Genotoxicity of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides in Normal and Cancer Cells In Vitro
Abstract
1. Introduction
2. Results
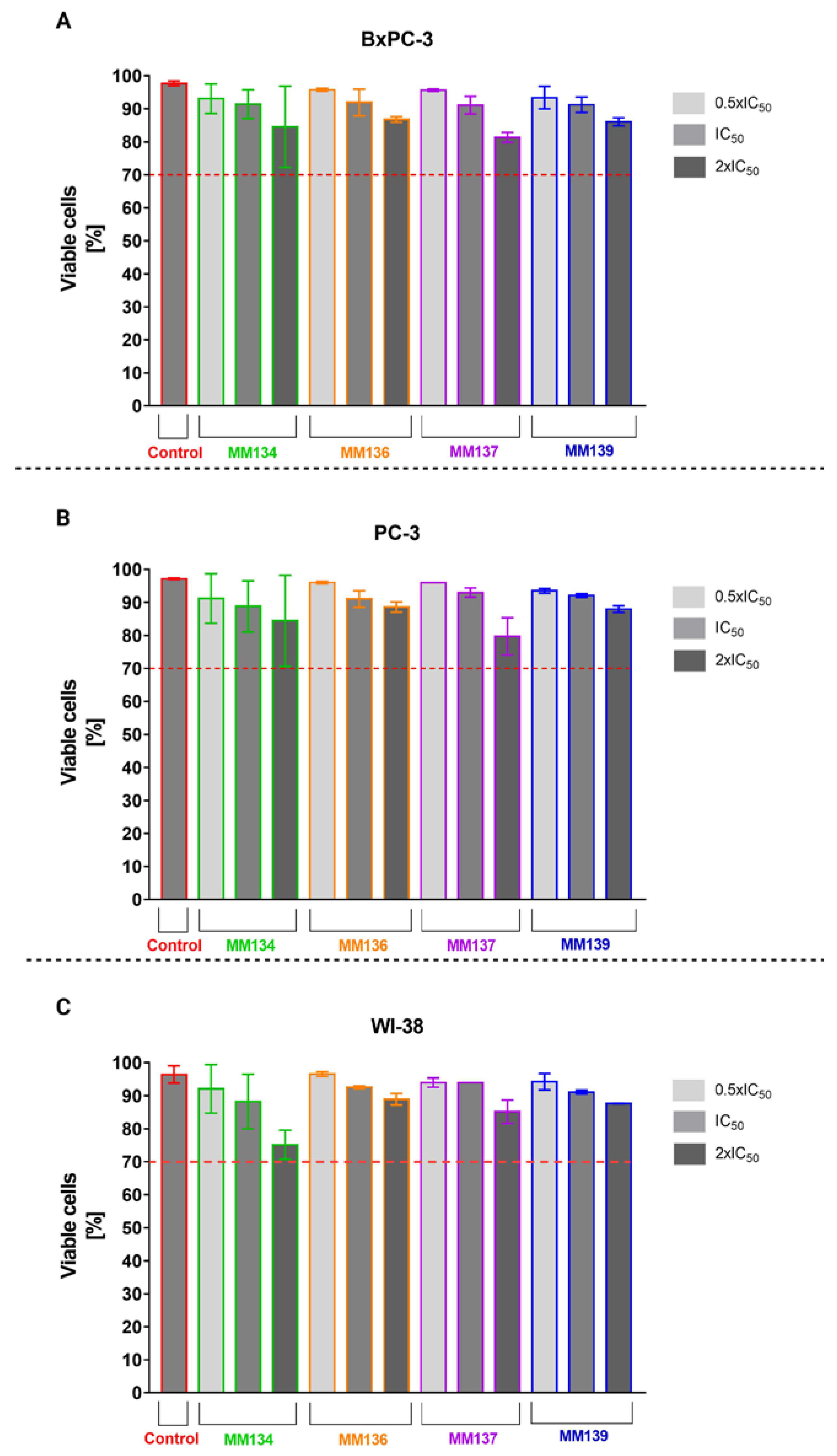
2.1. Trypan Blue Staining
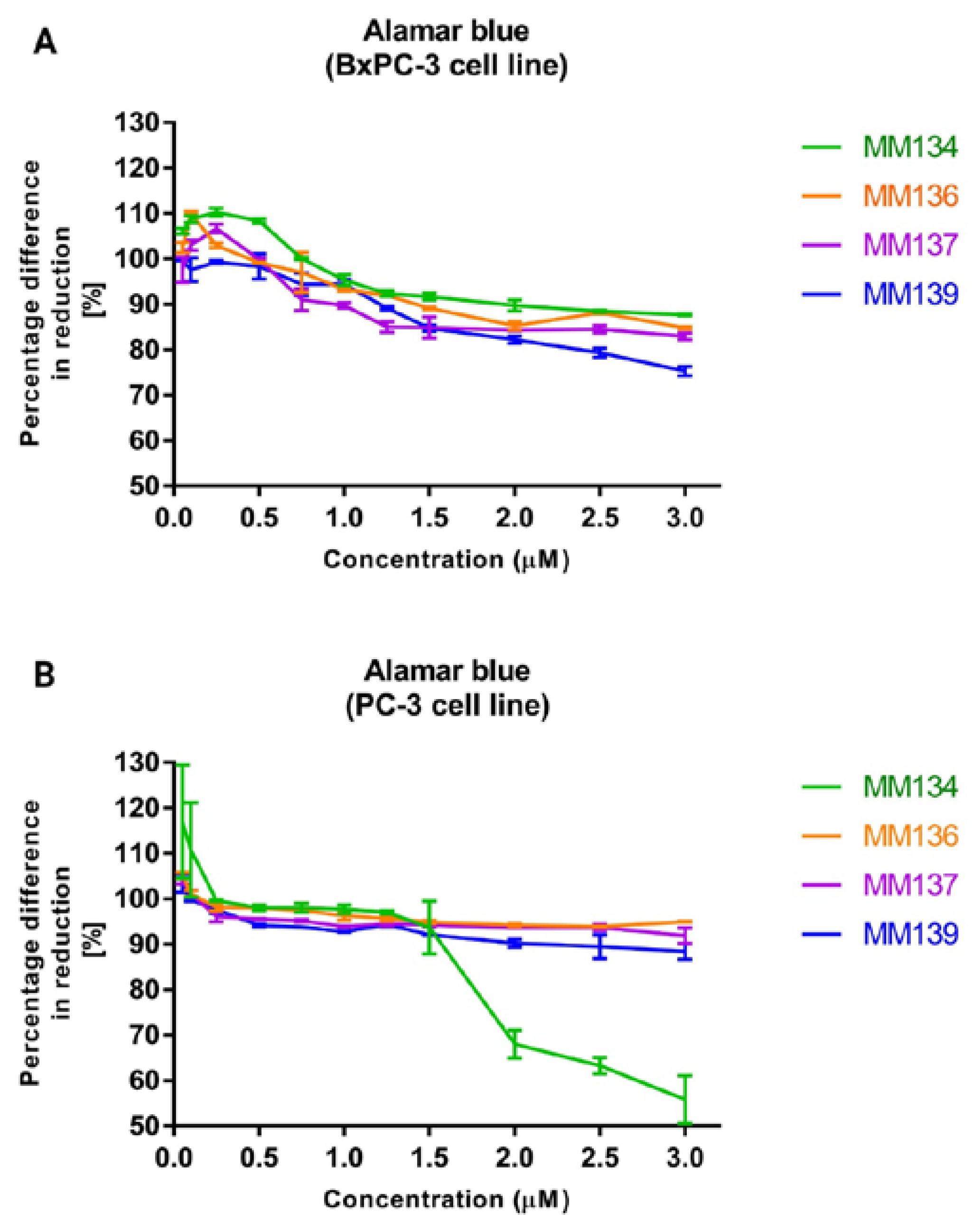
2.2. Alamar Blue
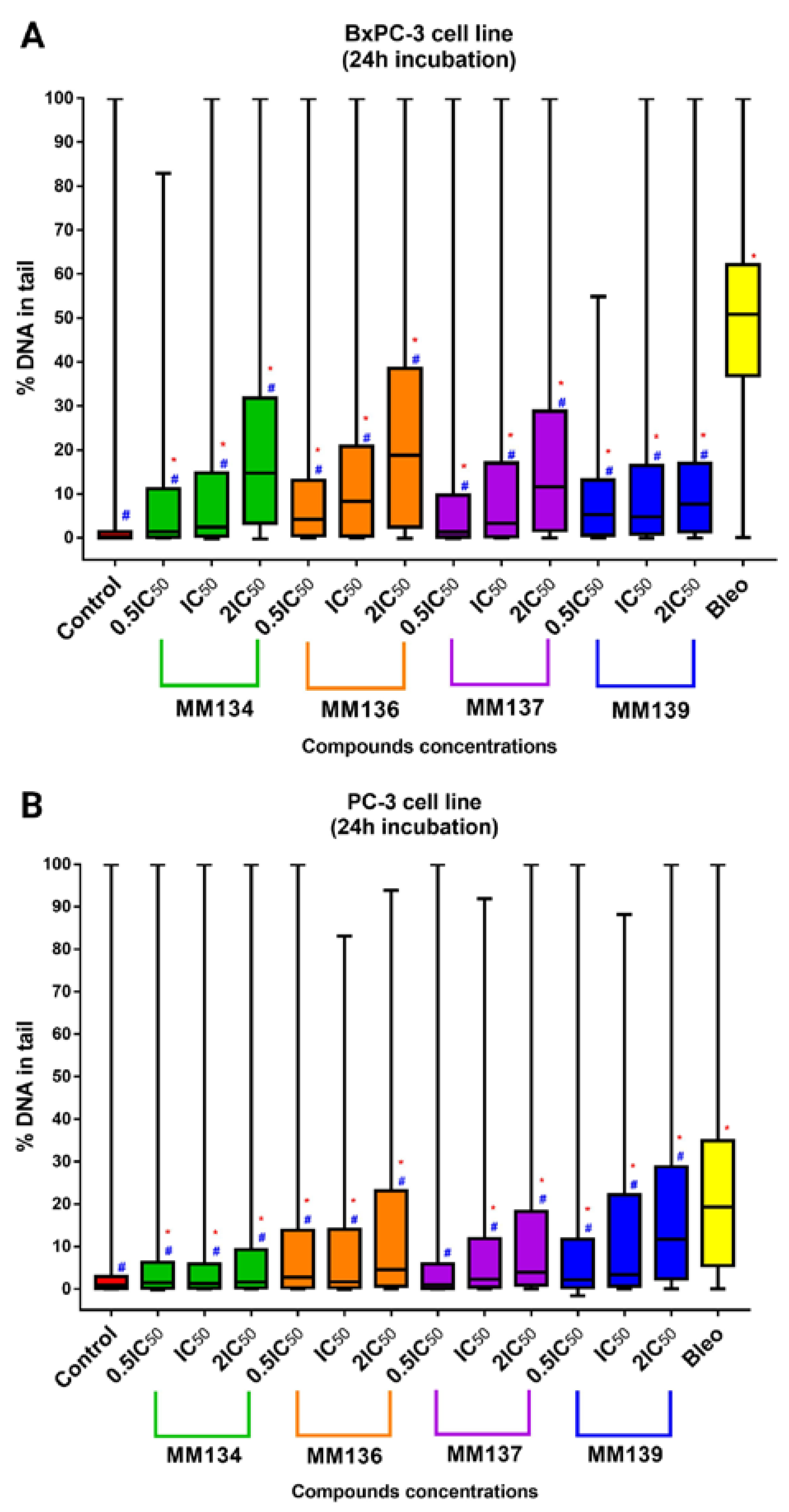
2.3. Alkaline Comet Assay
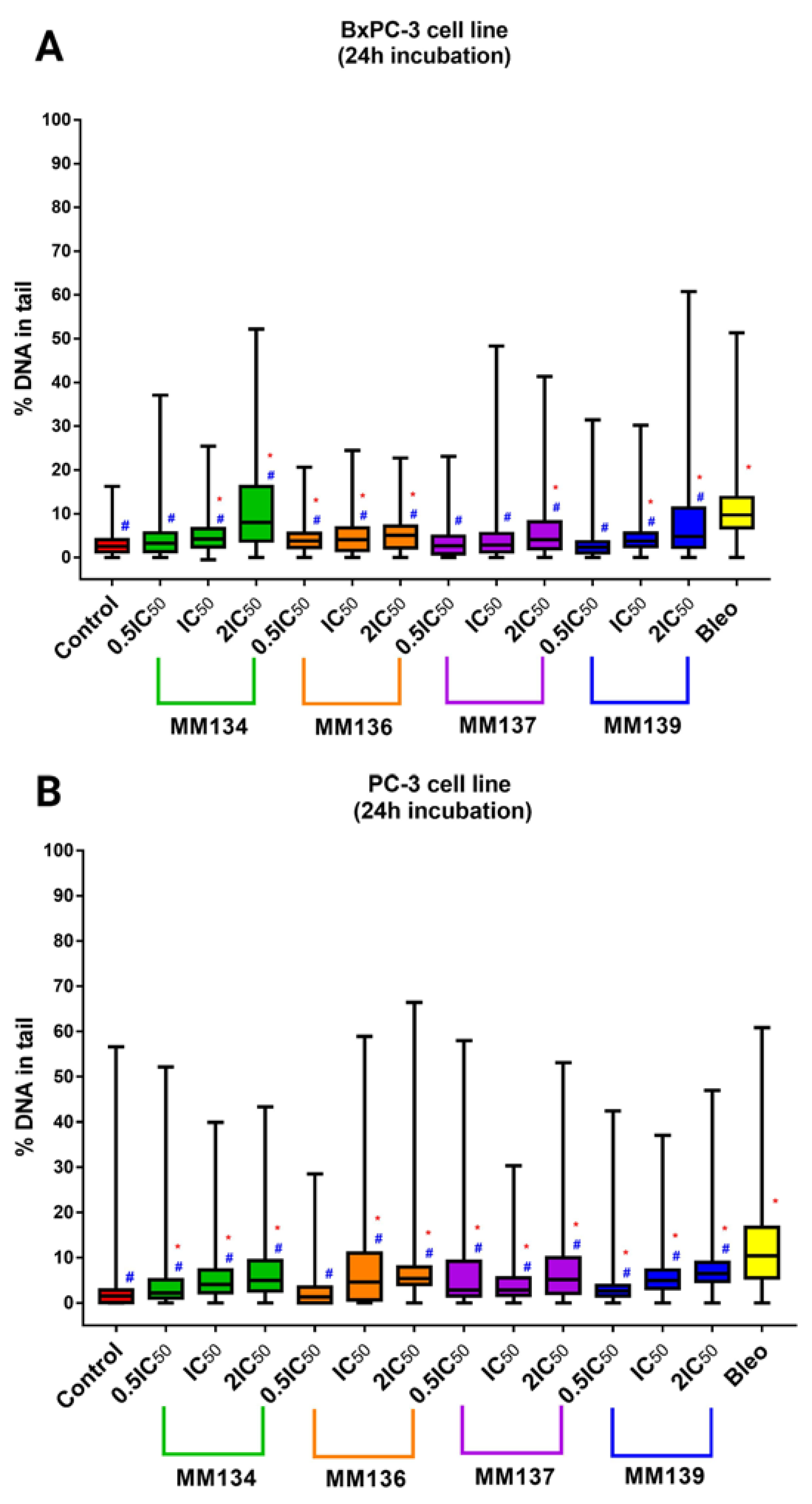
2.3.1. Cancer Cells
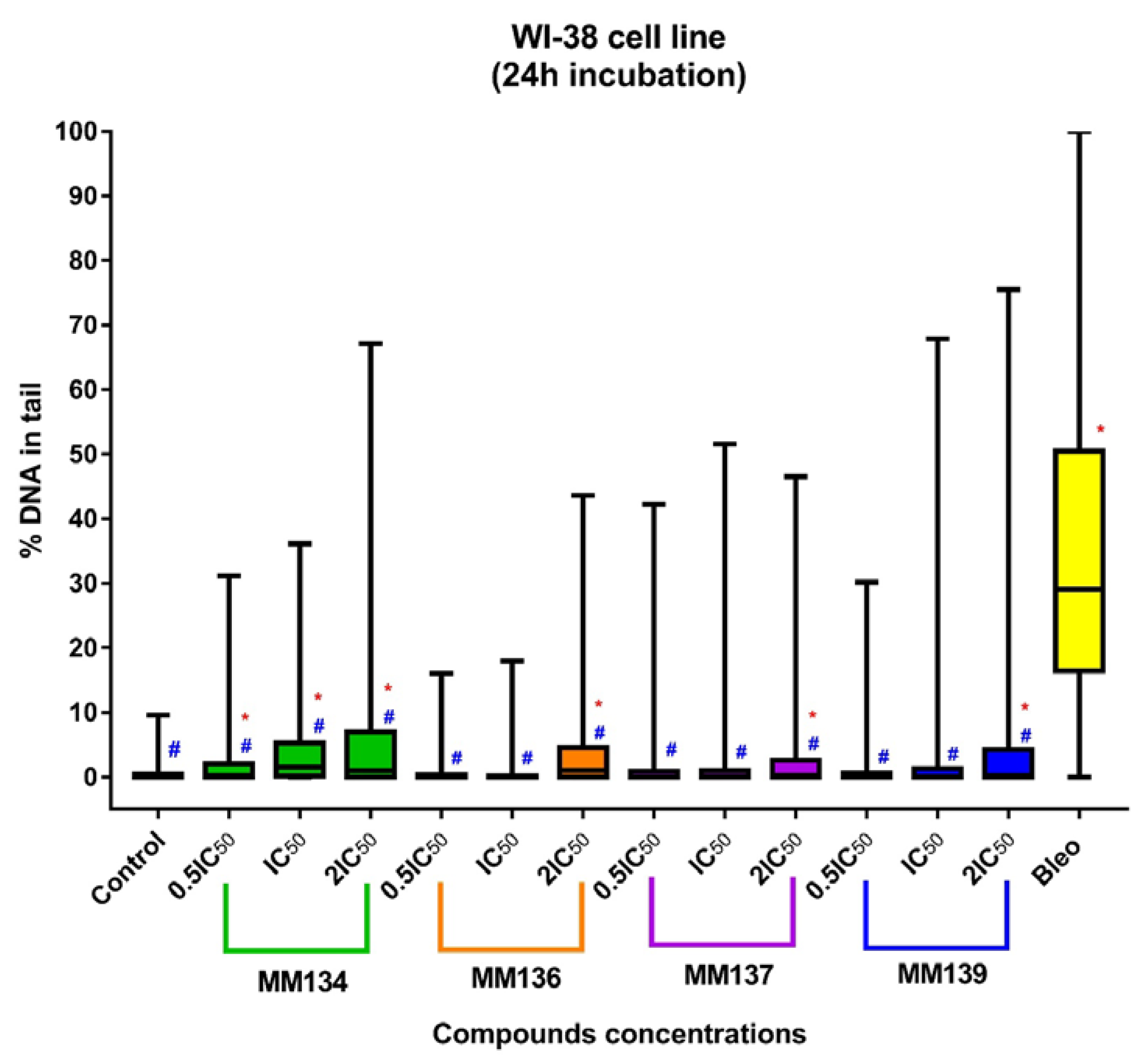
2.3.2. Normal Human Fibroblasts (WI-38 Cell Line)
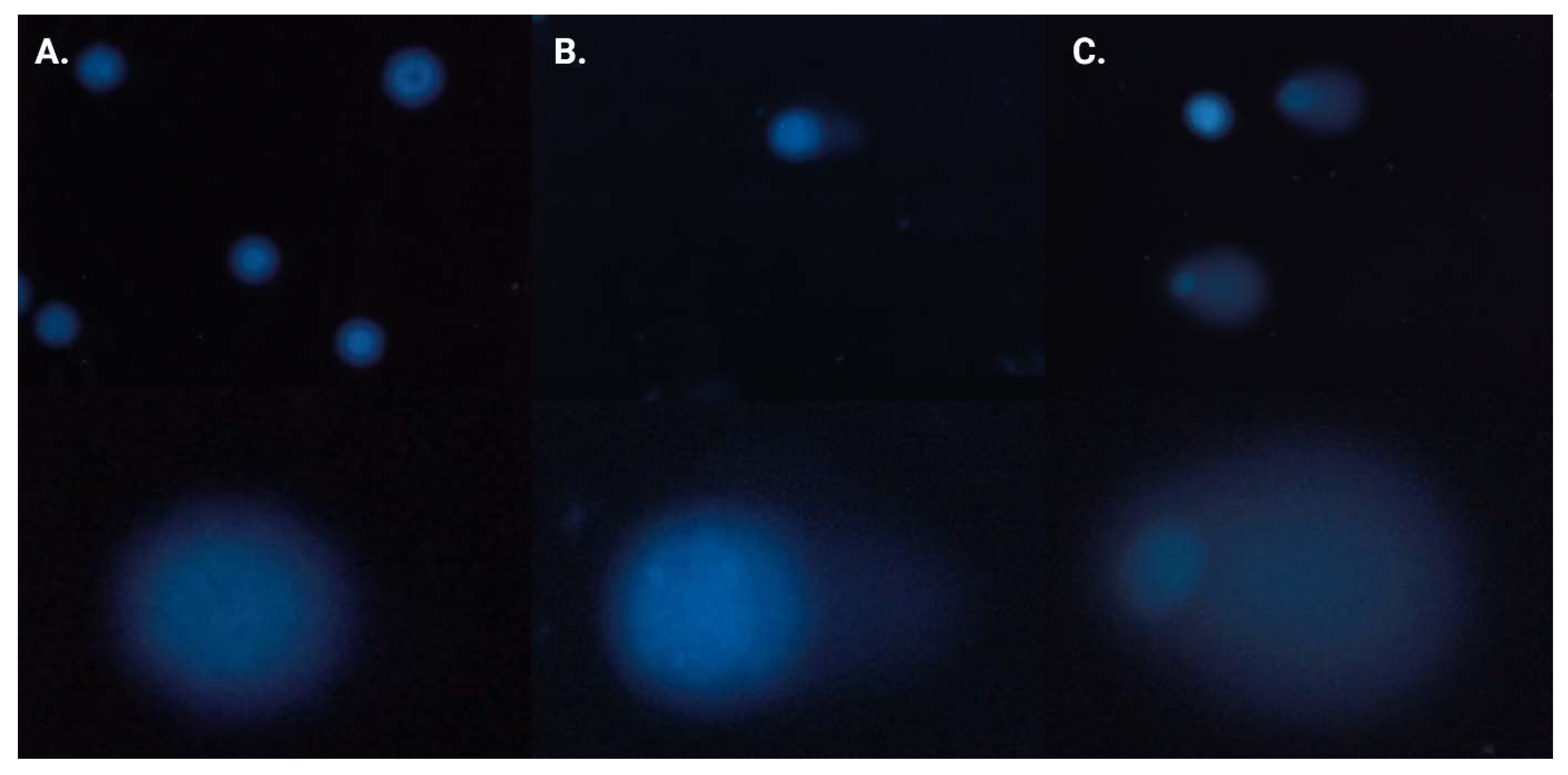
2.4. Neutral Comet Assay
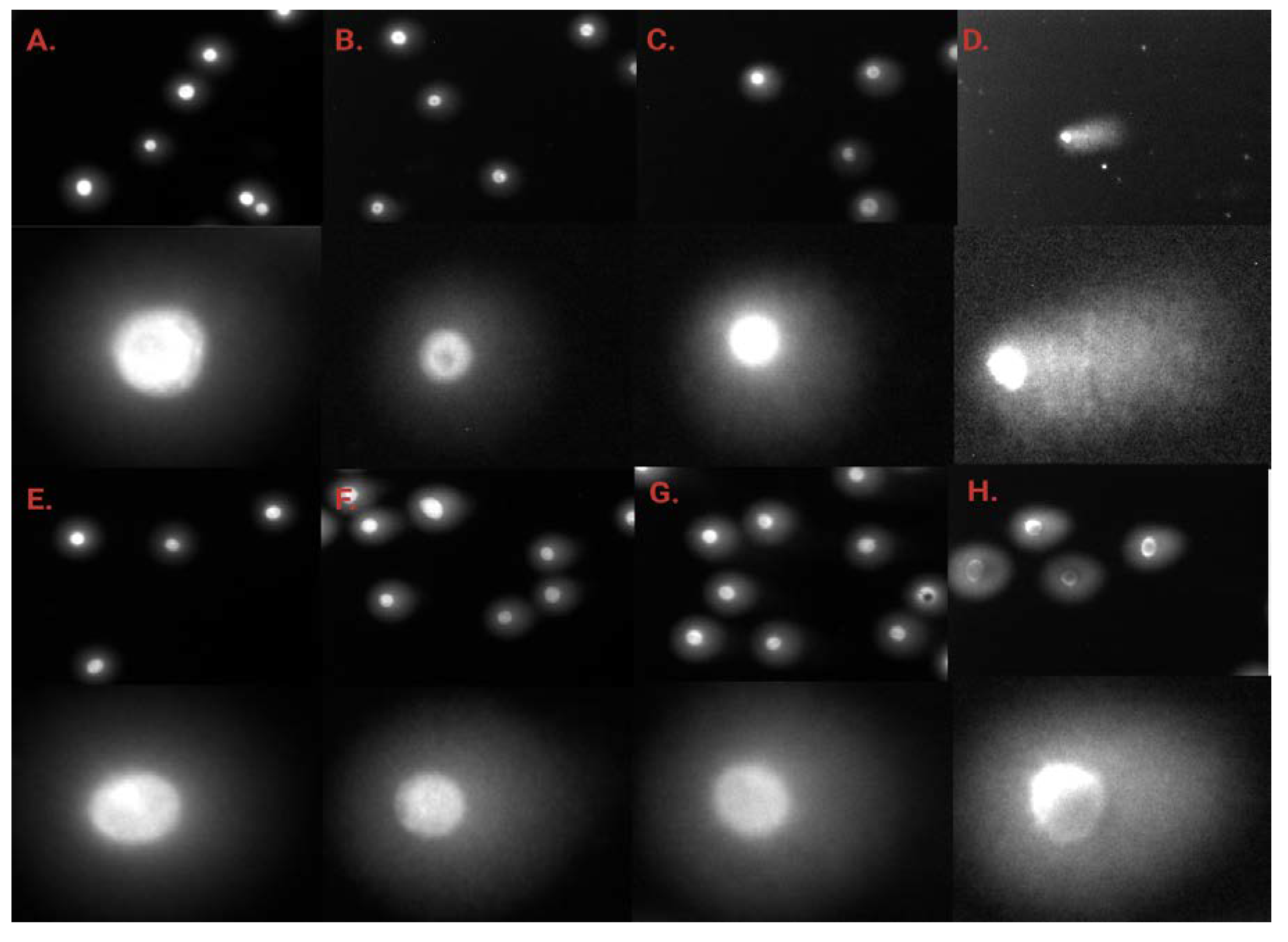
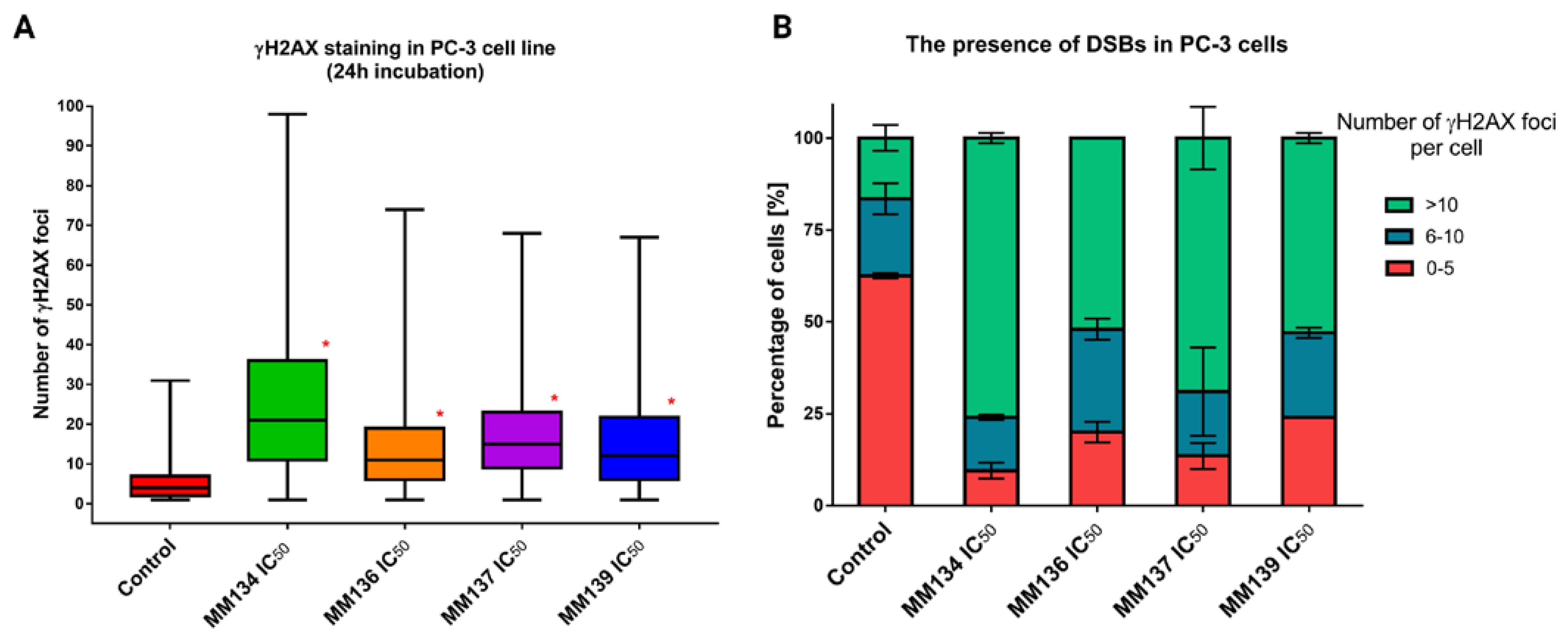
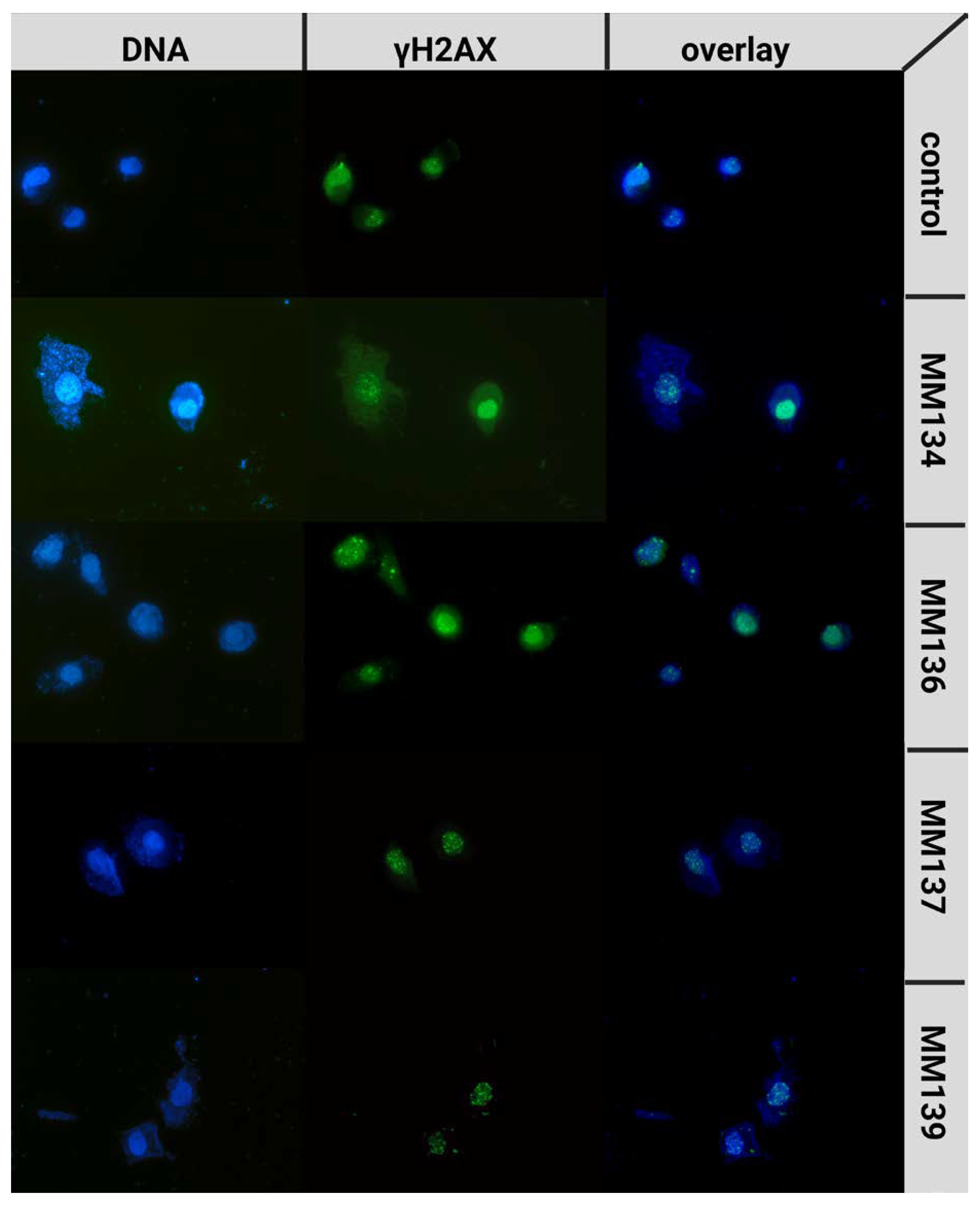
2.5. γH2AX Staining
2.6. Computational Studies
2.6.1. Molecular Docking
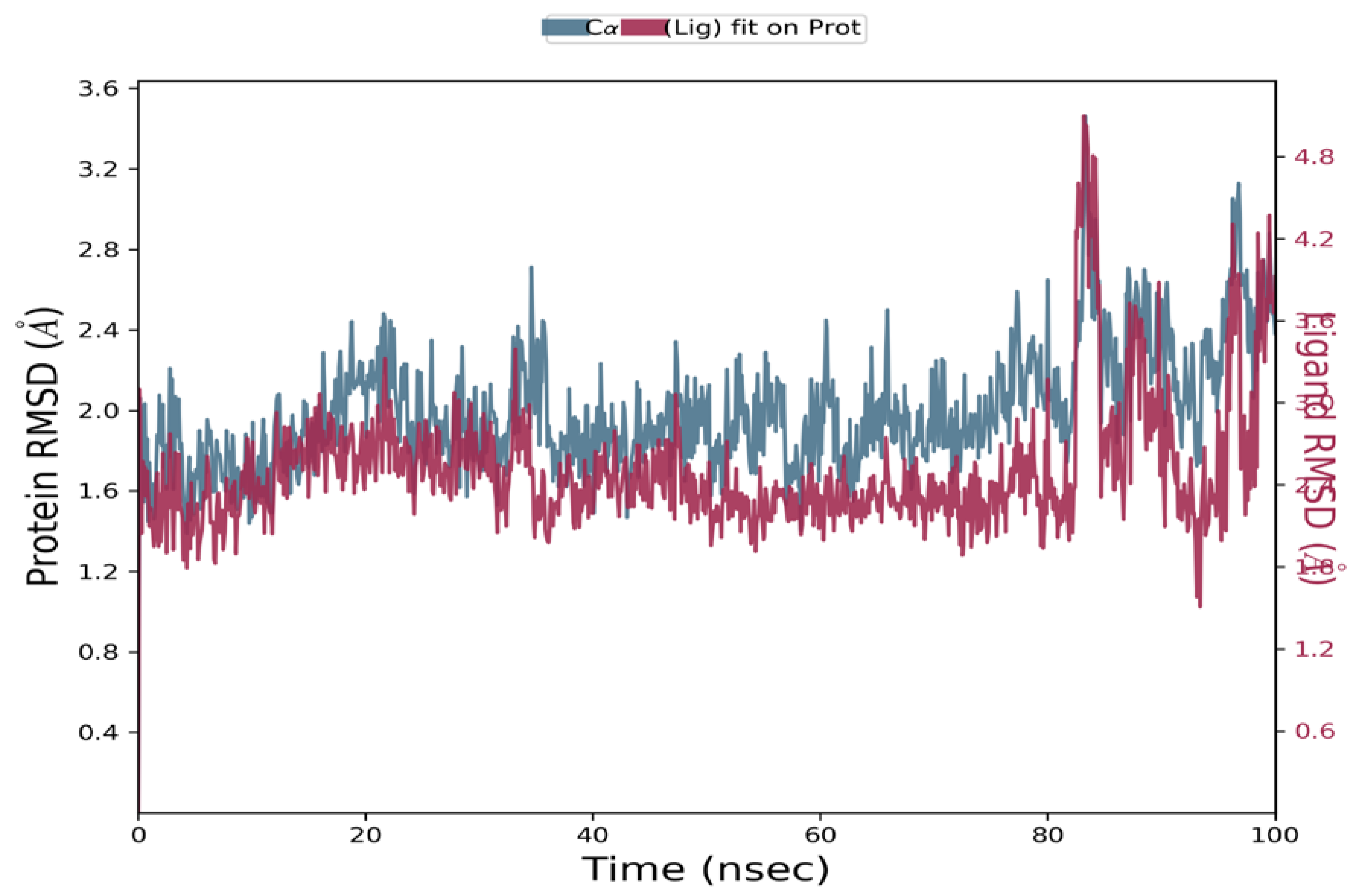
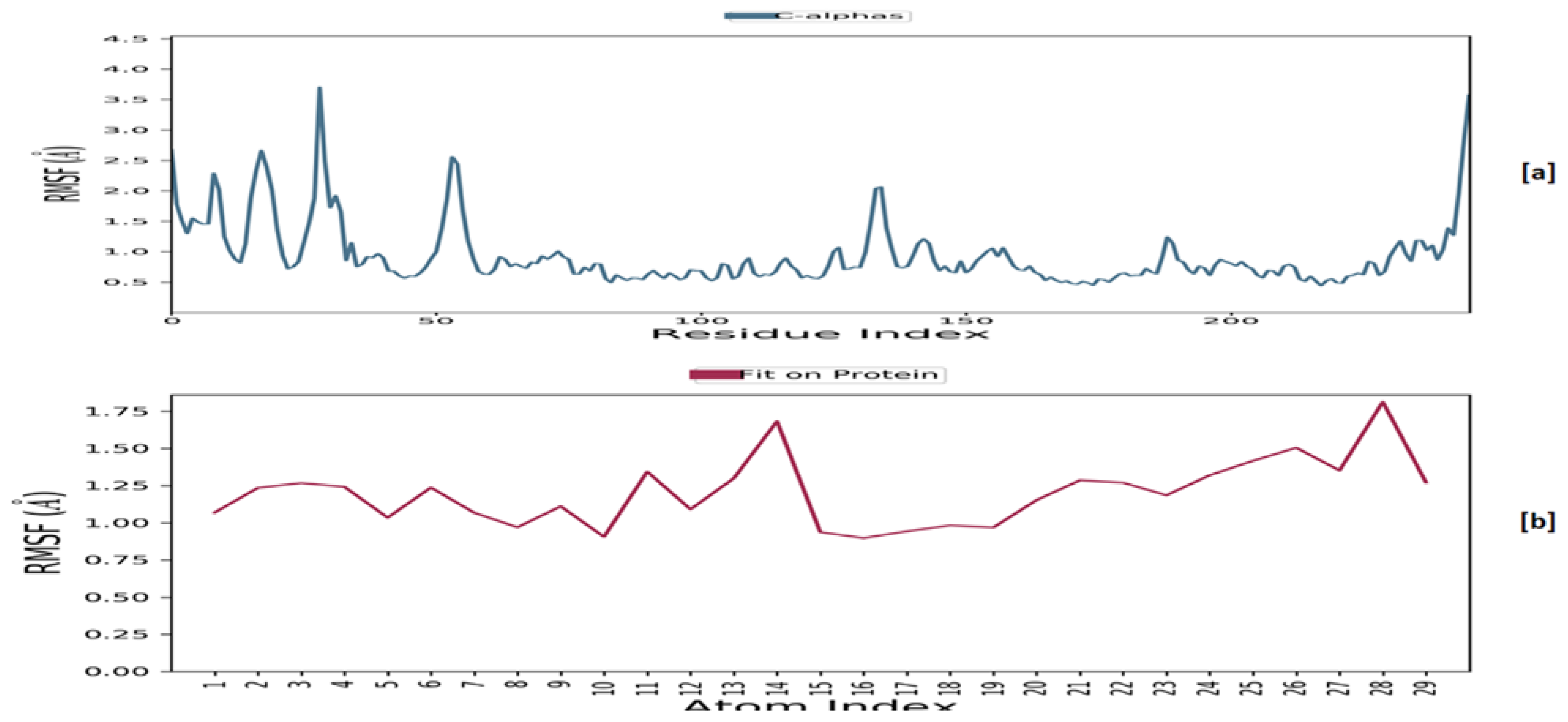
2.6.2. Molecular Dynamics
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Cell Treatment
4.4. Trypan Blue Staining
4.5. Alamar Blue
4.6. Comet Assays
4.6.1. Alkaline Comet Assay
4.6.2. Neutral Comet Assay
4.7. γH2AX Staining
4.8. Data Analysis
4.8.1. Alkaline and Neutral Comet Assays
4.8.2. γH2AX Staining
4.9. Computational Studies
4.9.1. Molecular Docking
4.9.2. Molecular Dynamics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABL | ABL protein kinase |
| AKT | RAC-alpha serine/threonine-protein kinase |
| ATM | serine/threonine protein kinase ATM |
| ATR | serine/threonine protein kinase ATR |
| BAX/BAK | pro-apoptotic protein BAX/BAK |
| BSA | bovine serum albumin |
| BTK | Bruton’s tyrosine kinase |
| CA | carbonic anhydrase |
| CDK | cyclin-dependent kinase |
| CHK1 | serine/threonine protein kinase CHK1 |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DDR | DNA damage response |
| DSB | double-strand breaks |
| FBS | fetal bovine serum |
| IRF1 | Interferon regulatory factor 1 |
| JAK | tyrosine-protein kinase JAK |
| MMP | mitochondria membrane potential |
| mTOR | mammalian target of rapamycin |
| NMP | normal melting point agarose |
| PARP1 | poly[ADP-ribose] polymerase 1 |
| PBS | buffered saline |
| PD-1 | programmed death 1 |
| PD-L1 | programmed death ligand 1 |
| PHA | paraformaldehyde |
| PI3K | phosphatidylinositol-4,5-bisphosphate 3-kinase |
| s-ICAM-1 | soluble intercellular adhesion molecule-1 |
| SSBs | single-strand breaks |
| STAT1/3 | signal transducer and activator of transcription 1/3 |
| TP53 | cellular tumor antigen p53 |
| TRIS | tris(hydroxymethyl)aminomethane |
References
- Choi, J.-S.; Berdis, A. Combating resistance to DNA damaging agents. Oncoscience 2018, 5, 134–136. [Google Scholar] [CrossRef]
- Mukhtar, M.; Bilal, M.; Rahdar, A.; Barani, M.; Arshad, R.; Behl, T.; Brisc, C.; Banica, F.; Bungau, S. Nanomaterials for Diagnosis and Treatment of Brain Cancer: Recent Updates. Chemosensors 2020, 8, 117. [Google Scholar] [CrossRef]
- Swift, L.H.; Golsteyn, R.M. Genotoxic Anti-Cancer Agents and Their Relationship to DNA Damage, Mitosis, and Checkpoint Adaptation in Proliferating Cancer Cells. Int. J. Mol. Sci. 2014, 15, 3403–3431. [Google Scholar] [CrossRef]
- Novak, M.; Žegura, B.; Modic, B.; Heath, E.; Filipič, M. Cytotoxicity and genotoxicity of anticancer drug residues and their mixtures in experimental model with zebrafish liver cells. Sci. Total. Environ. 2017, 601, 293–300. [Google Scholar] [CrossRef]
- Haystead, T.A.J. The Purinome, a Complex Mix of Drug and Toxicity Targets. Curr. Top. Med. Chem. 2006, 6, 1117–1127. [Google Scholar] [CrossRef]
- Lee, C.-I.; Lee, C.-L.; Hwang, J.-F.; Lee, Y.-H.; Wang, J.-J. Monascus-fermented red mold rice exhibits cytotoxic effect and induces apoptosis on human breast cancer cells. Appl. Microbiol. Biotechnol. 2012, 97, 1269–1278. [Google Scholar] [CrossRef]
- Lang, D.K.; Kaur, D.; Arora, R.; Saini, B.; Arora, S. Nitrogen-Containing Heterocycles as Anticancer Agents: An Overview. Anti-Cancer Agents Med. Chem. 2020, 20, 2150–2168. [Google Scholar] [CrossRef]
- Hatse, S.; De Clercq, E.; Balzarini, J. Role of antimetabolites of purine and pyrimidine nucleotide metabolism in tumor cell differentiation. Biochem. Pharmacol. 1999, 58, 539–555. [Google Scholar] [CrossRef]
- Parker, W.B. Enzymology of Purine and Pyrimidine Antimetabolites Used in the Treatment of Cancer. Chem. Rev. 2009, 109, 2880–2893. [Google Scholar] [CrossRef]
- Pizzorno, G.; Diasio, R.B.; Cheng, Y.-C. Pyrimidine and Purine Antimetabolites. In Holland-Frei Cancer Medicine, 6th ed.; Kufe, D.W., Pollock, R.E., Weichselbaum, R.R., Bast, R.C., Gansler, T.S., Holland, J.F., Frei, E., III, Eds.; BC Decker: Hamilton, ON, Canada, 2003; Chapter 50. Available online: https://www.ncbi.nlm.nih.gov/books/NBK13028/ (accessed on 1 January 2023).
- Saad, H.A.; Moustafa, A.H. Synthesis and Anticancer Activity of Some New S-Glycosyl and S-Alkyl 1,2,4-Triazinone Derivatives. Molecules 2011, 16, 5682–5700. [Google Scholar] [CrossRef]
- Kabbaj, Y.; Lazrek, H.B.; Barascut, J.L.; Imbach, J.L. Synthesis and Biological Activity of Some Unsaturated 6-Azauracil Acyclonucleosides. Nucleosides Nucleotides Nucleic Acids 2005, 24, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Mojzych, M.; Ceruso, M.; Bielawska, A.; Bielawski, K.; Fornal, E.; Supuran, C.T. New pyrazolo[4,3-e][1,2,4]triazine sulfonamides as carbonic anhydrase inhibitors. Bioorganic Med. Chem. 2015, 23, 3674–3680. [Google Scholar] [CrossRef] [PubMed]
- Mojzych, M.; Bielawska, A.; Bielawski, K.; Ceruso, M.; Supuran, C.T. Pyrazolo[4,3-e][1,2,4]triazine sulfonamides as carbonic anhydrase inhibitors with antitumor activity. Bioorganic Med. Chem. 2014, 22, 2643–2647. [Google Scholar] [CrossRef] [PubMed]
- Mojzych, M.; Dolashki, A.; Voelter, W. Synthesis of pyrazolo[4,3-e][1,2,4]triazine sulfonamides, novel Sildenafil analogs with tyrosinase inhibitory activity. Bioorganic Med. Chem. 2014, 22, 6616–6624. [Google Scholar] [CrossRef] [PubMed]
- Mojzych, M.; Tarasiuk, P.; Kotwica-Mojzych, K.; Rafiq, M.; Seo, S.-Y.; Nicewicz, M.; Fornal, E. Synthesis of chiral pyrazolo[4,3-e][1,2,4]triazine sulfonamides with tyrosinase and urease inhibitory activity. J. Enzym. Inhib. Med. Chem. 2016, 32, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Bernat, Z.; Szymanowska, A.; Kciuk, M.; Kotwica-Mojzych, K.; Mojzych, M. Review of the Synthesis and Anticancer Properties of Pyrazolo[4,3-e][1,2,4]triazine Derivatives. Molecules 2020, 25, 3948. [Google Scholar] [CrossRef]
- Byth, K.F.; Cooper, N.; Culshaw, J.D.; Heaton, D.W.; Oakes, S.E.; Minshull, C.A.; Norman, R.A.; Pauptit, R.A.; Tucker, J.A.; Breed, J.; et al. Imidazo[1,2-b]pyridazines: A potent and selective class of cyclin-dependent kinase inhibitors. Bioorganic Med. Chem. Lett. 2004, 14, 2249–2252. [Google Scholar] [CrossRef]
- Kciuk, M.; Mujwar, S.; Szymanowska, A.; Marciniak, B.; Bukowski, K.; Mojzych, M.; Kontek, R. Preparation of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides and Their Experimental and Computational Biological Studies. Int. J. Mol. Sci. 2022, 23, 5892. [Google Scholar] [CrossRef]
- Końca, K.; Lankoff, A.; Banasik, A.; Lisowska, H.; Kuszewski, T.; Góźdź, S.; Koza, Z.; Wojcik, A. A cross-platform public domain PC image-analysis program for the comet assay. Mutat. Res. Toxicol. Environ. Mutagen. 2002, 534, 15–20. [Google Scholar] [CrossRef]
- Luo, M.; Bao, Z.; Xu, F.; Wang, X.; Li, F.; Li, W.; Chen, Z.; Ying, S.; Shen, H. Unrepaired DNA damage in macrophages causes elevation of particulate matter-induced airway inflammatory response. Aging 2018, 10, 549–560. [Google Scholar] [CrossRef]
- Duez, P.; Dehon, G.; Kumps, A.; Dubois, J. Statistics of the Comet assay: A key to discriminate between genotoxic effects. Mutagenesis 2003, 18, 159–166. [Google Scholar] [CrossRef]
- Lorenzo, Y.; Costa, S.; Collins, A.; Azqueta, A. The comet assay, DNA damage, DNA repair and cytotoxicity: Hedgehogs are not always dead. Mutagenesis 2013, 28, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, D.W.; Olive, P.L.; O’Neill, K.L. The comet assay: A comprehensive review. Mutat. Res. Genet. Toxicol. 1995, 339, 37–59. [Google Scholar] [CrossRef]
- Nagelkerke, A.; Span, P.N. Staining Against Phospho-H2AX (γ-H2AX) as a Marker for DNA Damage and Genomic Instability in Cancer Tissues and Cells. Adv. Exp. Med. Biol. 2016, 899, 1–10. [Google Scholar] [CrossRef]
- Phillips, D.H.; Arlt, V.M. Genotoxicity: Damage to DNA and Its Consequences. EXS 2009, 99, 87–110. [Google Scholar] [CrossRef]
- Barabadi, H.; Najafi, M.; Samadian, H.; Azarnezhad, A.; Vahidi, H.; Mahjoub, M.A.; Koohiyan, M.; Ahmadi, A. A Systematic Review of the Genotoxicity and Antigenotoxicity of Biologically Synthesized Metallic Nanomaterials: Are Green Nanoparticles Safe Enough for Clinical Marketing? Medicina 2019, 55, 439. [Google Scholar] [CrossRef] [PubMed]
- Corvi, R.; Madia, F. In vitro genotoxicity testing–Can the performance be enhanced? Food Chem. Toxicol. 2017, 106, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Kubara, P.M.; Kernéis-Golsteyn, S.; Studény, A.; Lanser, B.B.; Meijer, L.; Golsteyn, R.M. Human cells enter mitosis with damaged DNA after treatment with pharmacological concentrations of genotoxic agents. Biochem. J. 2012, 446, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Hermanowicz, J.M.; Szymanowska, A.; Sieklucka, B.; Czarnomysy, R.; Pawlak, K.; Bielawska, A.; Bielawski, K.; Kalafut, J.; Przybyszewska, A.; Surazynski, A.; et al. Exploration of novel heterofused 1,2,4-triazine derivative in colorectal cancer. J. Enzym. Inhib. Med. Chem. 2021, 36, 535–548. [Google Scholar] [CrossRef]
- Hermanowicz, J.M.; Pawlak, K.; Sieklucka, B.; Czarnomysy, R.; Kwiatkowska, I.; Kazberuk, A.; Surazynski, A.; Mojzych, M.; Pawlak, D. MM-129 as a Novel Inhibitor Targeting PI3K/AKT/mTOR and PD-L1 in Colorectal Cancer. Cancers 2021, 13, 3203. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Gornowicz, A.; Szymanowska, A.; Mojzych, M.; Czarnomysy, R.; Bielawski, K.; Bielawska, A. The Anticancer Action of a Novel 1,2,4-Triazine Sulfonamide Derivative in Colon Cancer Cells. Molecules 2021, 26, 2045. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, K.; Marciniak, B.; Kciuk, M.; Mojzych, M.; Kontek, R. Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro. Molecules 2022, 27, 3761. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, S.; Liu, Y.; Li, X.; Wu, J.; Sun, Y.; Liu, G. DNA damage response and PD-1/PD-L1 pathway in ovarian cancer. DNA Repair 2021, 102, 103112. [Google Scholar] [CrossRef]
- Sato, H.; Jeggo, P.A.; Shibata, A. Regulation of programmed death-ligand 1 expression in response to DNA damage in cancer cells: Implications for precision medicine. Cancer Sci. 2019, 110, 3415–3423. [Google Scholar] [CrossRef]
- Sun, S.-Y. Searching for the real function of mTOR signaling in the regulation of PD-L1 expression. Transl. Oncol. 2020, 13, 100847. [Google Scholar] [CrossRef]
- Nikolova, T.; Marini, F.; Kaina, B. Genotoxicity testing: Comparison of the γH2AX focus assay with the alkaline and neutral comet assays. Mutat. Res. Toxicol. Environ. Mutagen. 2017, 822, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.F.; Walton, M.I.; Cherry, D.L.; Collins, I.; Clarke, P.A.; Garrett, M.D.; Workman, P. CHK1 Inhibition is Synthetically Lethal with Loss of B-Family DNA Polymerase Function in Human Lung and Colorectal Cancer Cells. Cancer Res. 2020, 80, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.; Scriven, K.; Massey, A.J. Inhibition of the checkpoint kinase Chk1 induces DNA damage and cell death in human Leukemia and Lymphoma cells. Mol. Cancer 2014, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Syljuåsen, R.G.; Sørensen, C.S.; Hansen, L.T.; Fugger, K.; Lundin, C.; Johansson, F.; Helleday, T.; Sehested, M.; Lukas, J.; Bartek, J. Inhibition of Human Chk1 Causes Increased Initiation of DNA Replication, Phosphorylation of ATR Targets, and DNA Breakage. Mol. Cell Biol. 2005, 25, 3553–3562. [Google Scholar] [CrossRef]
- Tran, S.-L.; Puhar, A.; Ngo-Camus, M.; Ramarao, N. Trypan Blue Dye Enters Viable Cells Incubated with the Pore-Forming Toxin HlyII of Bacillus cereus. PLoS ONE 2011, 6, e22876. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N. Multiple Applications of Alamar Blue as an Indicator of Metabolic Function and Cellular Health in Cell Viability Bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef] [PubMed]
- Czubaszek, M.; Szostek, M.; Wójcik, E.; Andraszek, K. The comet assay as a method of identifying chromosomes instability. Postep. Hig. Med. Dosw. 2014, 68, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Faust, F.; Kassie, F.; Knasmüller, S.; Boedecker, R.H.; Mann, M.; Mersch-Sundermann, V. The use of the alkaline comet assay with lymphocytes in human biomonitoring studies. Mutat. Res. Mol. Mech. Mutagen. 2004, 566, 209–229. [Google Scholar] [CrossRef] [PubMed]
- Horváthová, E.; Slamenová, D.; Gabelova, A. Use of single cell gel electrophoresis (comet assay) modifications for analysis of DNA damage. Gen. Physiol. Biophys. 1999, 18, 70–74. [Google Scholar]
- Moller, P. Assessment of reference values for DNA damage detected by the comet assay in human blood cell DNA. Mutat. Res. Mol. Mech. Mutagen. 2006, 612, 84–104. [Google Scholar] [CrossRef]
- Hartmann, A.; Agurell, E.; Beevers, C.; Brendler-Schwaab, S.; Burlinson, B.; Clay, P.; Collins, A.; Smith, A.; Speit, G.; Thybaud, V.; et al. Recommendations for Conducting the in Vivo Alkaline Comet Assay. 4th International Comet Assay Workshop. Mutagenesis 2003, 18, 45–51. [Google Scholar] [CrossRef]
- Liao, W.; McNutt, M.A.; Zhu, W.-G. The comet assay: A sensitive method for detecting DNA damage in individual cells. Methods 2009, 48, 46–53. [Google Scholar] [CrossRef]
- De Lapuente, J.; Lourenço, J.; Mendo, S.A.; Borràs, M.; Martins, M.G.; Costa, P.M.; Pacheco, M. The Comet Assay and its applications in the field of ecotoxicology: A mature tool that continues to expand its perspectives. Front. Genet. 2015, 6, 180. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Kontek, R.; Nowicka, H. The modulatory effect of melatonin on genotoxicity of irinotecan in healthy human lymphocytes and cancer cells. Drug Chem. Toxicol. 2012, 36, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R. The Comet Assay: Principles, Applications, and Limitations. Methods Mol. Biol. 2002, 203, 163–177. [Google Scholar] [CrossRef] [PubMed]
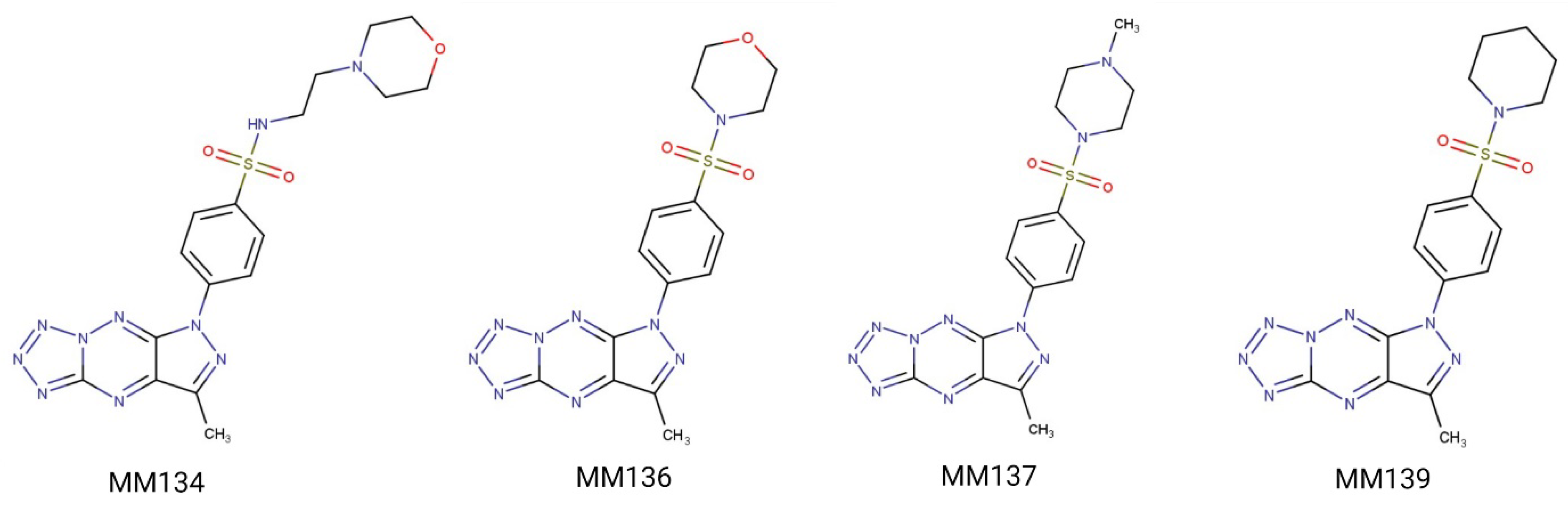

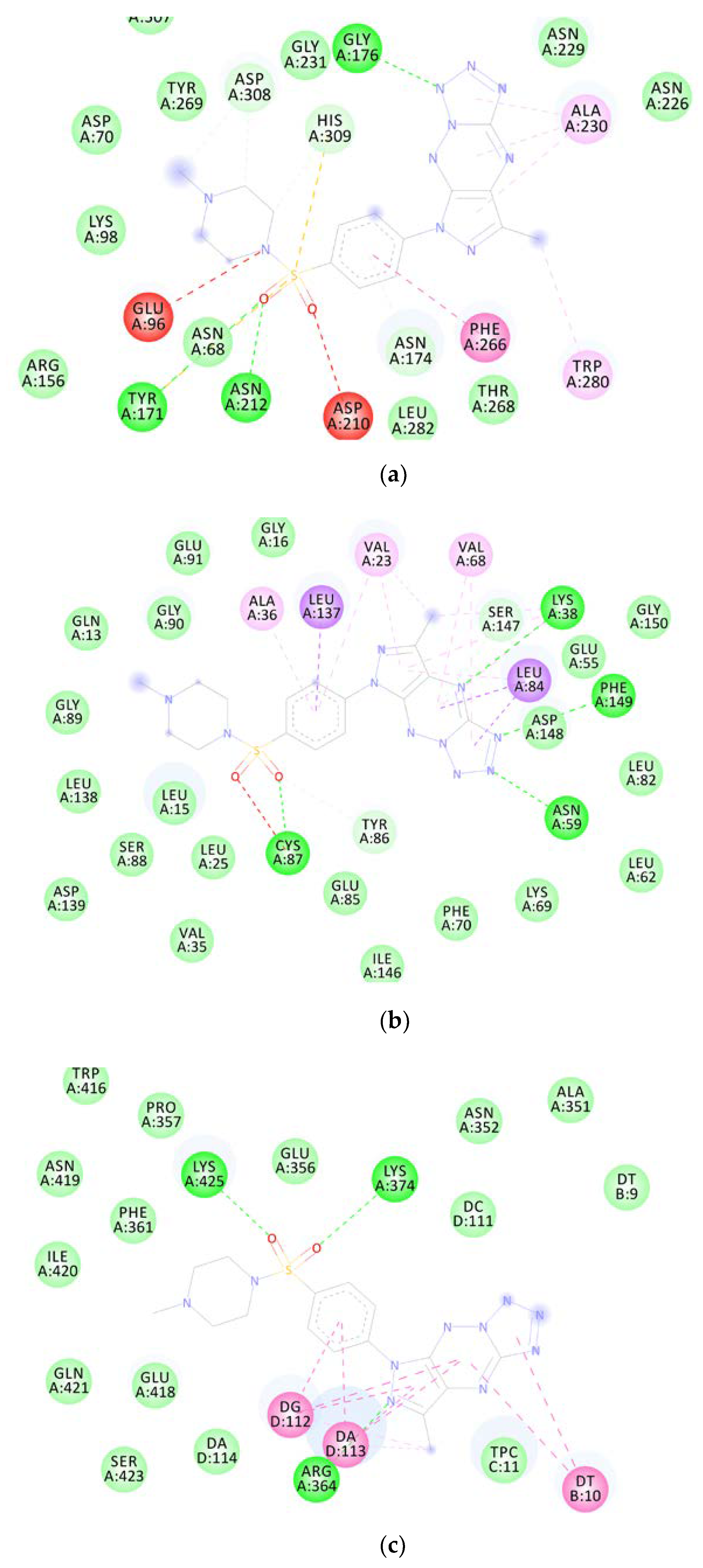
| MM Compound | Binding Energy (Kcal/mol) | Binding Residues | Reference Ligand | Binding Energy (Kcal/mol) | Binding Residues |
|---|---|---|---|---|---|
| APE-1 DNA-(apurinic or apyrimidinic site) endonuclease 6MKO | |||||
| MM134 | −9.18 | Asp308, His309, Gly176, Gly178, Arg177, Ala230, Trp280, Asn174 | Methoxyamine (No native ligand was complexed) | −5.74 | Asn212, Asp210, His309, Asn68, Asp308, Glu96 |
| MM136 | −8.07 | Gly178, Arg177, Gly176, Asp308, Trp280, His309, Leu282, Phe266 | |||
| MM137 | −9.64 | Asp308, His309, Gly176, Ala230, Trp280, Phe266, Asn212, Tyr171 | |||
| MM139 | −7.63 | Tyr269, Phe266, Met270, Thr268, Ala230, Asn174, Gly231 | |||
| ATR Serine/threonine-protein kinase ATR 4WAF | |||||
| MM134 | −10.06 | Asp933, Ile848, Tyr836, Ile932, Val850, Met922, Trp780, Thr856, His855 | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide | −10.62 | Ile848, Ile932, Val850, Val851, Met922, Glu849, Thr856, His855, Gln859, Lys802, Met800 |
| MM136 | −10.32 | Tyr836, Ile932, Val850, Arg852, Val851, Met922, Asn853, Ser854, Trp780 | |||
| MM137 | −10.95 | Tyr836, Val851, Met922, Val850, Ile932, Trp780, Ser854, Arg852 | |||
| MM139 | −10.34 | Ile848, Ile932, Val850, Trp780, Thr856, His855 | |||
| ATM Serine/threonine-protein kinase ATR 7NI5 | |||||
| MM134 | −9.02 | Pro2775, Gln2874, Trp2769, Leu2767, Leu2715, Ile2888, Tyr2755 | KU-55933 | −10.58 | Tyr2755, Cys2770, Glu2768, Leu2767, Lys2717, Trp2769, Leu2715, Pro2699, Ala2693, Pro2775, Leu2877 |
| MM136 | −9.17 | Asp2720, Lys2717, Leu2877, Trp2769, Thr2773, Cys2770, Tyr2755, Leu2767, Ile2888, Asn2697 | |||
| MM137 | −9.16 | Ala2693, Glu2778, Ile2888, Lys2717, Tyr2755, Leu2767, Cys2770, Trp2769, Leu2877, Pro2775, Val2774 | |||
| MM139 | −10.05 | Trp2769, Leu2877, Tyr2755, Ile2888, Gly2694, Pro2699, Leu2715, Cys2770 | |||
| CHK1 Serine/threonine-protein kinase Chk1 2YM8 | |||||
| MM134 | −9.75 | Cys87, Ala36, Leu137, Leu15, Val23, Leu84, Lys38, Phe149 | (R)-5-(8-chloroisoquinolin-3-ylamino)-3-(1-(dimethylamino)propan-2-yloxy)pyrazine-2-carbonitrile | −9.14 | Ala36, Leu84, Glu85, Val68, Lys38, Glu134, Asn135, Leu137, Val23, Cys87, Tyr86, Leu15 |
| MM136 | −10.44 | Cys87, Tyr86, Leu137, Leu84, Ala36, Val68, Phe149, Lys38, Val23, Gly90 | |||
| MM137 | −10–82 | Cys87, Ala36, Leu137, Val23, Val68, Lys38, Leu84, Phe149, Asn59, Tyr86 | |||
| MM139 | −10.61 | Cys87, Phe149, Lys38, Val23, Ala36, Leu137, Leu15 | |||
| CHK2 Serine/threonine-protein kinase Chk2 2W0J | |||||
| MM134 | −9.72 | Leu301, Lys249, Asn352, Ile299, Glu308, Val234, Leu354, Leu226 | 4,4′-diacetyldiphenylurea-bis(guanyl-hydrazone) | −9.76 | Glu273, Thr367, Ile299, Lys249, Leu301, Val234, Leu303, Leu226, Glu305 |
| MM136 | −9.64 | Leu226, Met304, Ala247, Leu301, Ile299, Lys249, Thr367, Leu354, Val234 | |||
| MM137 | −9.72 | Leu226, Met304, Ala247, Val234, Leu354, Lys249, Leu301, Ile299 | |||
| MM139 | −10.23 | Phe369, Lys249, Ala247, Leu354, Leu226, Met304, Leu303, Val234, Thr367 | |||
| PARP-1 Poly [ADP-ribose] polymerase 1 7ONS | |||||
| MM134 | −10.51 | Glu763, Tyr889, Asp766, Ala880, Leu877, Arg878, Leu769, Pro881, Tyr896 | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one | −10.36 | Gly863, Arg878, Leu769, Pro881, Asp770, Asn767, Tyr896, Tyr907, Lys903, His862, Ala898 |
| MM136 | −9.88 | Tyr907, His862, Asp766, Asp770, Leu877, Ile872, Arg878, Ala880 | |||
| MM137 | −10.29 | Arg878, Ala898, Tyr907, Tyr896, His862, Ile879 | |||
| MM139 | −10.36 | Ala880, Leu769, Pro881, Arg878, Asn767, Arg865, His909, Tyr907, His862, Asp770 | |||
| RPA70 Replication protein A 70 kDa DNA-binding subunit 4IJL | |||||
| MM134 | −5.81 | Arg41, Ile95, Ile83 | {[5-(3-chloro-1-benzothiophen-2-yl)-4-phenyl-4H-1,2,4-triazol-3-yl]sulfanyl}acetic acid | −5.97 | Leu87, Met57, Arg41, Ala59, Thr60, Ile95, Val93, Asn85 |
| MM136 | −6.16 | Met97, Ala59, Gln61, Ile83, Ile95 | |||
| MM137 | −5.71 | Arg41, Met92, Ala59, Ile95, Met57, Ile83 | |||
| MM139 | −7.08 | Met57, Ile95, Ile83 | |||
| TOP1 Topoisomerase I 1TL8 | |||||
| MM134 | −9.81 | Met428, Asn352, Lys436, Arg364, Lys425, Thrr426, DA13, DA14 | 2,3-dimethoxy-12H-[1,3]dioxolo[5,6]indeno[1,2]isoquinolin-6-ium | −9.53 | Asn722, Thr718, Arg364, DT10, DG12, DA13 |
| MM136 | −10.44 | Lys425, Arg364, Glu356, Asn722, DT10, DG12, DA13 | |||
| MM137 | −10.69 | Lys425, Lys374, Arg364, DT10, DG12, DA13 | |||
| MM139 | −10.34 | Lys532, DT10 | |||
| TOP2B Topoisomerase II 3QX3 | |||||
| MM134 | −8.54 | Glu477, Gly478, DC8, DA12, DG13 | Etoposide | −9.98 | Asp479, Arg503, Gly478, Met782, DC8, DA12, DG13 |
| MM136 | −8.24 | Arg503, Ala779, Met782, DA12, DG13 | |||
| MM137 | −8.32 | Ser480, Leu502, Asp559, Asp557 | |||
| MM139 | −7.76 | Gly478, Arg503, DC8, DA12, DG13 | |||
| WEE1 Wee1-like protein kinase 2IN6 | |||||
| MM134 | −9.14 | Asp386, Ile305, Cys379, Phe433, Val313, Ala326, Ser307 | PD311839 | −9.45 | Gly382, Cys379, Phe433, Ala326, Glu377, Val313, Val360, Asn376, Lys328, Ile305, Gly306, Ser307 |
| MM136 | −9.99 | Cys379, Asn376, Ile374, His350, Gly382, Val313, Ala326, Phe433, Val360, Lys328, Asp463 | |||
| MM137 | −10.07 | Ile305, Val313, Lys328, Asp463, Ile374, Asn376, Phe433, Val360, Ala326, Cys379 | |||
| MM139 | −9.92 | Lys328, Val313, Ala326, Phe433, Ile305, Tyr378, Gly382, Ser383 | |||
| Compound | Cell Line | ||
|---|---|---|---|
| IC50 Value ± SD | |||
| BxPC-3 | PC-3 | WI-28 | |
| MM134 | 0.32 ± 0.1 | 0.16 ± 0.02 | 0.65 ± 0.07 |
| MM136 | 0.25 ± 0.08 | 0.13 ± 0.01 | 0.48 ± 0.09 |
| MM137 | 0.16 ± 0.04 | 0.11 ± 0.007 | 0.27 ± 0.04 |
| MM139 | 0.33 ± 0.14 | 0.17 ± 0.003 | 0.64 ± 0.06 |
| Target | Full Name | PDB Code | x-D | y-D | z-D | Spacing (Ả) | X-Center | Y-Center | Z-Center |
|---|---|---|---|---|---|---|---|---|---|
| APE-1 | DNA-(apurinic or apyrimidinic site) endonuclease | 6MKO | 50 | 50 | 50 | 0.381 | 20.283 | 22.147 | 20.508 |
| ATR | Serine/threonine-protein kinase ATR | 4WAF | 40 | 40 | 46 | 0.397 | −1.215 | 8.294 | −17.439 |
| ATM | Serine/threonine-protein kinase ATR | 7NI5 | 40 | 40 | 40 | 0.397 | 111.559 | 150.337 | 210.394 |
| CHK1 | Serine/threonine-protein kinase Chk1 | 2YM8 | 40 | 40 | 40 | 0.414 | 15.394 | −1.219 | 11.745 |
| CHK2 | Serine/threonine-protein kinase Chk2 | 2W0J | 40 | 40 | 46 | 0.408 | 37.373 | −31.962 | 9.087 |
| PARP-1 | Poly [ADP-ribose] polymerase 1 | 7ONS | 40 | 40 | 46 | 0.397 | 10.962 | 42.743 | 7.819 |
| RPA70 | Replication protein A 70 kDa DNA-binding subunit | 4IJL | 40 | 40 | 46 | 0.369 | −5.944 | −9.334 | 1.963 |
| TOP1 | Topoisomerase I | 1TL8 | 40 | 40 | 40 | 0.392 | 22.245 | −4.32 | 27.329 |
| TOP2B | Topoisomerase II | 3QX3 | 40 | 40 | 40 | 0.553 | 32.884 | 95.413 | 50.785 |
| WEE1 | Wee1-like protein kinase | 2IN6 | 40 | 40 | 40 | 0.408 | 3.987 | 52.579 | 26.054 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kciuk, M.; Mujwar, S.; Marciniak, B.; Gielecińska, A.; Bukowski, K.; Mojzych, M.; Kontek, R. Genotoxicity of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides in Normal and Cancer Cells In Vitro. Int. J. Mol. Sci. 2023, 24, 4053. https://doi.org/10.3390/ijms24044053
Kciuk M, Mujwar S, Marciniak B, Gielecińska A, Bukowski K, Mojzych M, Kontek R. Genotoxicity of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides in Normal and Cancer Cells In Vitro. International Journal of Molecular Sciences. 2023; 24(4):4053. https://doi.org/10.3390/ijms24044053
Chicago/Turabian StyleKciuk, Mateusz, Somdutt Mujwar, Beata Marciniak, Adrianna Gielecińska, Karol Bukowski, Mariusz Mojzych, and Renata Kontek. 2023. "Genotoxicity of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides in Normal and Cancer Cells In Vitro" International Journal of Molecular Sciences 24, no. 4: 4053. https://doi.org/10.3390/ijms24044053
APA StyleKciuk, M., Mujwar, S., Marciniak, B., Gielecińska, A., Bukowski, K., Mojzych, M., & Kontek, R. (2023). Genotoxicity of Novel Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides in Normal and Cancer Cells In Vitro. International Journal of Molecular Sciences, 24(4), 4053. https://doi.org/10.3390/ijms24044053






