under Low-Temperature Stress in Chilling Sensitive and Tolerant Cultivars. Plant Cell Rep. 2020, 39, 693–708. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, H.; Cheng, Z.; Ke, Y.; Liu, J.; Ma, H. Evolution Analysis of the Fasciclin-like Arabinogalactan Proteins in Plants Shows Variable Fasciclin-AGP Domain Constitutions. Int. J. Mol. Sci. 2019, 20, 1945. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; MacMillan, C.P.; de Vries, L.; Mansfield, S.D.; Hao, P.; Ratcliffe, J.; Bacic, A.; Johnson, K.L. FLA11 and FLA12 Glycoproteins Fine-Tune Stem Secondary Wall Properties in Response to Mechanical Stresses. New Phytol. 2022, 233, 1750–1767. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, X.; Lai, Y.; Lv, L.; Ji, M.; Ji, M.; Han, K.; Yan, D.; Lu, Y.; Peng, J.; et al. Fasciclin-like Arabinogalactan Gene Family in Nicotiana benthamiana: Genome-Wide Identification, Classification and Expression in Response to Pathogens. BMC Plant Biol. 2020, 20, 305. [Google Scholar] [CrossRef] [PubMed]
- Veronico, P.; Rosso, L.C.; Melillo, M.T.; Fanelli, E.; De Luca, F.; Ciancio, A.; Colagiero, M.; Pentimone, I. Water Stress Differentially Modulates the Expression of Tomato Cell Wall Metabolism-Related Genes in Meloidogyne incognita Feeding Sites. Front. Plant Sci. 2022, 13, 817185. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, R.J.L.; Vílchez, J.I.; Zhang, S.; Kaushal, R.; He, D.; Zi, H.; Liu, R.; Niehaus, K.; Handa, A.K.; Zhang, H. Plant Transcriptome Reprograming and Bacterial Extracellular Metabolites Underlying Tomato Drought Resistance Triggered by a Beneficial Soil Bacteria. Metabolites 2021, 11, 369. [Google Scholar] [CrossRef]
- Hou, J.; Tian, S.; Yang, L.; Zhang, Z.; Liu, Y. A Systematic Review of the Uridine Diphosphate-Galactose/Glucose-4-Epimerase (UGE) in Plants. Plant Growth Regul. 2021, 93, 267–278. [Google Scholar] [CrossRef]
- Jung, Y.J.; Kyoung, J.H.; Nou, I.S.; Cho, Y.G.; Kang, K.K. Molecular Characterization of the UDP-Glucose 4-Epimerase (BrUGE) Gene Family in Response to Biotic and Abiotic Stress in Chinese Cabbage (Brassica rapa). Plant Biotechnol. Rep. 2015, 9, 339–350. [Google Scholar] [CrossRef]
- Rosli, H.G.; Zheng, Y.; Pombo, M.A.; Zhong, S.; Bombarely, A.; Fei, Z.; Collmer, A.; Martin, G.B. Transcriptomics-Based Screen for Genes Induced by Flagellin and Repressed by Pathogen Effectors Identifies a Cell Wall-Associated Kinase Involved in Plant Immunity. Genome Biol. 2013, 14, R139. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.; Lin, B.; Liao, J.; Zhuo, K. A Meloidogyne graminicola Pectate Lyase Is Involved in Virulence and Activation of Host Defense Responses. Front. Plant Sci. 2021, 12, 651627. [Google Scholar] [CrossRef]
- Schimmel, B.C.J.; Alba, J.M.; Wybouw, N.; Glas, J.J.; Meijer, T.T.; Schuurink, R.C.; Kant, M.R. Distinct Signatures of Host Defense Suppression by Plant-Feeding Mites. Int. J. Mol. Sci. 2018, 19, 3265. [Google Scholar] [CrossRef]
- Sun, K.; Wolters, A.M.A.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.H.M.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Silencing of Six Susceptibility Genes Results in Potato Late Blight Resistance. Transgenic Res. 2016, 25, 731–742. [Google Scholar] [CrossRef]
- Schwartz, A.R.; Morbitzer, R.; Lahaye, T.; Staskawicz, B.J. TALE-Induced BHLH Transcription Factors That Activate a Pectate Lyase Contribute to Water Soaking in Bacterial Spot of Tomato. Proc. Natl. Acad. Sci. USA 2017, 114, E897–E903. [Google Scholar] [CrossRef]
- Jiang, X.; Li, Y.; Li, R.; Gao, Y.; Liu, Z.; Yang, H.; Li, J.; Jiang, J.; Zhao, T.; Xu, X. Transcriptome Analysis of the Cf-13-Mediated Hypersensitive Response of Tomato to Cladosporium fulvum Infection. Int. J. Mol. Sci. 2022, 23, 4844. [Google Scholar] [CrossRef]
- Albaladejo, I.; Egea, I.; Morales, B.; Flores, F.B.; Capel, C.; Lozano, R.; Bolarin, M.C. Identification of Key Genes Involved in the Phenotypic Alterations of Res (Restored Cell Structure by Salinity) Tomato Mutant and Its Recovery Induced by Salt Stress through Transcriptomic Analysis. BMC Plant Biol. 2018, 18, 213. [Google Scholar] [CrossRef]
- Chattopadhyay, T.; Maurya, D.; Shree, B.; Akhtar, S.; Roy, C. Development of polymerase chain reaction-based marker for identification of a nonsense mutation in a Pseudomonas-responsive receptor like protein gene of tomato. J. Hort. Sci. Biol. 2022, 97, 113–121. [Google Scholar] [CrossRef]
- Święcicka, M.; Skowron, W.; Cieszyński, P.; Dąbrowska-Bronk, J.; Matuszkiewicz, M.; Filipecki, M.; Koter, M.D. The Suppression of Tomato Defence Response Genes upon Potato Cyst Nematode Infection Indicates a Key Regulatory Role of MiRNAs. Plant Physiol. Biochem. 2017, 113, 51–55. [Google Scholar] [CrossRef]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-Wide Identification, Phylogeny and Expression Analysis of the SPL Gene Family in Wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef]
- Nguyen, D.; Rieu, I.; Mariani, C.; van Dam, N.M. How Plants Handle Multiple Stresses: Hormonal Interactions Underlying Responses to Abiotic Stress and Insect Herbivory. Plant Mol. Biol. 2016, 91, 727–740. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-Wide Analysis of SAUR Gene Family in Solanaceae Species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef]
- Tognetti, V.B.; van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; de Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Jithesh, M.N.; Shukla, P.S.; Kant, P.; Joshi, J.; Critchley, A.T.; Prithiviraj, B. Physiological and Transcriptomics Analyses Reveal That Ascophyllum nodosum Extracts Induce Salinity Tolerance in Arabidopsis by Regulating the Expression of Stress Responsive Genes. J. Plant Growth Regul. 2019, 38, 463–478. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Martínez-Pérez, A.; Albacete, A.; Martínez-Melgarejo, P.A.; Dodd, I.C.; Thompson, A.J.; Mohareb, F.; Estelles-Lopez, L.; Kevei, Z.; Ferrández-Ayela, A.; et al. Overproduction of ABA in Rootstocks Alleviates Salinity Stress in Tomato Shoots. Plant Cell Environ. 2021, 44, 2966–2986. [Google Scholar] [CrossRef]
- Naeem, M.; Basit, A.; Ahmad, I.; Mohamed, H.I.; Wasila, H. Effect of Salicylic Acid and Salinity Stress on the Performance of Tomato Plants. Gesunde Pflanz. 2020, 72, 393–402. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of Water, Salinity, and Cold Stress Responses by Salicylic Acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef]
- Ahmad, I.; Maqsood, S.; Basra, A.; Wahid, A. Exogenous Application of Ascorbic Acid, Salicylic Acid and Hydrogen Peroxide Improves the Productivity of Hybrid Maize at Low Temperature Stress. Int. J. Agric. Biol. 2014, 16, 825–830. [Google Scholar]
- Klay, I.; Gouia, S.; Liu, M.; Mila, I.; Khoudi, H.; Bernadac, A.; Bouzayen, M.; Pirrello, J. Ethylene Response Factors (ERF) Are Differentially Regulated by Different Abiotic Stress Types in Tomato Plants. Plant Sci. 2018, 274, 137–145. [Google Scholar] [CrossRef]
- Wu, R.; Wang, T.; Richardson, A.C.; Allan, A.C.; Macknight, R.C.; Varkonyi-Gasic, E. Histone Modification and Activation by SOC1-like and Drought Stress-Related Transcription Factors May Regulate AcSVP2 Expression during Kiwifruit Winter Dormancy. Plant Sci. 2019, 281, 242–250. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.; Zhang, D.; Li, Q.; Zhao, X.; Chen, S. Genome-Wide Analysis of NAC Gene Family in Betula pendula. Forests 2019, 10, 741. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R. Heterologous Expression of Rice Calnexin (OsCNX) Confers Drought Tolerance in Nicotiana tabacum. Mol. Biol. Rep. 2013, 40, 5451–5464. [Google Scholar] [CrossRef]
- Ahsan, N.; Donnart, T.; Nouri, M.Z.; Komatsu, S. Tissue-Specific Defense and Thermo-Adaptive Mechanisms of Soybean Seedlings under Heat Stress Revealed by Proteomic Approach. J. Proteome Res. 2010, 9, 4189–4204. [Google Scholar] [CrossRef]
- Lee, H.J.; Seo, P.J. Ca2+talyzing Initial Responses to Environmental Stresses. Trends Plant Sci. 2021, 26, 849–870. [Google Scholar] [CrossRef] [PubMed]
- Asins, M.J.; Albacete, A.; Martínez-Andújar, C.; Celiktopuz, E.; Solmaz, İ.; Sarı, N.; Pérez-Alfocea, F.; Dodd, I.C.; Carbonell, E.A.; Topcu, S. Genetic Analysis of Root-to-Shoot Signaling and Rootstock-Mediated Tolerance to Water Deficit in Tomato. Genes 2021, 12, 10. [Google Scholar] [CrossRef]
- Yu, C.; Song, L.; Song, J.; Ouyang, B.; Guo, L.; Shang, L.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. ShCIGT, a Trihelix Family Gene, Mediates Cold and Drought Tolerance by Interacting with SnRK1 in Tomato. Plant Sci. 2018, 270, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hong, X.; Zhang, H.; Wang, Y.; Li, X.; Zhu, J.K.; Gong, Z. Disruption of the Cellulose Synthase Gene, AtCesA8/IRX1, Enhances Drought and Osmotic Stress Tolerance in Arabidopsis. Plant J. 2005, 43, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of Cellulose Synthesis in Response to Stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L.; et al. Cellulose Synthase-like Protein OsCSLD4 Plays an Important Role in the Response of Rice to Salt Stress by Mediating Abscisic Acid Biosynthesis to Regulate Osmotic Stress Tolerance. Plant Biotechnol. J. 2022, 20, 468–484. [Google Scholar] [CrossRef]
- Choe, S.; Choi, B.; Kang, J.H.; Seo, J.K. Tolerance to Tomato Yellow Leaf Curl Virus in Transgenic Tomato Overexpressing a Cellulose Synthase-like Gene. Plant Biotechnol. J. 2021, 19, 657–659. [Google Scholar] [CrossRef]
- Rui, Y.; Dinneny, J.R. A Wall with Integrity: Surveillance and Maintenance of the Plant Cell Wall under Stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef]
- Pasquet, J.C.; Changenet, V.; Macadré, C.; Boex-Fontvieille, E.; Soulhat, C.; Bouchabké-Coussa, O.; Dalmais, M.; Atanasova-Pénichon, V.; Bendahmane, A.; Saindrenan, P.; et al. A Brachypodium UDP-Glycosyltransferase Confers Root Tolerance to Deoxynivalenol and Resistance to Fusarium Infection. Plant Physiol. 2016, 172, 559–574. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Zuk, M.; Kulma, A.; Czemplik, M.; Kostyn, K.; Skala, J.; Starzycki, M.; Szopa, J. Engineering Flax with the GT Family 1 Solanum sogarandinum Glycosyltransferase SsGT1 Confers Increased Resistance to Fusarium Infection. J. Agric. Food Chem. 2009, 57, 6698–6705. [Google Scholar] [CrossRef] [PubMed]
- Saidi, A.; Hajibarat, Z.; Hajibarat, Z. Transcriptome Analysis of Phytophthora infestans and Colletotrichum coccodes in Tomato to Reveal Resistance Mechanisms. Asia-Pacific J. Mol. Biol. Biotechnol. 2020, 28, 39–51. [Google Scholar] [CrossRef]
- Milc, J.; Bagnaresi, P.; Aragona, M.; Valente, M.T.; Biselli, C.; Infantino, A.; Francia, E.; Pecchioni, N. Comparative Transcriptome Profiling of the Response to Pyrenochaeta lycopersici in Resistant Tomato Cultivar Mogeor and Its Background Genotype—Susceptible Moneymaker. Funct. Integr. Genom. 2019, 19, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Rupp, O.; Fahrentrapp, J. Pathogen Recognition in Compatible Plant-Microbe Interactions. Sci. Rep. 2017, 7, 6383. [Google Scholar] [CrossRef]
- Shi, R.; Panthee, D.R. Transcriptome-Based Analysis of Tomato Genotypes Resistant to Bacterial Spot (Xanthomonas perforans) Race T4. Int. J. Mol. Sci. 2020, 21, 4070. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, T.; Jiang, J.; Chen, X.; Zhang, H.; Liu, G.; Zhang, D.; Du, C.; Wang, S.; Xu, X.; et al. Transcriptome Analysis of the Sm-Mediated Hypersensitive Response to Stemphylium lycopersici in Tomato. Front. Plant Sci. 2017, 8, 1257. [Google Scholar] [CrossRef]
- Xue, D.Q.; Chen, X.L.; Zhang, H.; Chai, X.F.; Jiang, J.B.; Xu, X.Y.; Li, J.F. Transcriptome Analysis of the Cf-12-Mediated Resistance Response to Cladosporium fulvum in Tomato. Front. Plant Sci. 2017, 7, 2012. [Google Scholar] [CrossRef]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative Transcriptome Profiling of a Resistant vs. Susceptible Tomato (Solanum lycopersicum) Cultivar in Response to Infection by Tomato Yellow Leaf Curl Virus. PLoS ONE 2013, 8, e80816. [Google Scholar] [CrossRef]
- Tiwari, P.; Indoliya, Y.; Chauhan, A.S.; Singh, P.; Singh, P.K.; Singh, P.C.; Srivastava, S.; Pande, V.; Chakrabarty, D. Auxin-Salicylic Acid Crosstalk Ameliorates OsMYB–R1 Mediated Defense towards Heavy Metal, Drought and Fungal Stress. J. Hazard. Mater. 2020, 399, 122811. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, Z.; Li, S.; Zhou, X.; Li, J.; Su, X.; Zhang, L.; Zhang, Z.; Dong, J. Diterpenoid Compounds from Wedelia trilobata Induce Resistance to Tomato spotted wilt virus via the JA Signal Pathway in Tobacco Plants. Sci. Rep. 2019, 9, 2763. [Google Scholar] [CrossRef]
- Balyan, S.; Rao, S.; Jha, S.; Bansal, C.; Das, J.R.; Mathur, S. Characterization of Novel Regulators for Heat Stress Tolerance in Tomato from Indian Sub-Continent. Plant Biotechnol. J. 2020, 18, 2118–2132. [Google Scholar] [CrossRef]
- Landi, S.; Nurcato, R.; De Lillo, A.; Lentini, M.; Grillo, S.; Esposito, S. Glucose-6-Phosphate Dehydrogenase Plays a Central Role in the Response of Tomato (Solanum lycopersicum) Plants to Short and Long-Term Drought. Plant Physiol. Biochem. 2016, 105, 79–89. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Li, Z.; Peng, R.; Yao, Q. SlMYB14 Promotes Flavonoids Accumulation and Confers Higher Tolerance to 2,4,6-Trichlorophenol in Tomato. Plant Sci. 2021, 303, 110796. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Zhou, X.; Hou, X.; Yang, G.; Meng, J.; Luan, Y. Tomato MYB49 Enhances Resistance to Phytophthora infestans and Tolerance to Water Deficit and Salt Stress. Planta 2018, 248, 1487–1503. [Google Scholar] [CrossRef]
- Wong, G.R.; Fatihah Binti Abd Latif, S.N.; Mazumdar, P. Genome-Wide Investigation and Comparative Expression Profiling Reveal R2R3-MYB Genes Involved in Sclerotinia sclerotiorum Defence in Tomato. Physiol. Mol. Plant Pathol. 2022, 121, 101873. [Google Scholar] [CrossRef]
- Dong, X. NPR1, All Things Considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef]
- Shi, W.; Hao, L.; Li, J.; Liu, D.; Guo, X.; Li, H. The Gossypium hirsutum WRKY Gene GhWRKY39-1 Promotes Pathogen Infection Defense Responses and Mediates Salt Stress Tolerance in Transgenic Nicotiana benthamiana. Plant Cell Rep. 2014, 33, 483–498. [Google Scholar] [CrossRef]
- Roylawar, P.; Panda, S.; Kamble, A. Comparative Analysis of BABA and Piriformospora indica Mediated Priming of Defence-Related Genes in Tomato against Early Blight. Physiol. Mol. Plant Pathol. 2015, 91, 88–95. [Google Scholar] [CrossRef]
- Panda, S.; Busatto, N.; Hussain, K.; Kamble, A. Piriformospora indica-Primed Transcriptional Reprogramming Induces Defense Response against Early Blight in Tomato. Sci. Hortic. (Amst.) 2019, 255, 209–219. [Google Scholar] [CrossRef]
- Liu, A.C.; Cheng, C.P. Pathogen-Induced ERF68 Regulates Hypersensitive Cell Death in Tomato. Mol. Plant Pathol. 2017, 18, 1062–1074. [Google Scholar] [CrossRef]
- Luan, Y.; Cui, J.; Li, J.; Jiang, N.; Liu, P.; Meng, J. Effective Enhancement of Resistance to Phytophthora infestans by Overexpression of MiR172a and b in Solanum lycopersicum. Planta 2018, 247, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. The Sequence Read Archive: A decade more of explosive growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef]
- Kodama, Y.; Shumway, M.; Leinonen, R. International Nucleotide Sequence Database CollaborationThe Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Vara, C.; Paytuví-Gallart, A.; Cuartero, Y.; Le Dily, F.; Garcia, F.; Salvà-Castro, J.; Gómez-H, L.; Julià, E.; Moutinho, C.; Aiese Cigliano, R.; et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermatogenesis. Cell Rep. 2019, 28, 352–367.e9. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A Fast and Simple Web Server for Genome Scale Functional Annotation of Plant Sequence Data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).