Biophysical Parameters Can Induce Epithelial-to-Mesenchymal Phenotypic and Genotypic Changes in HT-29 Cells: A Preliminary Study
Abstract
1. Introduction
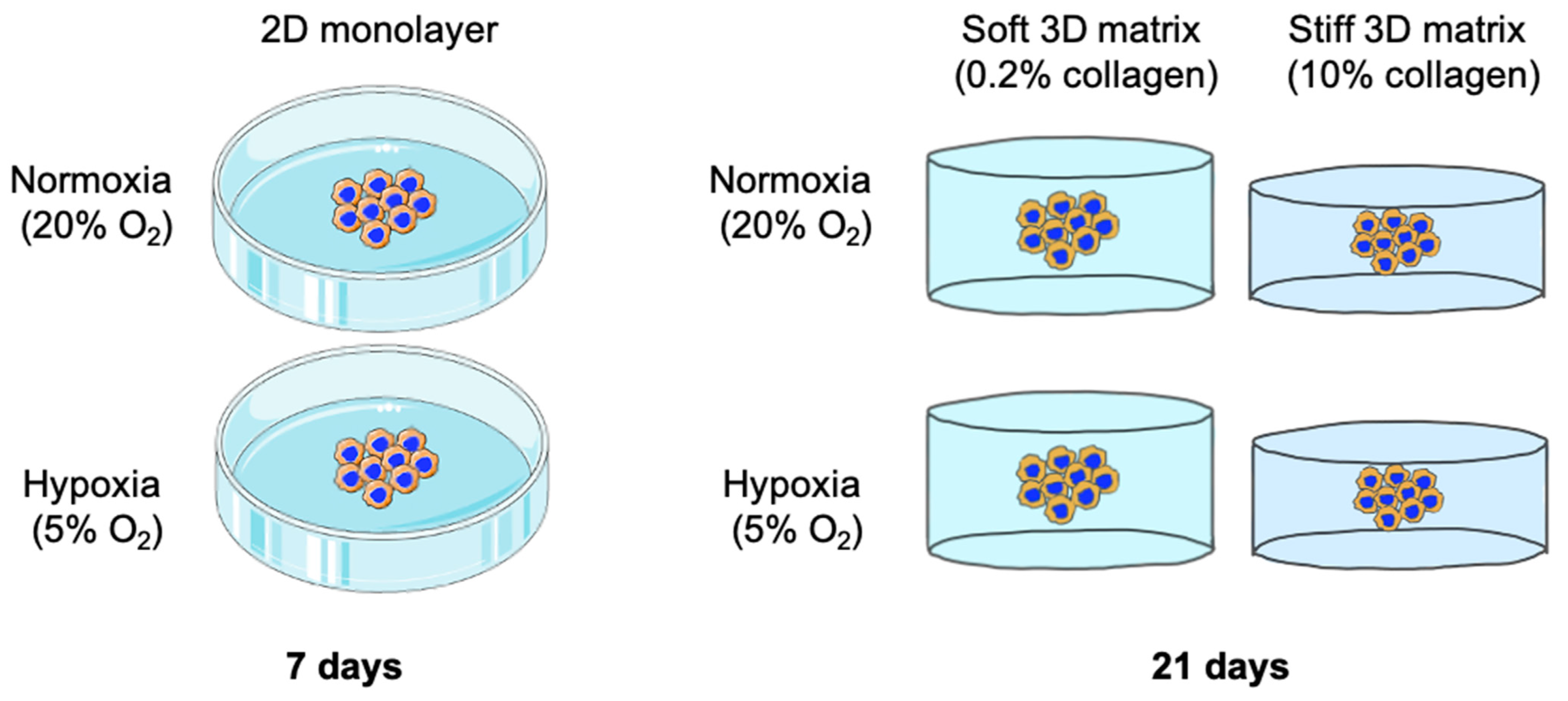
2. Results
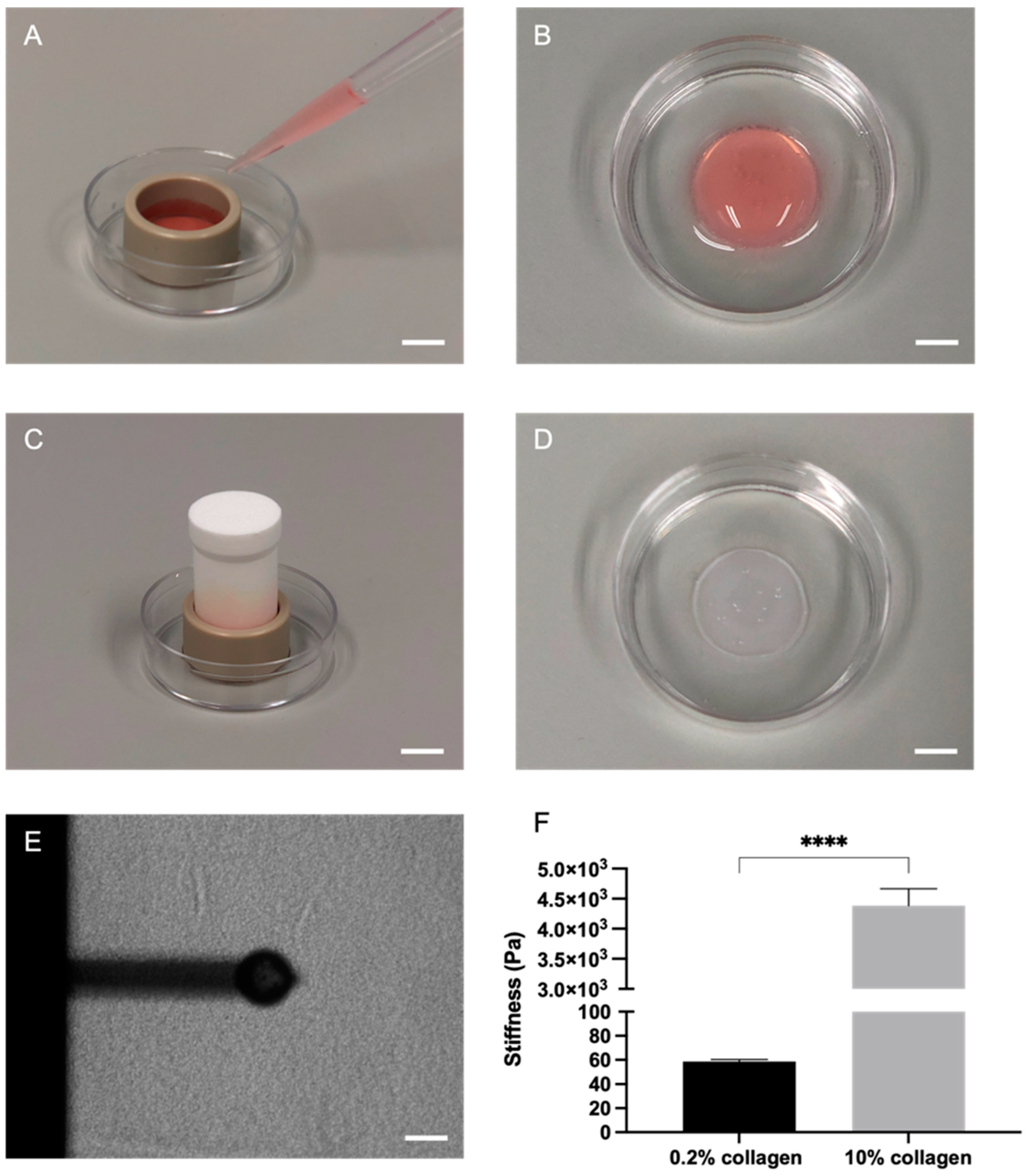
2.1. Plastic Compression of Collagen Significantly Increased Stiffness as Measured through Atomic Force Microscopy (AFM)
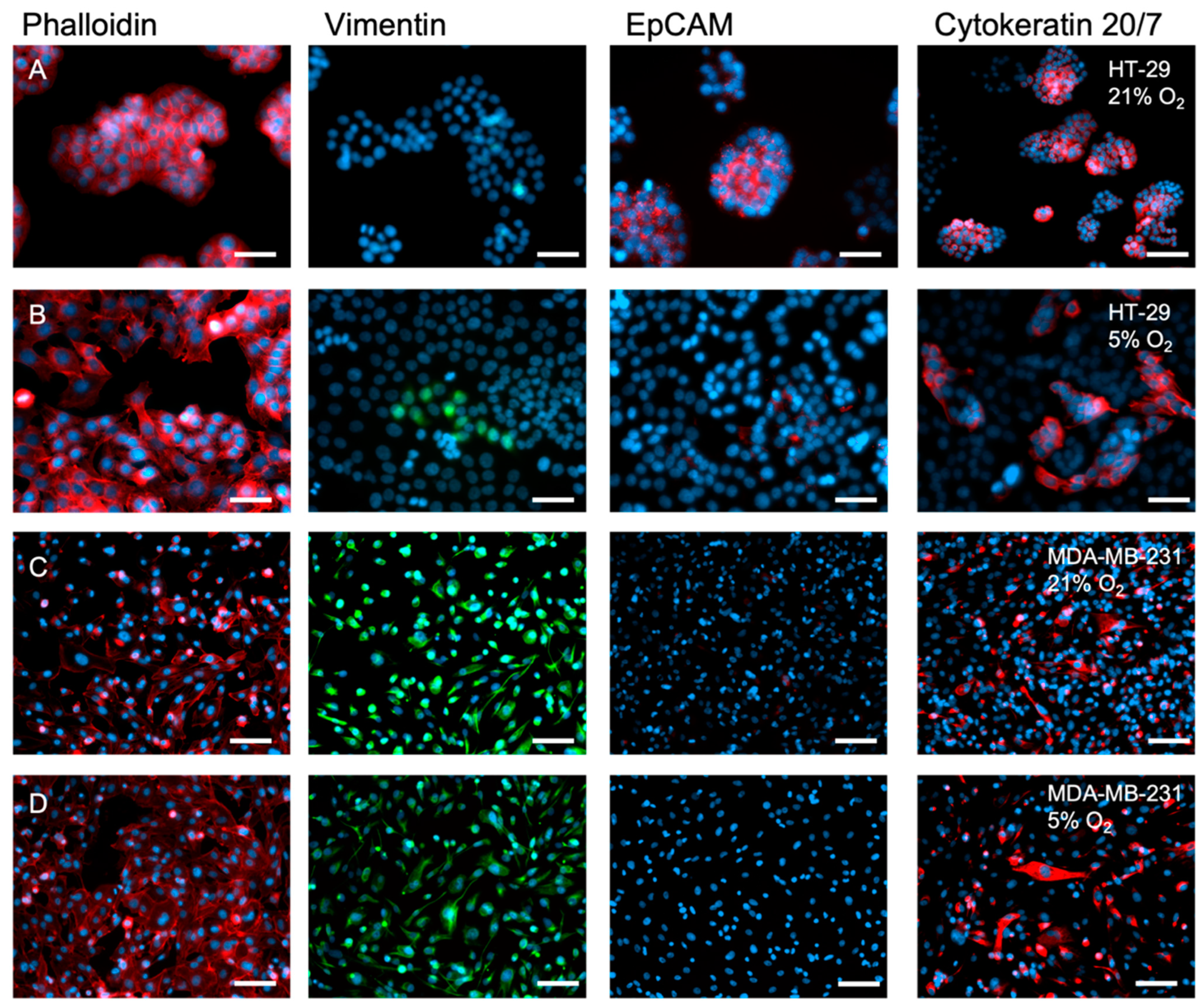
2.2. The Inherent EMT Status of HT-29 Cells and MDA-MB-231 Epithelial Cancer Cells
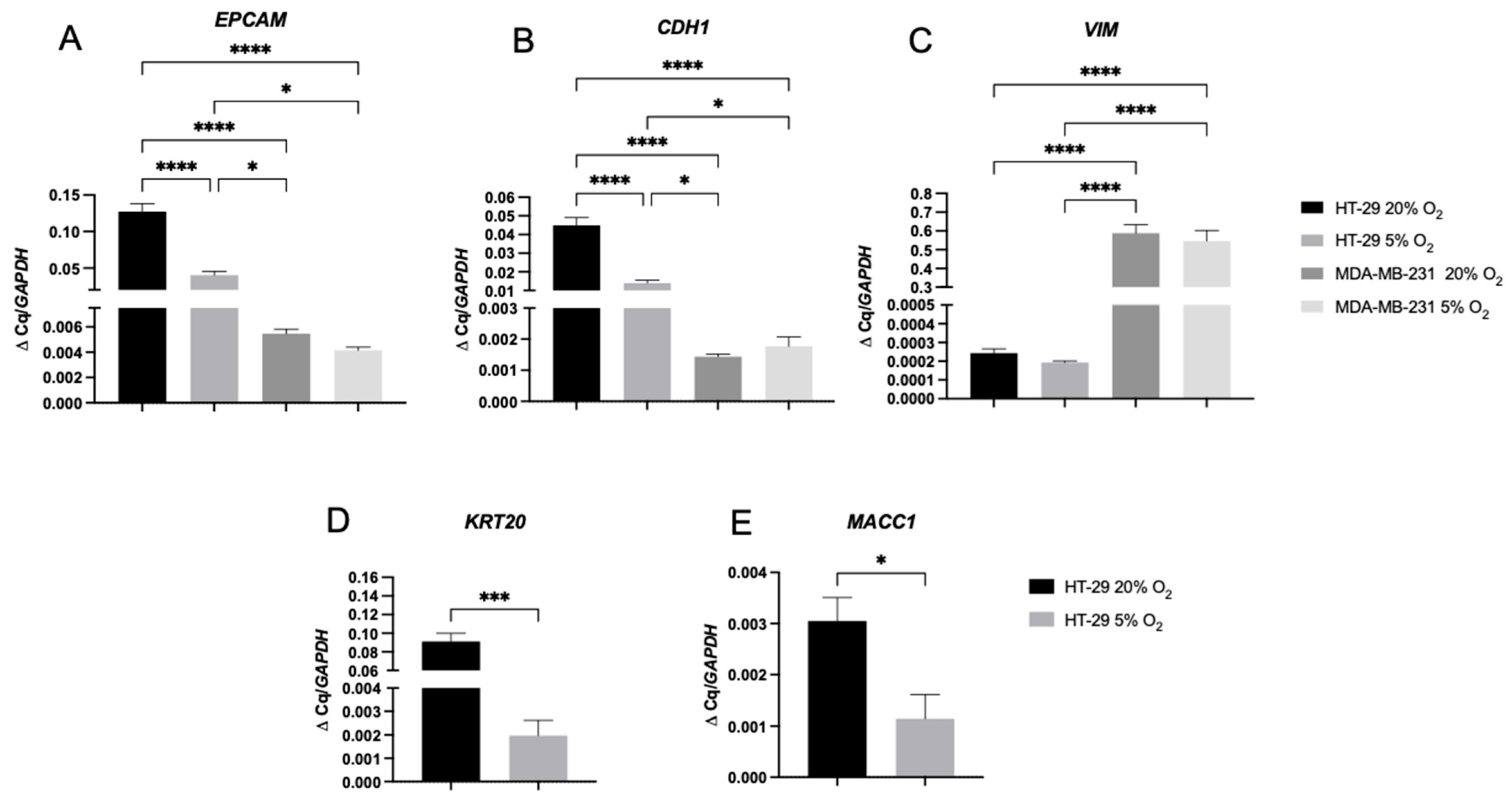
2.3. A 2D Hypoxic Environment Triggered EMT in HT-29 Evidenced by Decreased EPCAM and KRT20 Expression
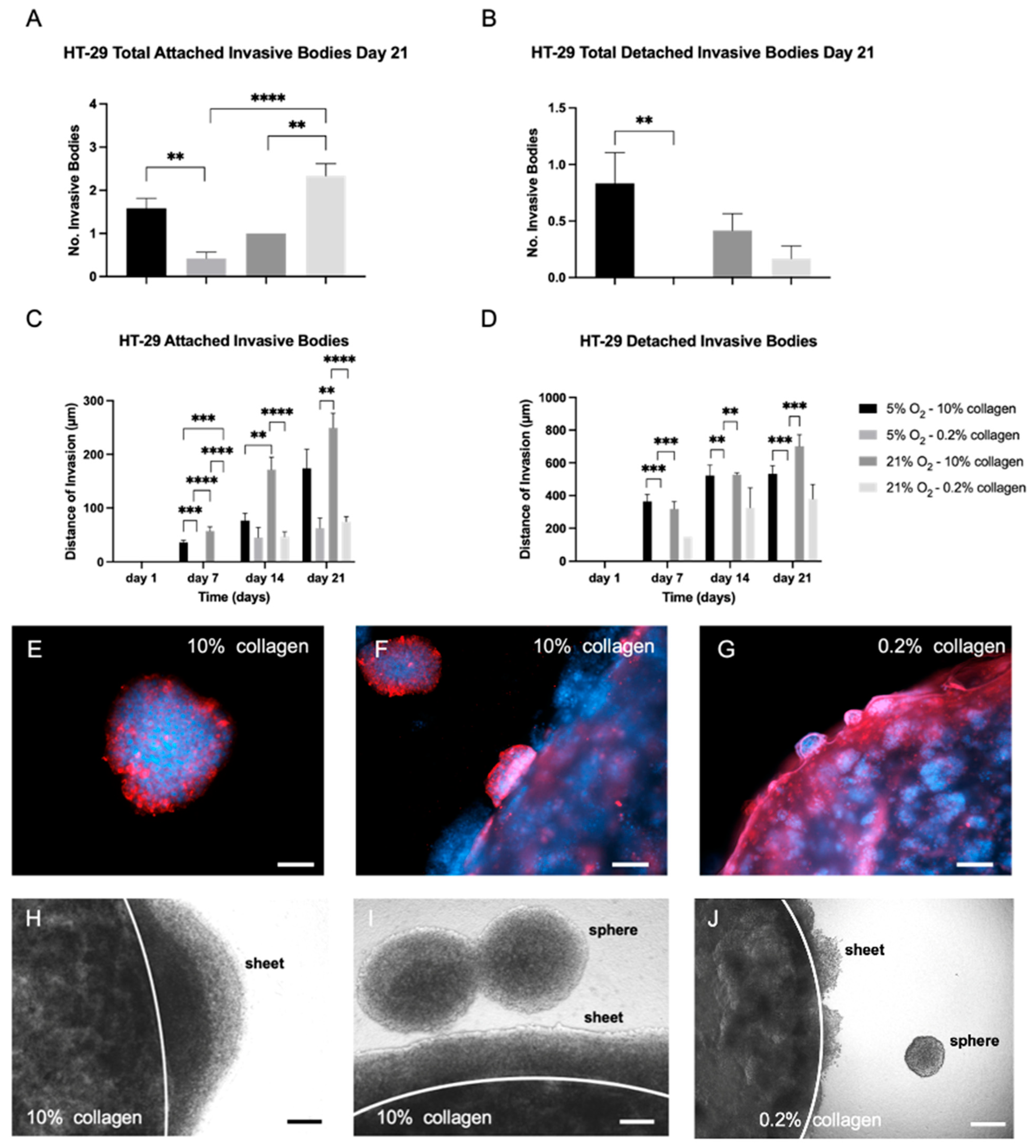
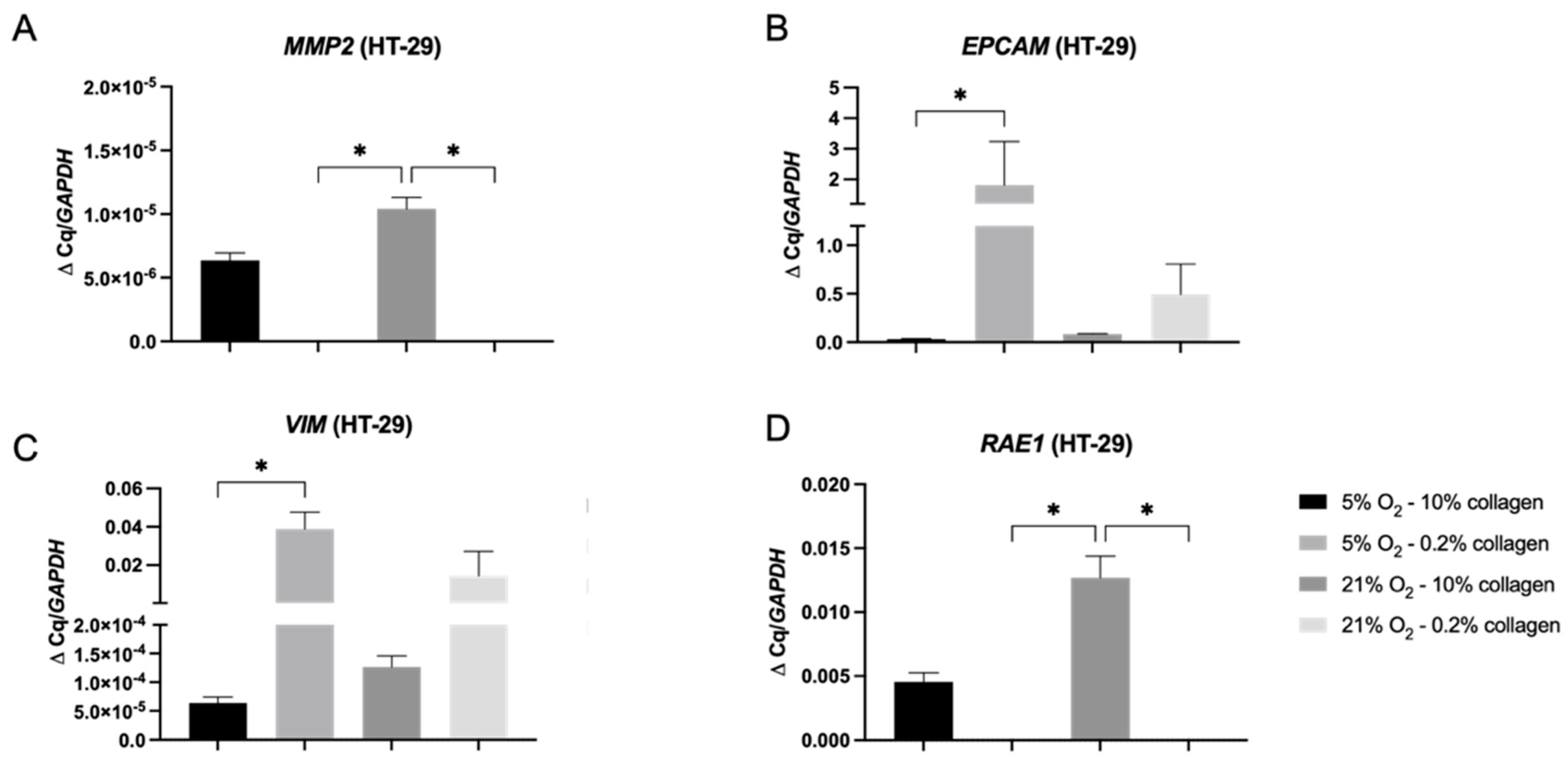
2.4. A 3D Stiff Environment at Physiological Hypoxia Induced an Invasive Phenotype and Genotype in HT-29 Cells
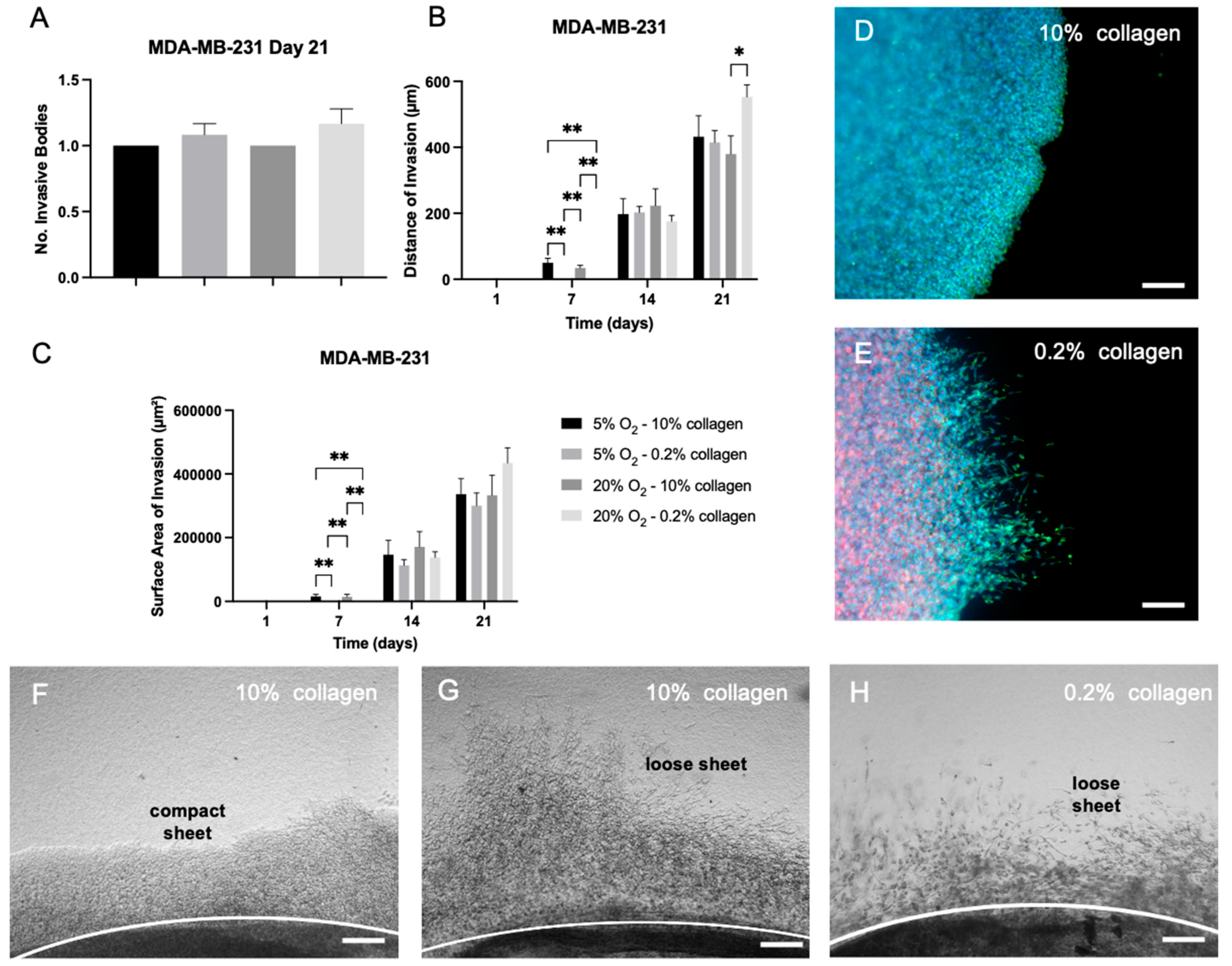
2.5. EMT Was Not Reversed by Biophysical Stimuli, Evidenced by MDA-MB-231 Cells, Which Showed Insignificant Changes in Invasion in Different 3D Microenvironments
3. Discussion
4. Materials and Methods
4.1. Atomic Force Microscopy (AFM)
4.2. Cancer Cell Culture
4.3. D Tumouroid Set-Up
4.4. Immunostaining
4.5. Measurements of the Invasive Phenotype
4.6. RNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018, 7, e37294. [Google Scholar] [CrossRef] [PubMed]
- Pillar, N.; Polsky, A.L.; Weissglas-Volkov, D.; Shomron, N. Comparison of breast cancer metastasis models reveals a possible mechanism of tumor aggressiveness. Cell Death Dis. 2018, 9, 1040. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.P.; Roozitalab, R.; Gibson, S.V.; Grose, R.P. Tumour microenvironment 3D-modelling: Simplicity to complexity and back again. Trends Cancer 2021, 7, 1033–1046. [Google Scholar] [CrossRef] [PubMed]
- Micalet, A.; Moeendarbary, E.; Cheema, U. 3D In Vitro Models for Investigating the Role of Stiffness in Cancer Invasion. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pape, J.; Emberton, M.; Cheema, U. 3D Cancer Models: The Need for a Complex Stroma, Compartmentalization and Stiffness. Front. Bioeng. Biotechnol. 2021, 9, 660502. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, R.H.; Lackmann, M.; Achen, M.G.; Stacker, S.A. Vascular remodeling in cancer. Oncogene 2014, 33, 3496–3505. [Google Scholar] [CrossRef] [PubMed]
- Leask, A. A centralized communication network: Recent insights into the role of the cancer associated fibroblast in the development of drug resistance in tumors. Semin Cell Dev. Biol. 2020, 101, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Magdeldin, T.; López-Dávila, V.; Pape, J.; Cameron, G.W.; Emberton, M.; Loizidou, M.; Cheema, U. Engineering a vascularised 3D in vitro model of cancer progression. Sci. Rep. 2017, 7, 44045. [Google Scholar] [CrossRef]
- Pape, J.; Magdeldin, T.; Stamati, K.; Nyga, A.; Loizidou, M.; Emberton, M.; Cheema, U. Cancer-associated fibroblasts mediate cancer progression and remodel the tumouroid stroma. Br. J. Cancer 2020, 123, 1178. [Google Scholar] [CrossRef]
- Pozzi, S.; Scomparin, A.; Dangoor, S.I.; Ajamil, D.R.; Ofek, P.; Neufeld, L.; Krivitsky, A.; Vaskovich-Koubi, D.; Kleiner, R.; Dey, P.; et al. Meet me halfway: Are in vitro 3D cancer models on the way to replace in vivo models for nanomedicine development? Adv. Drug Deliv. Rev. 2021, 175, 113760. [Google Scholar] [CrossRef]
- Pape, J.; Stamati, K.; Al Hosni, R.; Uchegbu, I.F.; Schatzlein, A.G.; Loizidou, M.; Emberton, M.; Cheema, U. Tissue-Engineering the Fibrous Pancreatic Tumour Stroma Capsule in 3D Tumouroids to Demonstrate Paclitaxel Response. Int. J. Mol. Sci. 2021, 22, 4289. [Google Scholar] [CrossRef] [PubMed]
- Barbazán, J.; Matic Vignjevic, D. Cancer associated fibroblasts: Is the force the path to the dark side? Curr. Opin. Cell Biol. 2019, 56, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Micalet, A.; Pape, J.; Bakkalci, D.; Javanmardi, Y.; Hall, C.; Cheema, U.; Moeendarbary, E. Evaluating the Impact of a Biomimetic Mechanical Environment on Cancer Invasion and Matrix Remodeling. Adv. Healthc. Mater. 2022, 2022, 2201749. [Google Scholar] [CrossRef] [PubMed]
- Cheema, U.; Rong, Z.; Kirresh, O.; MacRobert, A.J.; Vadgama, P.; Brown, R.A. Oxygen diffusion through collagen scaffolds at defined densities: Implications for cell survival in tissue models. J. Tissue Eng. Regen. Med. 2012, 6, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Hee, M.; Steven, K.; Lin, C. Engineering Tools for Regulating Hypoxia in Tumour Models. J. Cell. Mol. Med. 2021, 25, 7581–7592. [Google Scholar] [CrossRef]
- Biddle, A.; Mackenzie, I.C. Cancer stem cells and EMT in carcinoma. Cancer Metastasis Rev. 2012, 31, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Celià-Terrassa, T.; Jolly, M.K. Cancer stem cells and epithelial-to-mesenchymal transition in cancer metastasis. Cold Spring Harb. Perspect. Med. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Yadav, U.P.; Singh, T.; Kumar, P.; Sharma, P.; Kaur, H.; Sharma, S.; Singh, S.; Kumar, S.; Mehta, K. Metabolic Adaptations in Cancer Stem Cells. Front Oncol. 2020, 10, 1010. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Druzhkova, I.; Shirmanova, M.; Ignatova, N.; Dudenkova, V.; Lukina, M.; Zagaynova, E.; Safina, D.; Kostrov, S.; Didych, D.; Kuzmich, A.; et al. Expression of emt-related genes in hybrid e/m colorectal cancer cells determines fibroblast activation and collagen remodeling. Int. J. Mol. Sci. 2020, 21, 8119. [Google Scholar] [CrossRef]
- Cheema, U.; Brown, R.A. Rapid Fabrication of Living Tissue Models by Collagen Plastic Compression: Understanding Three-Dimensional Cell Matrix Repair In Vitro. Adv. Wound Care 2013, 2, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.A.; Wiseman, M.; Chuo, C.B.; Cheema, U.; Nazhat, S.N. Ultrarapid engineering of biomimetic materials and tissues: Fabrication of nano- and microstructures by plastic compression. Adv. Funct. Mater. 2005, 15, 1762–1770. [Google Scholar] [CrossRef]
- Boon, K. The Mesenchymal-Like Phenotype of the MDA-MB-231 Cell Line. In Breast Cancer—Focus Tumor Microenviron Stem Cells Metastasis; InTech Open: London, UK, 2011. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, W.; Lu, D.; Wu, Z.; Duan, H.; Luo, Y.; Feng, J.; Yang, D.; Fu, L.; Yan, X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.H.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Park, S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015, 46, 1267–1274. [Google Scholar] [CrossRef]
- Liu, K.; Newbury, P.A.; Glicksberg, B.S.; Zeng, W.Z.D.; Paithankar, S.; Andrechek, E.R.; Chen, B. Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nat. Commun. 2019, 10, 16022. [Google Scholar] [CrossRef]
- Vannier, C.; Mock, K.; Brabletz, T.; Driever, W. Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion molecule) expression to control cell behavior in early zebrafish development. J. Biol. Chem. 2013, 288, 18643–18659. [Google Scholar] [CrossRef]
- Rios De La Rosa, J.M.; Wubetu, J.; Tirelli, N.; Tirella, A. Colorectal tumor 3D in vitro models: Advantages of biofabrication for the recapitulation of early stages of tumour development. Biomed. Phys. Eng. Express 2018, 4, 045010. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Masuda, T.; Fujii, A.; Shimizu, D.; Sato, K.; Kitagawa, A.; Tobo, T.; Ozato, Y.; Saito, H.; Kuramitsu, S.; et al. Mitotic checkpoint regulator RAE1 promotes tumor growth in colorectal cancer. Cancer Sci. 2021, 112, 3173–3189. [Google Scholar] [CrossRef]
- Oh, J.H.; Hur, H.; Lee, J.Y.; Kim, Y.; Seo, Y.; Kim, M.H. The mitotic checkpoint regulator RAE1 induces aggressive breast cancer cell phenotypes by mediating epithelial-mesenchymal transition. Sci. Rep. 2017, 7, 42256. [Google Scholar] [CrossRef]
- Oh, J.H.; Lee, J.Y.; Yu, S.; Cho, Y.; Hur, S.; Nam, K.T.; Kim, M.H. RAE1 mediated ZEB1 expression promotes epithelial–mesenchymal transition in breast cancer. Sci. Rep. 2019, 9, 2977. [Google Scholar] [CrossRef]
- Tripathi, S.; Chakraborty, P.; Levine, H.; Jolly, M.K. A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells. PLoS Comput. Biol. 2020, 16, e1007619. [Google Scholar] [CrossRef] [PubMed]
- Spennati, G.; Horowitz, L.F.; McGarry, D.J.; Rudzka, D.A.; Armstrong, G.; Olson, M.F.; Folch, A.; Yin, H. Organotypic platform for studying cancer cell metastasis. Exp. Cell. Res. 2021, 401, 112527. [Google Scholar] [CrossRef] [PubMed]
- Vian, L.; Pękowska, A.; Rao, S.S.; Kieffer-Kwon, K.R.; Jung, S.; Baranello, L.; Huang, S.C.; El Khattabi, L.; Dose, M.; Pruett, N.; et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893.e13. [Google Scholar] [CrossRef]
- Krakhmal, N.V.; Zavyalova, M.V.; Denisov, E.V.; Vtorushin, S.V.; Perelmuter, V.M. Cancer Invasion: Patterns and Mechanisms. Acta Naturae 2015, 7, 17. [Google Scholar] [CrossRef]
- Brauchle, E.; Kasper, J.; Daum, R.; Schierbaum, N.; Falch, C.; Kirschniak, A.; Schäffer, T.E. Biomechanical and biomolecular characterization of extracellular matrix structures in human colon carcinomas. Matrix Biol. 2018, 68–69, 180–193. [Google Scholar] [CrossRef]
- Pape, J.; Magdeldin, T.; Ali, M.; Walsh, C.; Lythgoe, M.; Emberton, M.; Cheema, U. Cancer invasion regulates vascular complexity in a three-dimensional biomimetic model. Eur. J. Cancer 2019, 119, 179–193. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb-prot543. [Google Scholar] [CrossRef] [PubMed]
- Shipley, G. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CTmethod. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pape, J.; Micalet, A.; Alsheikh, W.; Ezbakh, N.; Virjee, R.-I.; Al Hosni, R.; Moeendarbary, E.; Cheema, U. Biophysical Parameters Can Induce Epithelial-to-Mesenchymal Phenotypic and Genotypic Changes in HT-29 Cells: A Preliminary Study. Int. J. Mol. Sci. 2023, 24, 3956. https://doi.org/10.3390/ijms24043956
Pape J, Micalet A, Alsheikh W, Ezbakh N, Virjee R-I, Al Hosni R, Moeendarbary E, Cheema U. Biophysical Parameters Can Induce Epithelial-to-Mesenchymal Phenotypic and Genotypic Changes in HT-29 Cells: A Preliminary Study. International Journal of Molecular Sciences. 2023; 24(4):3956. https://doi.org/10.3390/ijms24043956
Chicago/Turabian StylePape, Judith, Auxtine Micalet, Wissal Alsheikh, Nadia Ezbakh, Rania-Iman Virjee, Rawiya Al Hosni, Emad Moeendarbary, and Umber Cheema. 2023. "Biophysical Parameters Can Induce Epithelial-to-Mesenchymal Phenotypic and Genotypic Changes in HT-29 Cells: A Preliminary Study" International Journal of Molecular Sciences 24, no. 4: 3956. https://doi.org/10.3390/ijms24043956
APA StylePape, J., Micalet, A., Alsheikh, W., Ezbakh, N., Virjee, R.-I., Al Hosni, R., Moeendarbary, E., & Cheema, U. (2023). Biophysical Parameters Can Induce Epithelial-to-Mesenchymal Phenotypic and Genotypic Changes in HT-29 Cells: A Preliminary Study. International Journal of Molecular Sciences, 24(4), 3956. https://doi.org/10.3390/ijms24043956







