Ran GTPase and Its Importance in Cellular Signaling and Malignant Phenotype
Abstract
1. Introduction
1.1. The Roles of Ran within the Cell
1.2. The Role of Ran in Nucleocytoplasmic Transport and Cell Cycle Progression
1.3. The Role of Ran in Cancer Progression
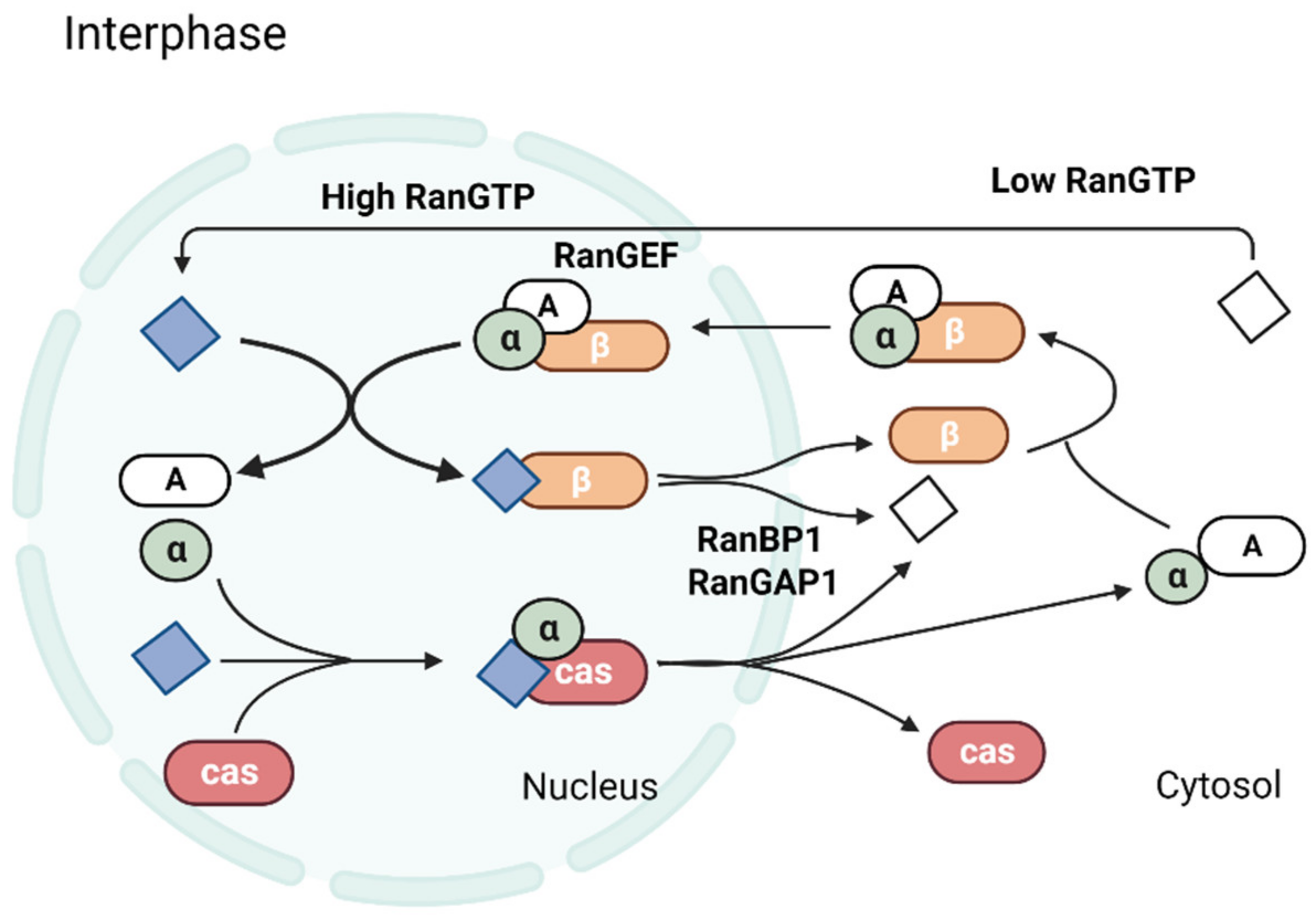
2. Ran Regulates Nucleocytoplasmic Transport
Molecules Transported
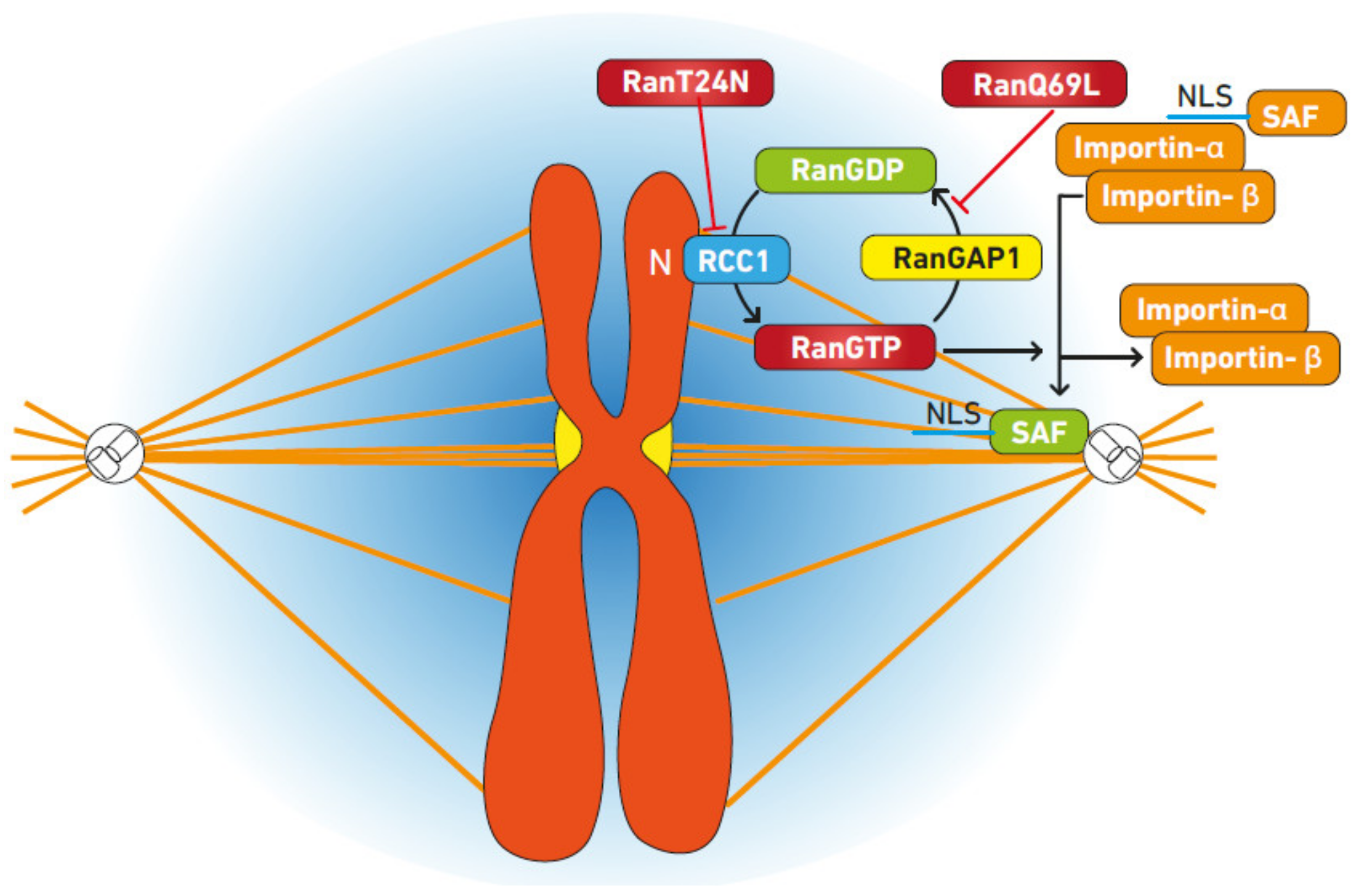
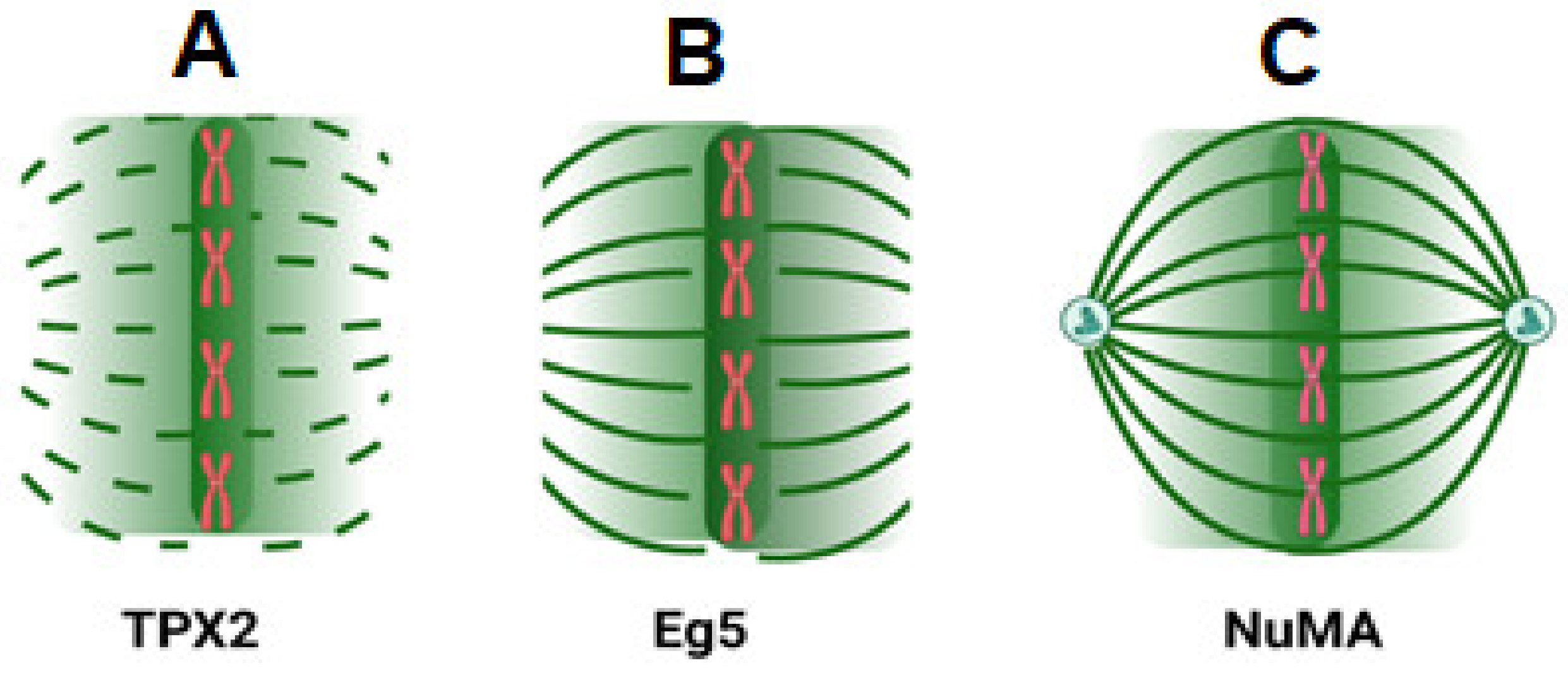
3. Ran Regulates Spindle Formation
3.1. Mechanism of Spindle Regulation by Ran
3.2. Ran Regulates Nuclear Envelope Reassembly
4. The Overexpression of Ran Alters Cellular Growth and Proliferation and Is Present in Cancer
4.1. Ran Overexpression
4.1.1. Ran Overexpression and Malignancy in Human Cancers
4.1.2. Ran Expression and Survival Time
5. Mechanism of Altered Expression—Ran Is a Downstream Effector of the PI3K/Akt and MEK/ERK Pathways
5.1. Aberrant Control of Pathways and Tumor Cell Dependence on Ran
5.2. Signaling Pathways Phosphorylation of Ran Binding Proteins and Control of Ran Expression
5.3. Altered Ran Expression and Regulation of Cellular Function and Nucleocytoplasmic Transport
5.4. Effect of Ran Expression on Spindle Formation and Tumor Cell Survival
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matchett, K.B.; McFarlane, S.; Hamilton, S.E.; Eltuhamy, Y.S.A.; Davidson, M.A.; Murray, J.T.; Faheem, A.M.; El-Tanani, M. Ran GTPase in Nuclear Envelope Formation and Cancer Metastasis. Adv. Exp. Med. Biol. 2014, 773, 323–351. [Google Scholar] [CrossRef] [PubMed]
- Dasso, M. Running on Ran: Nuclear transport and the mitotic spindle. Cell 2001, 104, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Boudhraa, Z.; Carmona, E.; Provencher, D.; Mes-Masson, A.-M. Ran GTPase: A Key Player in Tumor Progression and Metastasis. Front. Cell Dev. Biol. 2020, 8, 345. [Google Scholar] [CrossRef]
- Dasso, M. The Ran GTPase: Theme and Variations. Curr. Biol. 2002, 12, R502–R508. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, S.; Kouzoukakis, I.; Fielden, C.; Li, W.; Lashin, S.E.; Khair, N.; Raposo, T.P.; Fadhil, W.; Rudland, P.; Aleskandarany, M.; et al. Ran GTPase is an independent prognostic marker in malignant melanoma which promotes tumour cell migration and invasion. J. Clin. Pathol. 2022, 75, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.-F.; Chan, K.-K.; Grills, C.; Murray, J.T.; Platt-Higgins, A.; Eldin, O.S.; O’Byrne, K.; Janne, P.; Fennell, D.A.; Johnston, P.G.; et al. Ran Is a Potential Therapeutic Target for Cancer Cells with Molecular Changes Associated with Activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK Pathways. Clin. Cancer Res. 2012, 18, 380–391. [Google Scholar] [CrossRef]
- Yuen, H.-F.; Chan, K.-K.; Platt-Higgins, A.; Dakir, E.-H.; Matchett, K.B.; Haggag, Y.A.; Jithesh, P.V.; Habib, T.; Faheem, A.; Dean, F.A.; et al. Ran GTPase promotes cancer progression via Met receptor-mediated downstream signaling. Oncotarget 2016, 7, 75854–75864. [Google Scholar] [CrossRef]
- Yuen, H.-F.; Gunasekharan, V.-K.; Chan, K.-K.; Zhang, S.-D.; Platt-Higgins, A.; Gately, K.; O’Byrne, K.; Fennell, D.A.; Johnston, P.G.; Rudland, P.S.; et al. RanGTPase: A Candidate for Myc-Mediated Cancer Progression. J. Natl. Cancer Inst. 2013, 105, 475–488. [Google Scholar] [CrossRef]
- Köhler, A.; Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar] [CrossRef]
- Girdhar, A.; Guo, L. Regulating Phase Transition in Neurodegenerative Diseases by Nuclear Import Receptors. Biology 2022, 11, 1009. [Google Scholar] [CrossRef]
- Katahira, J. Nuclear export of messenger RNA. Genes 2015, 6, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Beaudet, D.; Akhshi, T.; Phillipp, J.; Law, C.; Piekny, A. Active Ran regulates anillin function during cytokinesis. Mol. Biol. Cell 2017, 28, 3517–3531. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.R.; Zhang, C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008, 9, 464–477. [Google Scholar] [CrossRef]
- Izaurralde, E.; Kutay, U.; von Kobbe, C.; Mattaj, I.; Görlich, D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997, 16, 6535–6547. [Google Scholar] [CrossRef] [PubMed]
- Forbes, D.J.; Travesa, A.; Nord, M.S.; Bernis, C. Nuclear transport factors: Global regulation of mitosis. Curr. Opin. Cell Biol. 2015, 35, 78–90. [Google Scholar] [CrossRef]
- Weaver, L.N.; Walczak, C.E. Spatial gradients controlling spindle assembly. Biochem. Soc. Trans. 2015, 43, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.; Zheng, Y. Stimulation of Microtubule Aster Formation and Spindle Assembly by the Small GTPase Ran. Science 1999, 284, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Gruss, O.J.; Vernos, I. The mechanism of spindle assembly: Functions of Ran and its target TPX2. J. Cell Biol. 2004, 166, 949–955. [Google Scholar] [CrossRef]
- Moore, J.D. The Ran-GTPase and cell-cycle control. Bioessays 2000, 23, 77–85. [Google Scholar] [CrossRef]
- Güttler, T.; Görlich, D. Ran-dependent nuclear export mediators: A structural perspective. EMBO J. 2011, 30, 3457–3474. [Google Scholar] [CrossRef]
- Xia, F.; Canovas, P.M.; Guadagno, T.M.; Altieri, D.C. A Survivin-Ran Complex Regulates Spindle Formation in Tumor Cells. Mol. Cell. Biol. 2008, 28, 5299–5311. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Lee, C.W.; Altieri, D.C. Tumor Cell Dependence on Ran-GTP–Directed Mitosis. Cancer Res. 2008, 68, 1826–1833. [Google Scholar] [CrossRef]
- Harel, A.; Forbes, D.J. Importin beta: Conducting a much larger cellular symphony. Mol. Cell 2004, 16, 319–330. [Google Scholar] [PubMed]
- Hetzer, M.; Cortes, D.B.; Walther, T.; Gruss, O.J.; Mattaj, I.W. GTP Hydrolysis by Ran Is Required for Nuclear Envelope Assembly. Mol. Cell 2000, 5, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Qin, R.-Y. Mechanism and its regulation of tumor-induced angiogenesis. World J. Gastroenterol. 2003, 9, 1144–1155. [Google Scholar] [CrossRef]
- Kurisetty, V.V.; Johnston, P.G.; Johnston, N.; Erwin, P.; Crowe, P.; Fernig, D.G.; Campbell, F.C.; Anderson, I.P.; Rudland, P.S.; El-Tanani, M.K. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene 2008, 27, 7139–7149. [Google Scholar] [CrossRef] [PubMed]
- Ly, T.K.; Wang, J.; Pereira, R.; Rojas, K.S.; Peng, X.; Feng, Q.; Cerione, R.A.; Wilson, K.F. Activation of the Ran GTPase Is Subject to Growth Factor Regulation and Can Give Rise to Cellular Transformation. J. Biol. Chem. 2010, 285, 5815–5826. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.T.; Minopoli, M.; Carriero, M.V. Tumor Associated Neutrophils. Their Role in Tumorigenesis, Metastasis, Prognosis and Therapy. Front. Oncol. 2019, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Qiu, J.; Wang, Y.; He, Z.; Wang, H.; Wang, Q.; Huang, Y.; Zhu, L.; Shi, F.; Chen, Y.; et al. Knockdown of Ran GTPase expression inhibits the proliferation and migration of breast cancer cells. Mol. Med. Rep. 2018, 18, 157–168. [Google Scholar] [CrossRef]
- Saxena, S.; Gandhi, A.; Lim, P.-W.; Relles, D.; Sarosiek, K.; Kang, C.; Chipitsyna, G.; Sendecki, J.; Yeo, C.J.; Arafat, H.A. RAN GTPase and Osteopontin in Pancreatic Cancer. Pancreat. Disord. Ther. 2013, 3, 113. [Google Scholar] [CrossRef]
- Kurisetty, V.V.; Johnston, P.G.; Rudland, P.S.; El-Tanani, M. Identification of genes differentially expressed between benign and osteopontin transformed rat mammary epithelial cells. BMC Res. Notes 2009, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Haggag, Y.; Abu Ras, B.; El-Tanani, Y.; Tambuwala, M.M.; McCarron, P.; Isreb, M.; El-Tanani, M. Co-delivery of a RanGTP inhibitory peptide and doxorubicin using dual-loaded liposomal carriers to combat chemotherapeutic resistance in breast cancer cells. Expert Opin. Drug Deliv. 2020, 17, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Dakir el, H.; Pickard, A.; Srivastava, K.; McCrudden, C.M.; Gross, S.R.; Lloyd, S.; Zhang, S.-D.; Margariti, A.; Morgan, R.; Rudland, P.S.; et al. The anti-psychotic drug pimozide is a novel chemotherapeutic for breast cancer. Oncotarget 2018, 9, 34889–34910. [Google Scholar] [CrossRef] [PubMed]
- Chavarri-Guerra, Y.; Sullivan, R.; Goss, P.E. Cancer control in Chile Reply. Lancet Oncol. 2013, 14, E337–E338. [Google Scholar] [CrossRef]
- Kuersten, S.; Ohno, M.; Mattaj, I.W. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2002, 11, 497–503. [Google Scholar] [CrossRef]
- Menyhárd, D.K.; Pálfy, G.; Orgován, Z.; Vida, I.; Keserű, G.M.; Perczel, A. Structural impact of GTP binding on downstream KRAS signaling. Chem. Sci. 2020, 11, 9272–9289. [Google Scholar] [CrossRef]
- Gray, J.L.; Von Delft, F.; Brennan, P.E. Targeting the Small GTPase Superfamily through Their Regulatory Proteins. Angew. Chem. Int. Ed. 2020, 59, 6342–6366. [Google Scholar] [CrossRef]
- Seewald, M.J.; Körner, C.; Wittinghofer, A.; Vetter, I.R. RanGAP mediates GTP hydrolysis without an arginine finger. Nature 2002, 415, 662–666. [Google Scholar] [CrossRef]
- Ren, X.; Jiang, K.; Zhang, F. The Multifaceted Roles of RCC1 in Tumorigenesis. Front. Mol. Biosci. 2020, 7, 225. [Google Scholar] [CrossRef]
- Yau, K.C.; Arnaoutov, A.; Aksenova, V.; Kaufhold, R.; Chen, S.; Dasso, M. RanBP1 controls the Ran pathway in mammalian cells through regulation of mitotic RCC1 dynamics. Cell Cycle 2020, 19, 1899–1916. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lyanguzova, M.; Kaufhold, R.; Haase, K.M.P.; Lee, H.; Arnaoutov, A.; Dasso, M. Association of RanGAP to nuclear pore complex component, RanBP2/Nup358, is required for pupal development in Drosophila. Cell Rep. 2021, 37, 110151. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Kutay, U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999, 15, 607–660. [Google Scholar] [CrossRef] [PubMed]
- Debler, E.W.; Blobel, G.; Hoelz, A. Nuclear transport comes full circle. Nat. Struct. Mol. Biol. 2009, 16, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Schwoebel, E.D.; Ho, T.; Moore, M.S. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 2002, 157, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Cavazza, T.; Vernos, I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 2016, 3, 82. [Google Scholar] [CrossRef]
- Sazer, S.; Dasso, M. The ran decathlon: Multiple roles of Ran. J. Cell Sci. 2000, 113, 1111–1118. [Google Scholar] [CrossRef]
- Matsuura, Y. Mechanistic Insights from Structural Analyses of Ran-GTPase-Driven Nuclear Export of Proteins and RNAs. J. Mol. Biol. 2016, 428, 2025–2039. [Google Scholar] [CrossRef]
- Cekan, P.; Hasegawa, K.; Pan, Y.; Tubman, E.; Odde, D.; Chen, J.-Q.; Herrmann, M.A.; Kumar, S.; Kalab, P. RCC1-dependent activation of Ran accelerates cell cycle and DNA repair, inhibiting DNA damage–induced cell senescence. Mol. Biol. Cell 2016, 27, 1346–1357. [Google Scholar] [CrossRef]
- Yaseen, N.R.; Blobel, G. GTP Hydrolysis Links Initiation and Termination of Nuclear Import on the Nucleoporin Nup358. J. Biol. Chem. 1999, 274, 26493–26502. [Google Scholar] [CrossRef]
- Oka, M.; Yoneda, Y. Importin α: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Dahlberg, J.E.; Lund, E. Functions of the GTPase Ran in RNA export from the nucleus. Curr. Opin. Cell Biol. 1998, 10, 400–408. [Google Scholar] [CrossRef]
- Björk, P.; Wieslander, L. Integration of mRNP formation and export. Cell. Mol. Life Sci. 2017, 74, 2875–2897. [Google Scholar] [CrossRef]
- DI Liegro, C.M.; Schiera, G.; DI Liegro, I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review). Int. J. Mol. Med. 2014, 33, 747–762. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Nigg, E.A. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature 1997, 386, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Cimica, V.; Chen, H.-C.; Iyer, J.K.; Reich, N.C. Dynamics of the STAT3 Transcription Factor: Nuclear Import Dependent on Ran and Importin-β1. PLoS ONE 2011, 6, e20188. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.C.; Barker, A.R.; Wakefield, J.G. The Ran Pathway in Drosophila melanogaster Mitosis. Front. Cell Dev. Biol. 2015, 3, 74. [Google Scholar] [CrossRef]
- Silverman-Gavrila, R.V.; Wilde, A. Ran Is Required before Metaphase for Spindle Assembly and Chromosome Alignment and after Metaphase for Chromosome Segregation and Spindle Midbody Organization. Mol. Biol. Cell 2006, 17, 2069–2080. [Google Scholar] [CrossRef]
- Hinchcliffe, E.H. The centrosome and bipolar spindle assembly: Does one have anything to do with the other? Cell Cycle 2011, 10, 3841–3848. [Google Scholar] [CrossRef] [PubMed]
- Goupil, A.; Nano, M.; Letort, G.; Gemble, S.; Edwards, F.; Goundiam, O.; Gogendeau, D.; Pennetier, C.; Basto, R. Chromosomes function as a barrier to mitotic spindle bipolarity in polyploid cells. J. Cell Biol. 2020, 219, e201908006. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Yoneda, Y. Small GTPase Ran and Ran-binding proteins. Biomol. Concepts 2012, 3, 703–718. [Google Scholar] [CrossRef]
- Carazo-Salas, R.E.; Gruss, O.J.; Mattaj, I.; Karsenti, E. Ran–GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nature 2001, 3, 228–234. [Google Scholar] [CrossRef]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef]
- Ozugergin, I.; Piekny, A. Complementary functions for the Ran gradient during division. Small GTPases 2020, 12, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Merdes, A.; Cleveland, D.W. Pathways of Spindle Pole Formation: Different Mechanisms; Conserved Components. J. Cell Biol. 1997, 138, 953–956. [Google Scholar] [CrossRef]
- O’Connell, C.B.; Khodjakov, A.L. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 2007, 120, 1717–1722. [Google Scholar] [CrossRef]
- Tsuchiya, K.; Hayashi, H.; Nishina, M.; Okumura, M.; Sato, Y.; Kanemaki, M.T.; Goshima, G.; Kiyomitsu, T. Ran-GTP Is Non-essential to Activate NuMA for Mitotic Spindle-Pole Focusing but Dynamically Polarizes HURP Near Chromosomes. Curr. Biol. 2020, 31, 115–127.e3. [Google Scholar] [CrossRef]
- Carazo-Salas, R.E.; Guarguaglini, G.; Gruss, O.J.; Segref, A.; Karsenti, E.; Mattaj, I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 1999, 400, 178–181. [Google Scholar] [CrossRef]
- Zhang, M.S.; Arnaoutov, A.; Dasso, M. RanBP1 Governs Spindle Assembly by Defining Mitotic Ran-GTP Production. Dev. Cell 2014, 31, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Roostalu, J.; Cade, N.I.; Surrey, T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nature 2015, 17, 1422–1434. [Google Scholar] [CrossRef]
- Safari, M.S.; King, M.R.; Brangwynne, C.P.; Petry, S. Interaction of spindle assembly factor TPX2 with importins-α/β inhibits protein phase separation. J. Biol. Chem. 2021, 297, 100998. [Google Scholar] [CrossRef] [PubMed]
- Kalab, P.; Weis, K.; Heald, R. Visualization of a Ran-GTP Gradient in Interphase and Mitotic Xenopus Egg Extracts. Science 2002, 295, 2452–2456. [Google Scholar] [CrossRef]
- Chang, C.-C.; Chen, C.-J.; Grauffel, C.; Pien, Y.-C.; Lim, C.; Tsai, S.-Y.; Hsia, K.-C. Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-α. Nat. Commun. 2019, 10, 4307. [Google Scholar] [CrossRef]
- Nachury, M.V.; Maresca, T.J.; Salmon, W.S.; Waterman-Storer, C.M.; Heald, R.; Weis, K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 2001, 104, 95–106. [Google Scholar] [CrossRef]
- Torosantucci, L.; De Luca, M.; Guarguaglini, G.; Lavia, P.; Degrassi, F. Localized RanGTP Accumulation Promotes Microtubule Nucleation at Kinetochores in Somatic Mammalian Cells. Mol. Biol. Cell 2008, 19, 1873–1882. [Google Scholar] [CrossRef]
- Wiese, C.; Wilde, A.; Moore, M.S.; Adam, S.A.; Merdes, A.; Zheng, Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 2001, 291, 653–656. [Google Scholar] [CrossRef]
- Ems-McClung, S.C.; Zheng, Y.; Walczak, C.E. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell 2004, 15, 46–57. [Google Scholar] [CrossRef]
- Radulescu, A.E.; Cleveland, D.W. NuMA after 30 years: The matrix revisited. Trends Cell Biol. 2010, 20, 214–222. [Google Scholar] [CrossRef]
- Koffa, M.D.; Casanova, C.M.; Santarella, R.; Köcher, T.; Wilm, M.; Mattaj, l.W. Faculty Opinions recommendation of HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006, 16, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Gruss, O.J.; Carazo-Salas, R.E.; Schatz, C.A.; Guarguaglini, G.; Kast, J.; Wilm, M.; Le Bot, N.; Vernos, I.; Karsenti, E.; Mattaj, I.W. Ran Induces Spindle Assembly by Reversing the Inhibitory Effect of Importin α on TPX2 Activity. Cell 2001, 104, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Roostalu, J.; Surrey, T.; Nogales, E. Structural insight into TPX2-stimulated microtubule assembly. Elife 2017, 6, e30959. [Google Scholar] [CrossRef] [PubMed]
- Heald, R.; Khodjakov, A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J. Cell Biol. 2015, 211, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, H.S.; Clarke, P.R. Cell Biology: Ran, Mitosis and the Cancer Connection. Curr. Biol. 2006, 16, R466–R468. [Google Scholar] [CrossRef]
- Tsai, M.-Y.; Wiese, C.; Cao, K.; Martin, O.; Donovan, P.; Ruderman, J.; Prigent, C.; Zheng, Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature 2003, 5, 242–248. [Google Scholar] [CrossRef]
- Silljé, H.H.; Nagel, S.; Körner, R.; Nigg, E. HURP Is a Ran-Importin β-Regulated Protein that Stabilizes Kinetochore Microtubules in the Vicinity of Chromosomes. Curr. Biol. 2006, 16, 731–742. [Google Scholar] [CrossRef]
- Brouhard, G.J.; Stear, J.H.; Noetzel, T.L.; Al-Bassam, J.; Kinoshita, K.; Harrison, S.C.; Howard, J.; Hyman, A.A. XMAP215 Is a Processive Microtubule Polymerase. Cell 2008, 132, 79–88. [Google Scholar] [CrossRef]
- Askjaer, P.; Galy, V.; Hannak, E.; Mattaj, I.W. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 2002, 13, 4355–4370. [Google Scholar] [CrossRef]
- Milano, S.K.; Kwon, W.; Pereira, R.; Antonyak, M.A.; Cerione, R.A. Characterization of a Novel Activated Ran GTPase Mutant and Its Ability to Induce Cellular Transformation. J. Biol. Chem. 2012, 287, 24955–24966. [Google Scholar] [CrossRef]
- Azuma, K.; Sasada, T.; Takedatsu, H.; Shomura, H.; Koga, M.; Maeda, Y.; Yao, A.; Hirai, T.; Takabayashi, A.; Shichijo, S.; et al. Ran, a small GTPase gene, encodes cytotoxic T lymphocyte (CTL) epitopes capable of inducing HLA-A33-restricted and tumor-reactive CTLs in cancer patients. Clin. Cancer Res. 2004, 10, 6695–6702. [Google Scholar] [CrossRef]
- Shevde, L.A.; Samant, R.S. Role of osteopontin in the pathophysiology of cancer. J. Int. Soc. Matrix Biol. 2014, 37, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Khodavirdi, A.C.; Song, Z.; Yang, S.; Zhong, C.; Wang, S.; Wu, H.; Pritchard, C.; Nelson, P.S.; Roy-Burman, P. Increased Expression of Osteopontin Contributes to the Progression of Prostate Cancer. Cancer Res. 2006, 66, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, W.; He, J.; Jiang, L.; Li, X. Osteopontin as a biomarker for osteosarcoma therapy and prognosis (Review). Oncol. Lett. 2019, 17, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Shang, Y.; Guo, S.; Liu, C.; Zhou, L.; Sun, Y.; Nie, Y.; Fan, D.; Lu, Y.; Guo, X. Ran GTPase protein promotes metastasis and invasion in pancreatic cancer by deregulating the expression of AR and CXCR4. Cancer Biol. Ther. 2014, 15, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Kamai, T.; Shirataki, H.; Oyama, T.; Arai, K.; Yoshida, K.-I. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int. J. Cancer 2008, 122, 2391–2397. [Google Scholar] [CrossRef] [PubMed]
- van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. Mol. Mech. Mutagen. 2011, 728, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, V.; Provencher, D.M.; Maugard, C.M.; Page, C.L.; Ren, F.; Lussier, C.; Novak, J.; Ge, B.; Hudson, T.J.; Tonin, P.N.; et al. Discrimination between serous low malignant potential and invasive epithelial ovarian tumors using molecular profiling. Oncogene 2005, 24, 4672–4687. [Google Scholar] [CrossRef]
- Cáceres-Gorriti, K.Y.; Carmona, E.; Barrès, V.; Rahimi, K.; Létourneau, I.J.; Tonin, P.N.; Provencher, D.; Mes-Masson, A.-M. RAN Nucleo-Cytoplasmic Transport and Mitotic Spindle Assembly Partners XPO7 and TPX2 Are New Prognostic Biomarkers in Serous Epithelial Ovarian Cancer. PLoS ONE 2014, 9, e91000. [Google Scholar] [CrossRef]
- Wei, Z.; Duan, X.; Li, Q.; Li, Q.; Wang, Y. High expression of Ran binding protein 1 predicts poor outcomes in hepatocellular carcinoma patients: A Cancer Genome Atlas database analysis. J. Gastrointest. Oncol. 2021, 12, 2966–2984. [Google Scholar] [CrossRef]
- Ning, S.; Li, H.; Qiao, K.; Wang, Q.; Shen, M.; Kang, Y.; Yin, Y.; Liu, J.; Liu, L.; Hou, S.; et al. Identification of long-term survival-associated gene in breast cancer. Aging 2020, 12, 20332–20349. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Wheler, J.J.; Westin, S.N.; Moulder, S.L.; Naing, A.; Tsimberidou, A.M.; Fu, S.; Falchook, G.S.; Hong, D.S.; Garrido-Laguna, I.; et al. PI3K/AKT/mTOR Inhibitors in Patients with Breast and Gynecologic Malignancies Harboring PIK3CA Mutations. J. Clin. Oncol. 2012, 30, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; Venet, D.; Brohée, S.; Pondé, N.; Sotiriou, C. pAKT pathway activation is associated with PIK3CA mutations and good prognosis in luminal breast cancer in contrast to p-mTOR pathway activation. NPJ Breast Cancer 2019, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Phillips, W.A.; Loi, S. PIK3CA mutations in breast cancer: Reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014, 16, 201. [Google Scholar] [CrossRef]
- Yip, P.Y. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl. Lung Cancer Res. 2015, 4, 165–176. [Google Scholar] [PubMed]
- Jančík, S.; Drábek, J.; Radzioch, D.; Hajdúch, M. Clinical Relevance of KRAS in Human Cancers. J. Biomed. Biotechnol. 2010, 2010, 150960. [Google Scholar] [CrossRef]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Condorelli, A.G.; Battaglia, R.; Tamburello, L.; Barbagallo, D.; Di Pietro, C.; Purrello, M. Non-coding landscapes of colorectal cancer. World J. Gastroenterol. 2015, 21, 11709–11739. [Google Scholar] [CrossRef]
- Rivas-Ortiz, C.I.; Lopez-Vidal, Y.; Arredondo-Hernandez, L.J.R.; Castillo-Rojas, G. Genetic Alterations in Gastric Cancer Associated with Helicobacter pylori Infection. Front. Med. 2017, 4, 47. [Google Scholar] [CrossRef]
- Martens-de Kemp, S.R.; Nagel, R.; Stigter-van Walsum, M.; Van Der Meulen, I.H.; Van Beusechem, V.W.; Braakhuis, B.J.; Brakenhoff, R.H. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin. Cancer Res. 2013, 19, 1994–2003. [Google Scholar] [CrossRef]
- Xia, F.; Dohi, T.; Martin, N.M.; Raskett, C.M.; Liu, Q.; Altieri, D.C. Essential Role of the Small GTPase Ran in Postnatal Pancreatic Islet Development. PLoS ONE 2011, 6, e27879. [Google Scholar] [CrossRef] [PubMed]
- Barrès, V.; Ouellet, V.; Lafontaine, J.; Tonin, P.N.; Provencher, D.M. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol. Cancer 2010, 9, 272. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Lu, Y.; Qin, H.; Zhou, Y.; Gu, Y.; Zhou, J.; Wang, X.; Fan, D. High Ran level is correlated with poor prognosis in patients with colorectal cancer. Int. J. Clin. Oncol. 2012, 18, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Bin Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Lui, G.Y.; Kovacevic, Z.; Richardson, V.; Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget 2015, 6, 18748–18779. [Google Scholar] [CrossRef]
- Radisavljevic, Z.M.; González-Flecha, B. TOR kinase and Ran are downstream from PI3K/Akt in H2O2-induced mitosis. J. Cell Biochem. 2004, 91, 1293–1300. [Google Scholar] [CrossRef]
- Pistritto, G.; Trisciuoglio, D.; Ceci, C.; Garufi, A.; D’Orazi, G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 2016, 8, 603–619. [Google Scholar] [CrossRef]
- Woo, S.-U.; Sangai, T.; Akcakanat, A.; Chen, H.; Wei, C.; Meric-Bernstam, F. Vertical inhibition of the PI3K/Akt/mTOR pathway is synergistic in breast cancer. Oncogenesis 2017, 6, e385. [Google Scholar] [CrossRef]
- Morgan-Lappe, S.E.; Tucker, L.A.; Huang, X.; Zhang, Q.; Sarthy, A.V.; Zakula, D.; Vernetti, L.; Schurdak, M.; Wang, J.; Fesik, S.W. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 2007, 67, 4390–4398. [Google Scholar] [CrossRef]
- Yu, C.; Luo, D.; Yu, J.; Zhang, M.; Zheng, X.; Xu, G.; Wang, J.; Wang, H.; Xu, Y.; Jiang, K.; et al. Genome-wide CRISPR-cas9 knockout screening identifies GRB7 as a driver for MEK inhibitor resistance in KRAS mutant colon cancer. Oncogene 2022, 41, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Manning, B.D. A Molecular Link between AKT Regulation and Chemotherapeutic Response. Cancer Cell 2009, 16, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Usatyuk, P.V.; Fu, P.; Mohan, V.; Epshtein, Y.; Jacobson, J.R.; Gomez-Cambronero, J.; Wary, K.K.; Bindokas, V.; Dudek, S.M.; Salgia, R.; et al. Role of c-Met/phosphatidylinositol 3-kinase (PI3k)/Akt signaling in hepatocyte growth factor (HGF)-mediated lamellipodia formation, reactive oxygen species (ROS) generation, and motility of lung endothelial cells. J. Biol. Chem. 2014, 289, 13476–13491. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, Y.; Liu, G.; Li, C.; Song, Y.; Cao, Z.; Li, W.; Hu, J.; Lu, C.; Liu, Y. PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis. 2020, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.G.; Burrows, J.; Salgia, R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005, 225, 797. [Google Scholar] [CrossRef]
- Beviglia, L.; Matsumoto, K.; Lin, C.S.; Ziober, B.L.; Kramer, R.H. Expression of the c-Met/HGF receptor in human breast carcinoma: Correlation with tumor progression. Int. J. Cancer 1997, 74, 301–309. [Google Scholar] [CrossRef]
- Ho-Yen, C.M.; Jones, J.L.; Kermorgant, S. The clinical and functional significance of c-Met in breast cancer: A review. Breast Cancer Res. 2015, 17, 52. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, M.; Jin, K.; Wang, S.; Wei, H.; Fan, C.; Wu, Y.; Li, X.; Li, X.; Li, G.; et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 2018, 17, 45. [Google Scholar] [CrossRef]
- Salgia, R. Role of c-Met in Cancer: Emphasis on Lung Cancer. Semin. Oncol. 2009, 36, S52–S58. [Google Scholar] [CrossRef]
- Tanizaki, J.; Okamoto, I.; Okamoto, K.; Takezawa, K.; Kuwata, K.; Yamaguchi, H.; Nakagawa, K. MET Tyrosine Kinase Inhibitor Crizotinib (PF-02341066) Shows Differential Antitumor Effects in Non-small Cell Lung Cancer According to MET Alterations. J. Thorac. Oncol. 2011, 6, 1624–1631. [Google Scholar] [CrossRef]
- Razeghian, E.; Suksatan, W.; Rahman, H.S.; Bokov, D.O.; Abdelbasset, W.K.; Hassanzadeh, A.; Marofi, F.; Yazdanifar, M.; Jarahian, M. Harnessing TRAIL-Induced Apoptosis Pathway for Cancer Immunotherapy and Associated Challenges. Front. Immunol. 2021, 12, 699746. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-O.; Shin, S.; Liu, Y.; Ballif, B.A.; Woo, M.S.; Gygi, S.P.; Blenis, J. Ran-Binding Protein 3 Phosphorylation Links the Ras and PI3-Kinase Pathways to Nucleocytoplasmic Transport. Mol. Cell 2008, 29, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, Y.; Liu, S.; Tao, T.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; He, Z.; Li, J.; et al. EGF-induced nuclear localization of SHCBP1 activates β-catenin signaling and promotes cancer progression. Oncogene 2018, 38, 747–764. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Franklin, R.A.; Montalto, G.; Cervello, M.; Libra, M.; Candido, S.; Malaponte, G.; et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR Cascade Inhibitors: How Mutations Can Result in Therapy Resistance and How to Overcome Resistance. Oncotarget 2012, 3, 1068–1111. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Zhang, N.; Xu, Y.; Li, X.; Shi, D.; Wang, Y.; Li, D.; Cai, S. TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J. Transl. Med. 2013, 11, 313. [Google Scholar] [CrossRef]
- Scrace, S.F.; O’Neill, E. RASSF Signalling and DNA Damage: Monitoring the Integrity of the Genome? Mol. Biol. Int. 2012, 2012, 141732. [Google Scholar] [CrossRef]
- Donninger, H.; Vos, M.D.; Clark, G.J. The RASSF1A tumor suppressor. J. Cell Sci. 2007, 120, 3163–3172. [Google Scholar] [CrossRef]
- Moore, W.J.; Zhang, C.; Clarke, P.R. Targeting of RCC1 to Chromosomes Is Required for Proper Mitotic Spindle Assembly in Human Cells. Curr. Biol. 2002, 12, 1442–1447. [Google Scholar] [CrossRef]
- Dallol, A.; Hesson, L.B.; Matallanas, D.; Cooper, W.N.; O’Neill, E.; Maher, E.R.; Kolch, W.; Latif, F. RAN GTPase Is a RASSF1A Effector Involved in Controlling Microtubule Organization. Curr. Biol. 2009, 19, 1227–1232. [Google Scholar] [CrossRef]
- Bai, H.; Lester, G.S.; Petishnok, L.C.; Dean, D.A. Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep. 2017, 37, BSR20160616. [Google Scholar] [CrossRef] [PubMed]
- Lüdke, D.; Rohmann, P.F.W.; Wiermer, M. Nucleocytoplasmic Communication in Healthy and Diseased Plant Tissues. Front. Plant Sci. 2021, 12, 719453. [Google Scholar] [CrossRef] [PubMed]
- Crouch, J.; Shvedova, M.; Thanapaul, R.J.R.S.; Botchkarev, V.; Roh, D. Epigenetic Regulation of Cellular Senescence. Cells 2022, 11, 672. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Tanani, M.; Nsairat, H.; Mishra, V.; Mishra, Y.; Aljabali, A.A.A.; Serrano-Aroca, Á.; Tambuwala, M.M. Ran GTPase and Its Importance in Cellular Signaling and Malignant Phenotype. Int. J. Mol. Sci. 2023, 24, 3065. https://doi.org/10.3390/ijms24043065
El-Tanani M, Nsairat H, Mishra V, Mishra Y, Aljabali AAA, Serrano-Aroca Á, Tambuwala MM. Ran GTPase and Its Importance in Cellular Signaling and Malignant Phenotype. International Journal of Molecular Sciences. 2023; 24(4):3065. https://doi.org/10.3390/ijms24043065
Chicago/Turabian StyleEl-Tanani, Mohamed, Hamdi Nsairat, Vijay Mishra, Yachana Mishra, Alaa A. A. Aljabali, Ángel Serrano-Aroca, and Murtaza M. Tambuwala. 2023. "Ran GTPase and Its Importance in Cellular Signaling and Malignant Phenotype" International Journal of Molecular Sciences 24, no. 4: 3065. https://doi.org/10.3390/ijms24043065
APA StyleEl-Tanani, M., Nsairat, H., Mishra, V., Mishra, Y., Aljabali, A. A. A., Serrano-Aroca, Á., & Tambuwala, M. M. (2023). Ran GTPase and Its Importance in Cellular Signaling and Malignant Phenotype. International Journal of Molecular Sciences, 24(4), 3065. https://doi.org/10.3390/ijms24043065









