Gluten Exorphins Promote Cell Proliferation through the Activation of Mitogenic and Pro-Survival Pathways
Abstract
1. Introduction
2. Results
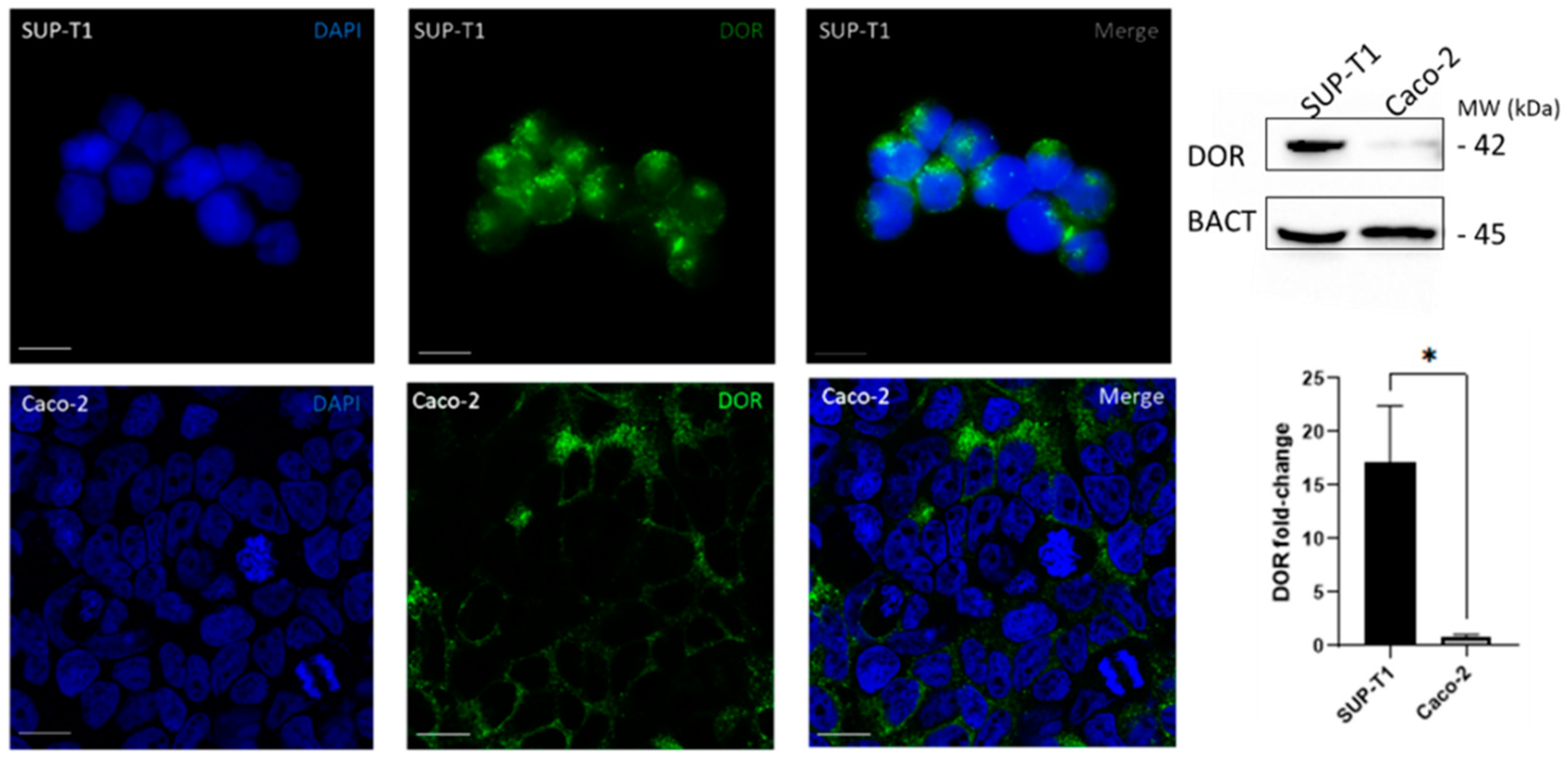
2.1. SUP-T1 and Caco-2 Cells Differentially Express DOR
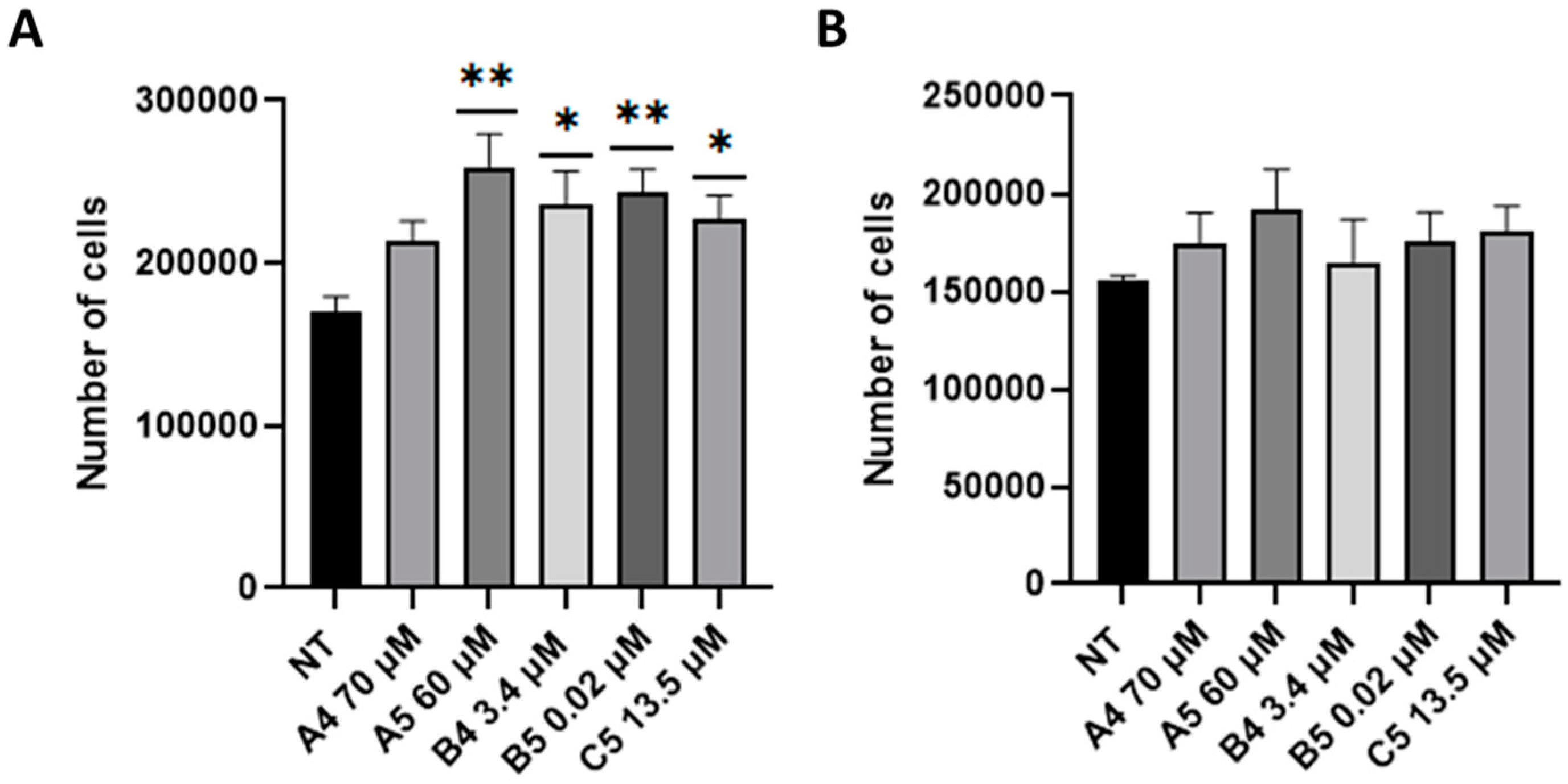
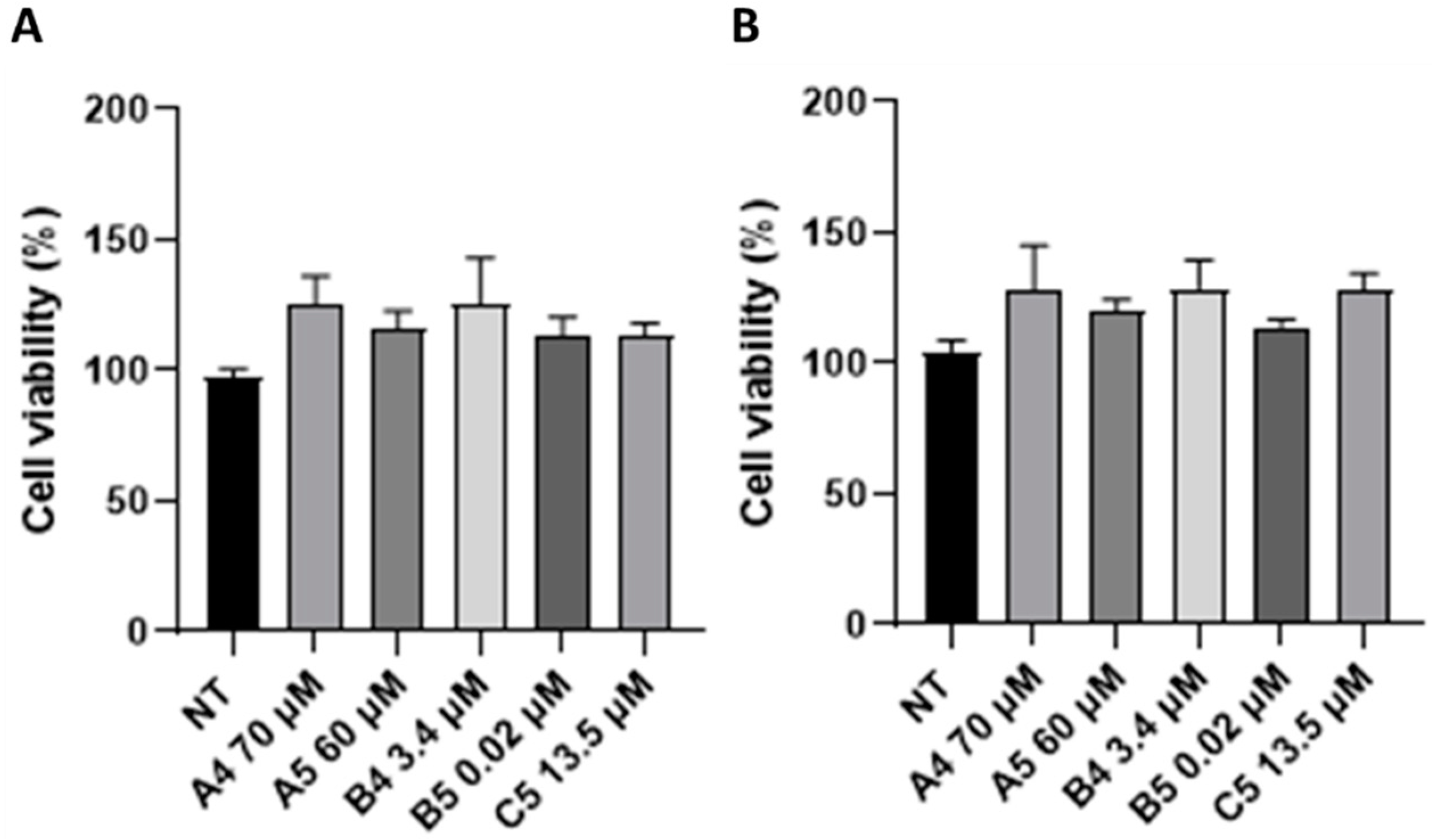
2.2. GEs Treatment Promoted SUP-T1 Cells Proliferation
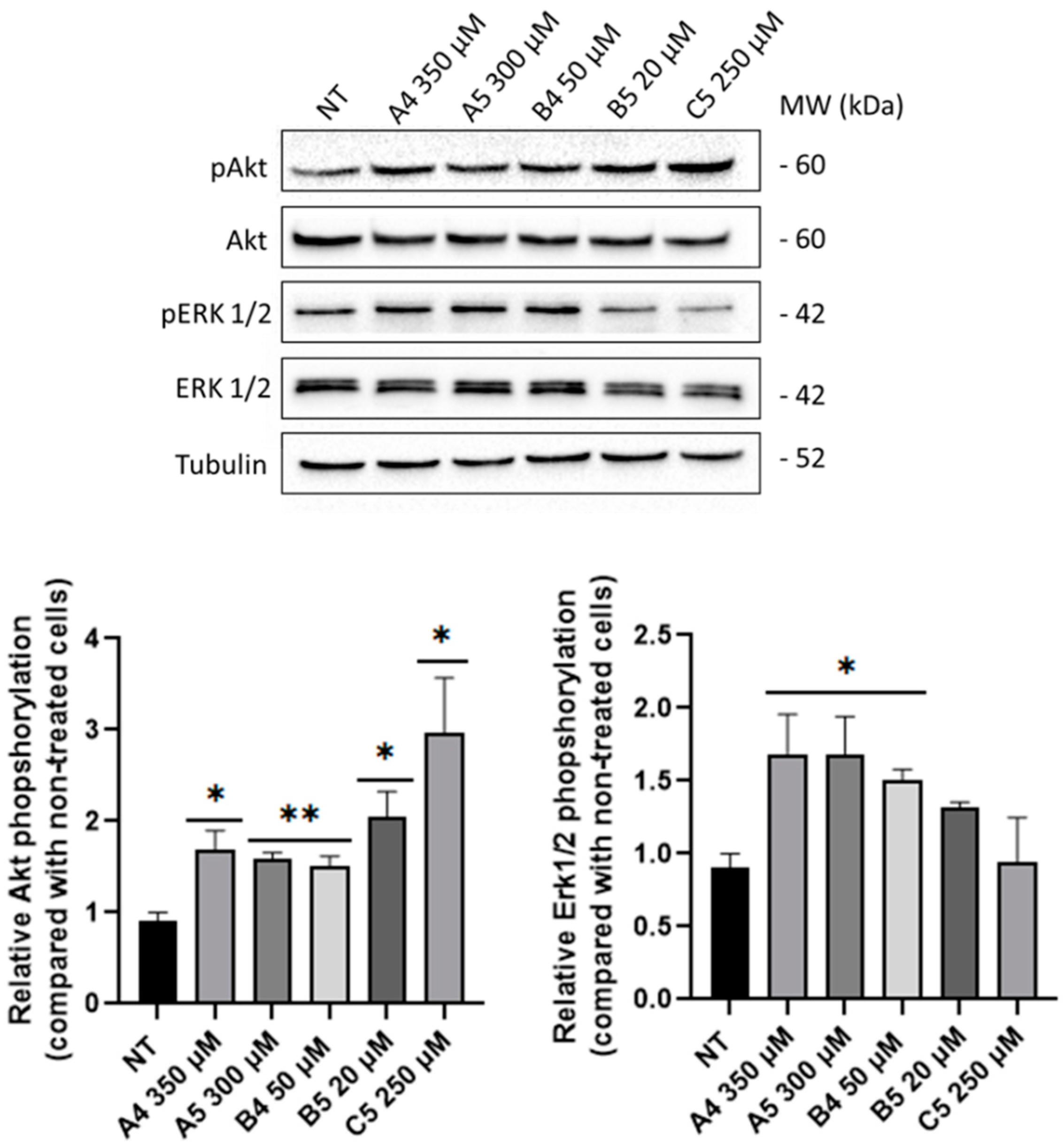
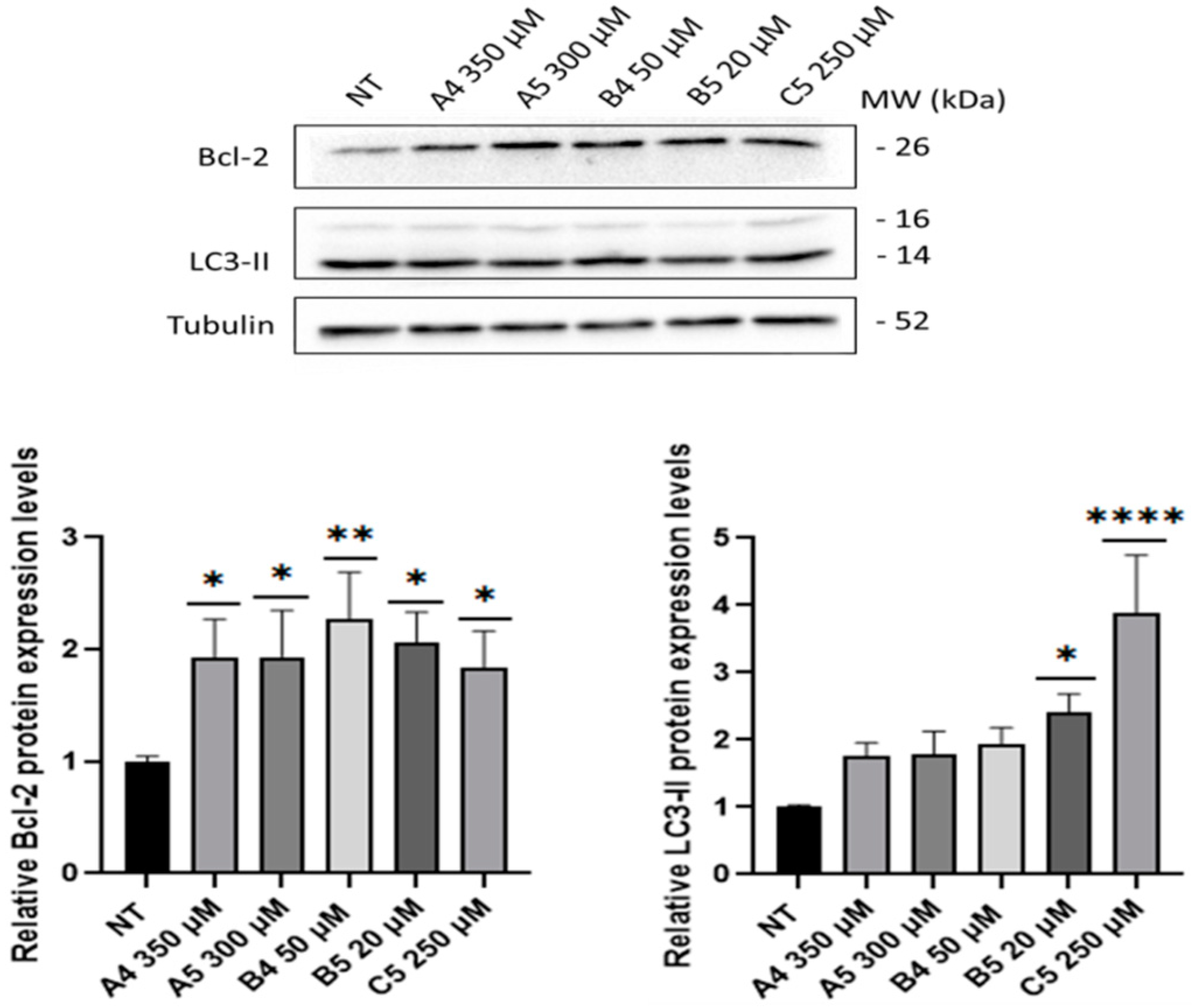
2.3. GEs Treatment Induced Mitogenic and Pro-Survival Pathways in SUP-T1 Cells
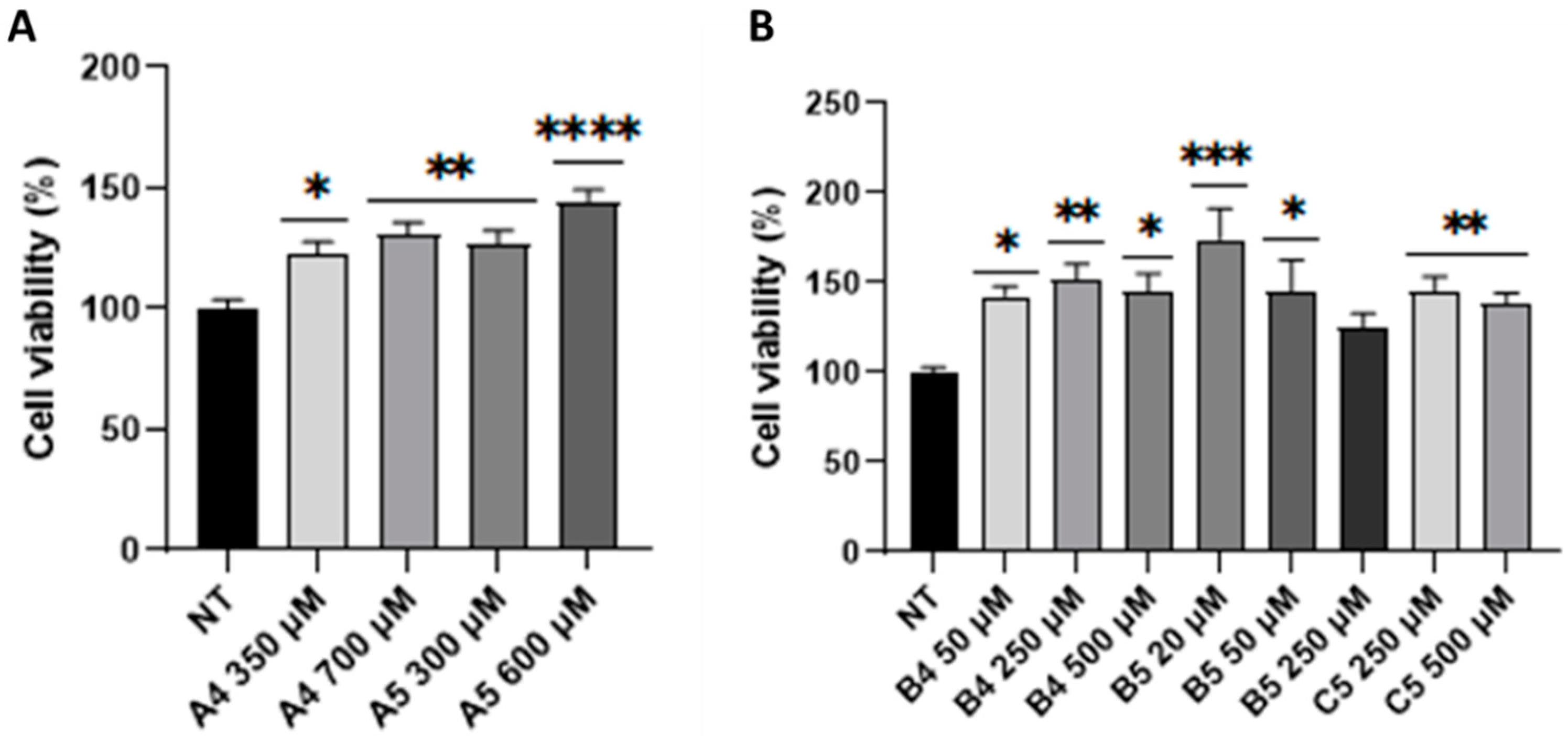
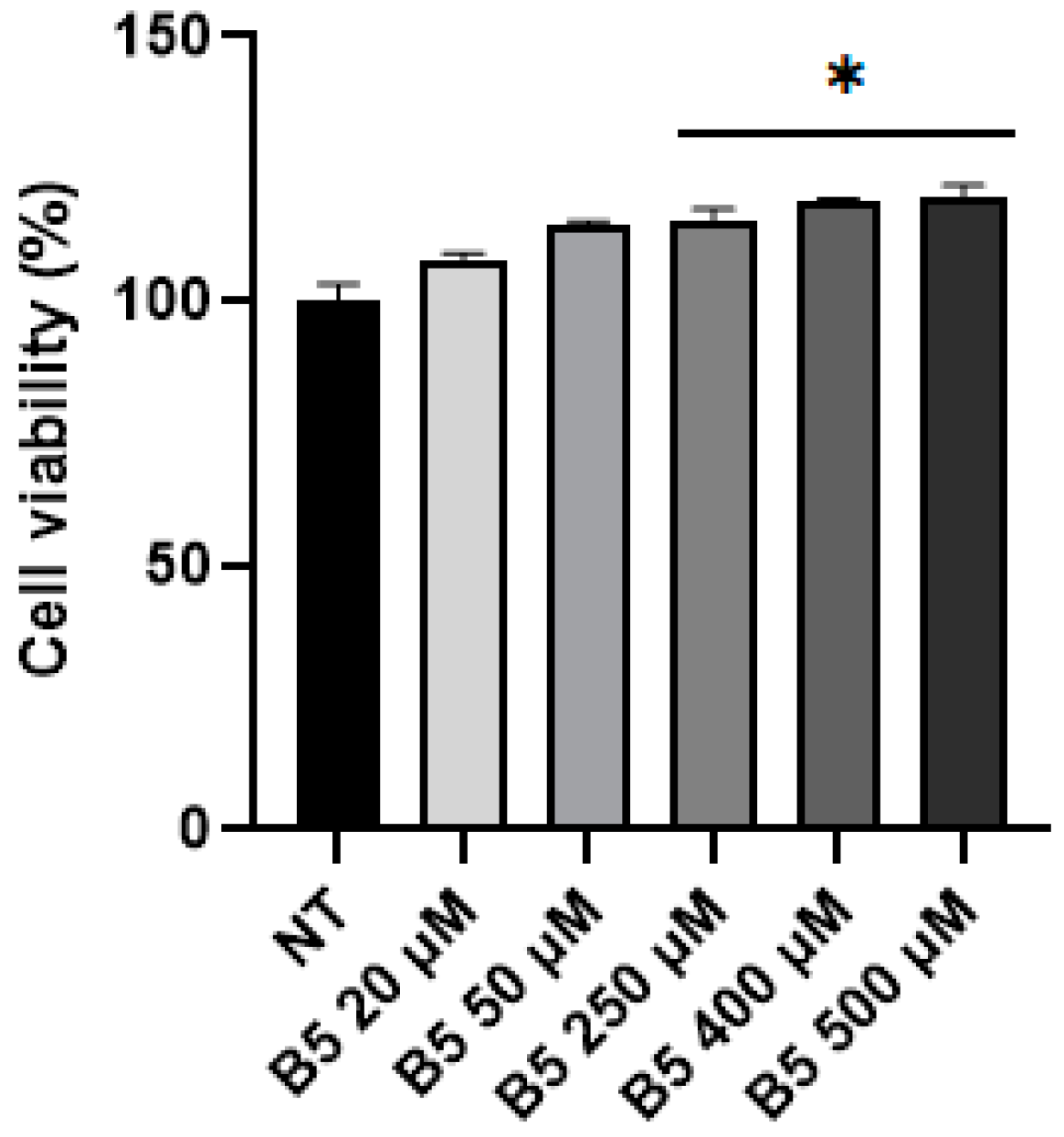
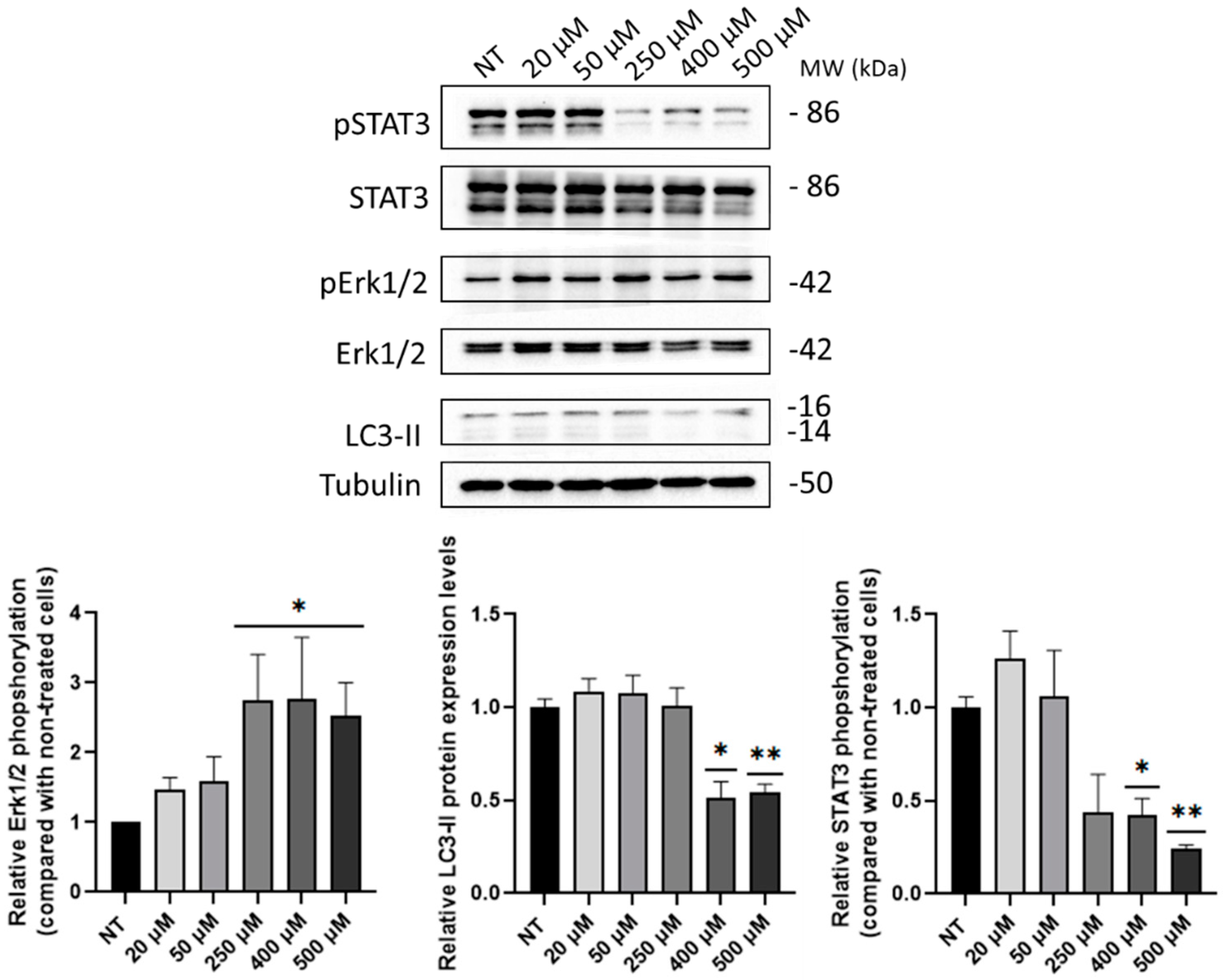
2.4. B5 Exorphin Induced Mitogenic and Pro-Survival Pathways in Caco-2 Cells
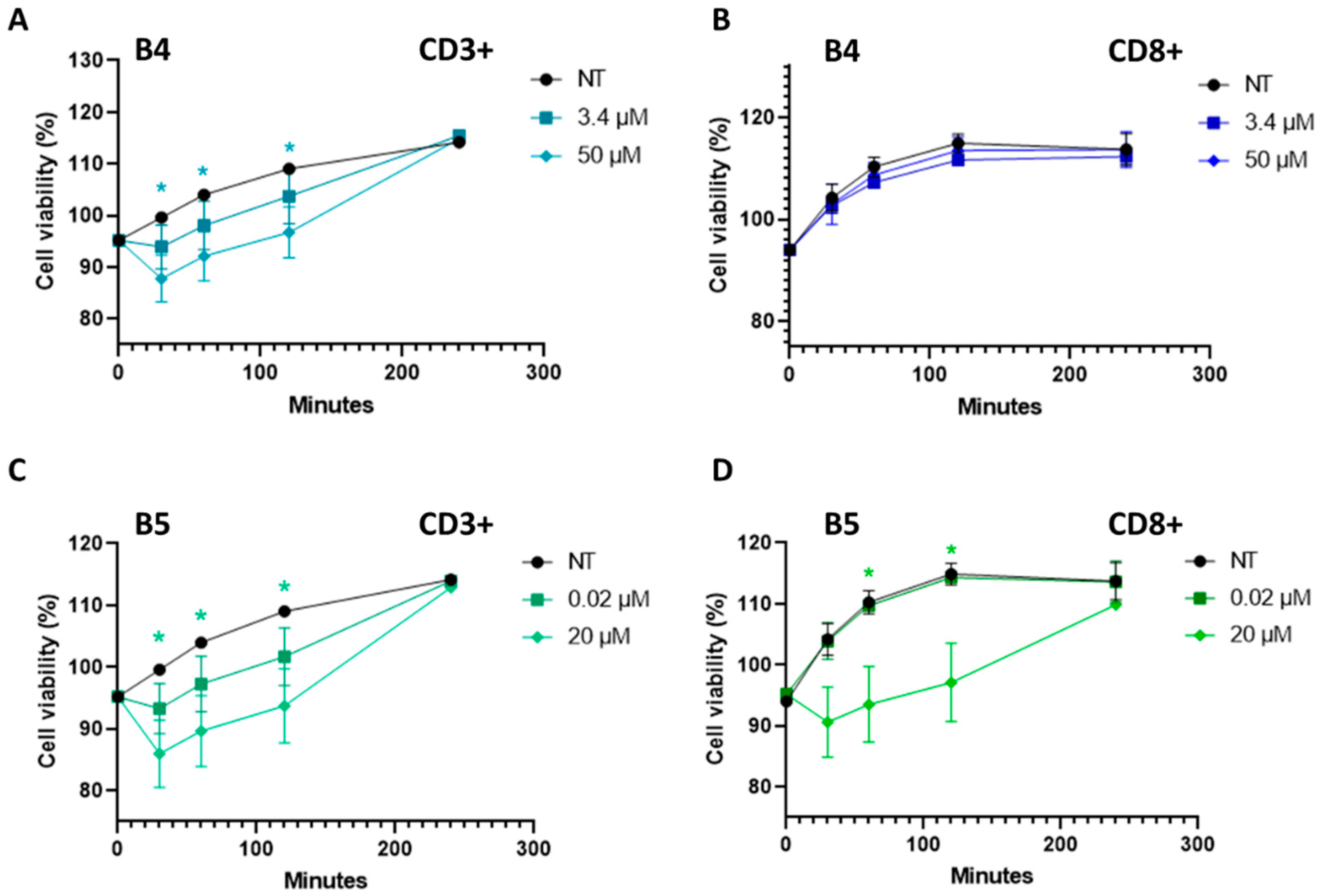
2.5. Gluten B Exorphins Did Not Induce Increase in Viability in Normal CD3+ and CD8+ T-Lymphocytes
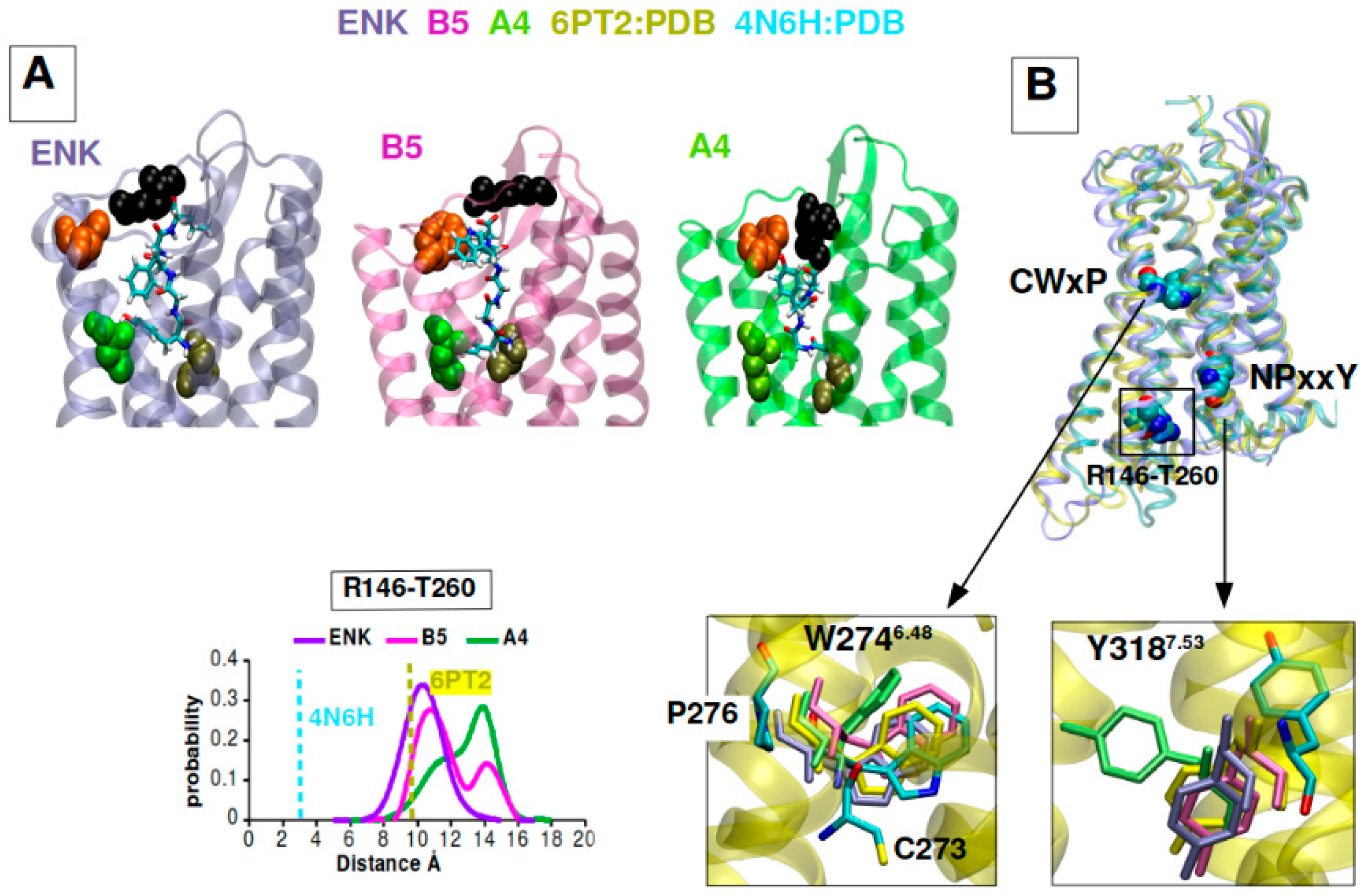
2.6. B5 and A4 Showed a Differential DOR Dynamic Profile of Activation in Computational Models
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and GEs Administration
4.2. Immunofluorescence Analysis
4.3. MTS Viability Assay
4.4. Cytofluorimetric Analysis
4.5. Immunoblotting Analysis
4.6. Molecular Dynamics Simulations
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Sabatino, A.; Corazza, G.R. Coeliac disease. Lancet 2009, 373, 1480–1493. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Di Sabatino, A.; Corazza, G.R. The immune recognition of gluten in coeliac disease. Clin. Exp. Immunol. 2005, 140, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Leffler, D.A.; Bai, J.C.; Biagi, F.; Fasano, A.; Green, P.H.; Hadjivassiliou, M.; Kaukinen, K.; Kelly, C.P.; Leonard, J.N.; et al. The Oslo definitions for coeliac disease and related terms. Gut 2013, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Di Sabatino, A.; Corazza, G.R. Some clarification is necessary on the Oslo definitions for coeliac disease-related terms. Gut 2013, 62, 182. [Google Scholar] [CrossRef]
- Mustalahti, K.; Catassi, C.; Reunanen, A.; Fabiani, E.; Heier, M.; McMillan, S.; Murray, L.; Metzger, M.H.; Gasparin, M.; Bravi, E.; et al. Coeliac EU Cluster, Project Epidemiology. The prevalence of celiac disease in Europe: Results of a centralized, international mass screening project. Ann. Med. 2010, 42, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, S.; Jinsmaa, Y.; Matsukawa, T.; Sasaki, R.; Yoshikawa, M. Release of opioid peptides, gluten exorphins by the action of pancreatic elastase. FEBS Lett. 1997, 412, 475–479. [Google Scholar] [CrossRef]
- Zioudrou, C.; Streaty, R.A.; Klee, W.A. Opioid peptides derived from food proteins. The exorphins. J. Biol. Chem. 1979, 254, 2446–2449. [Google Scholar] [CrossRef]
- Pruimboom, L.; de Punder, K. The opioid effects of gluten exorphins: Asymptomatic celiac disease. J. Health Popul. Nutr. 2015, 33, 24. [Google Scholar] [CrossRef]
- Schusdziarra, V.; Henrichs, I.; Holland, A.; Klier, M.; Pfeiffer, E.F. Evidence for an effect of exorphins on plasma insulin and glucagon levels in dogs. Diabetes 1981, 30, 362–364. [Google Scholar] [CrossRef]
- Morley, J.E.; Levine, A.S.; Yamada, T.; Gebhard, R.L.; Prigge, W.F.; Shafer, R.B.; Goetz, F.C.; Silvis, S.E. Effect of exorphins on gastrointestinal function, hormonal release, and appetite. Gastroenterology 1983, 84, 1517–1523. [Google Scholar] [CrossRef]
- Fukudome, S.; Yoshikawa, M. Opioid peptides derived from wheat gluten: Their isolation and characterization. FEBS Lett. 1992, 296, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Fukudome, S.; Yoshikawa, M. Gluten exorphin C. A novel opioid peptide derived from wheat gluten. FEBS Lett. 1993, 316, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Gendron, L.; Cahill, C.M.; von Zastrow, M.; Schiller, P.W.; Pineyro, G. Molecular Pharmacology of δ-Opioid Receptors. Pharmacol Rev. 2016, 68, 631–700. [Google Scholar] [CrossRef] [PubMed]
- Stuknyte, M.; Maggioni, M.; Cattaneo, S.; De Luca, P.; Fiorilli, A.; Ferraretto, A.; De Noni, I. Release of wheat gluten exorphins A5 and C5 during in vitro gastrointestinal digestion of bread and pasta and their absorption through an in vitro model of intestinal epithelium. Food Res. Intern. 2015, 72, 208–214. [Google Scholar] [CrossRef]
- Maggioni, M.; Stuknyte, M.; De Luca, P.; Cattaneo, S.; Fiorilli, A.; De Noni, I.; Ferraretto, A. Transport of wheat gluten exorphins A5 and C5 through an in vitro model of intestinal epithelium. Food Res. Intern. 2016, 88, 319–326. [Google Scholar] [CrossRef]
- DiCello, J.J.; Saito, A.; Rajasekhar, P.; Eriksson, E.M.; McQuade, R.M.; Nowell, C.J.; Sebastian, B.W.; Fichna, J.; Veldhuis, N.A.; Canals, M.; et al. Inflammation-associated changes in DOR expression and function in the mouse colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G544–G559. [Google Scholar] [CrossRef]
- Cahill, C.M.; Morinville, A.; Hoffert, C.; O’Donnell, D.; Beaudet, A. Up-regulation and trafficking of delta opioid receptor in a model of chronic inflammation: Implications for pain control. Pain 2003, 101, 199–208. [Google Scholar] [CrossRef]
- Pol, O.; Palacio, J.R.; Puig, M.M. The expression of delta- and kappa-opioid receptor is enhanced during intestinal inflammation in mice. J. Pharmacol. Exp. Ther. 2003, 306, 455–462. [Google Scholar] [CrossRef]
- Pol, O.; Puig, M.M. Expression of opioid receptors during peripheral inflammation. Curr. Top. Med. Chem. 2004, 4, 51–61. [Google Scholar] [CrossRef]
- Singleton, P.A.; Mirzapoiazova, T.; Hasina, R.; Salgia, R.; Moss, J. Increased μ-opioid receptor expression in metastatic lung cancer. Br. J. Anaesth. 2014, 113, i103–i108. [Google Scholar] [CrossRef]
- Chen, D.; Chen, Y.; Yan, Y.; Pan, J.; Xing, W.; Li, Q.; Zeng, W. Down-regulation of the tumour suppressor κ-opioid receptor predicts poor prognosis in hepatocellular carcinoma patients. BMC Cancer 2017, 17, 553. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.C.; Zhang, B.; Li, X.; Liu, X.M.; Zhang, J.; Lei, B.; Li, B.; Zhai, R.; Chen, Q.; Li, Y. Upregulation and activation of δ-opioid receptors promotes the progression of human breast cancer. Oncol. Rep. 2016, 36, 2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Li, Y.; Yuan, S.; Tomlinson, S.; He, S. Upregulation of the δ opioid receptor in liver cancer promotes liver cancer progression both in vitro and in vivo. Int. J. Oncol. 2013, 43, 1281–1290. [Google Scholar] [CrossRef]
- Debruyne, D.; Leroy, A.; De Wever, O.; Vakaet, L.; Mareel, M.; Bracke, M. Direct effects of delta opioid receptor agonists on invasion-associated activities of HCT-8/E11 colon cancer cells. Anticancer Res. 2010, 30, 9–17. [Google Scholar]
- Szczepaniak, A.; Fichna, J.; Zielińska, M. Opioids in Cancer Development, Progression and Metastasis: Focus on Colorectal Cancer. Curr. Treat. Options Oncol. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Chander, U.; Leeman-Neill, R.J.; Bhagat, G. Pathogenesis of enteropathy-associated T cell lymphoma. Curr. Hematol. Malig. Rep. 2018, 13, 308–317. [Google Scholar] [CrossRef]
- Marafini, I.; Monteleone, G.; Stolfi, C. Association Between Celiac Disease and Cancer. Int. J. Mol. Sci. 2020, 21, 4155. [Google Scholar] [CrossRef] [PubMed]
- Bayani, J.; Squire, J.A. Preparation of cytogenetic specimens from tissue samples. Current Protoc. Cell. Biol. 2004, 23, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Grundmann, M.; Kostenis, E. Temporal Bias: Time-Encoded Dynamic GPCR Signaling. Trends Pharmacol. Sci. 2017, 38, 1110–1124. [Google Scholar] [CrossRef]
- Ruvolo, P.P.; Deng, X.; May, W.S. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 2001, 4, 515–522. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [PubMed]
- Han, J.; Theiss, A.L. Stat3: Friend or foe in colitis and colitis-associated cancer? Inflamm. Bowel Dis. 2014, 20, 2405–2411. [Google Scholar] [CrossRef] [PubMed]
- Katritch, V.; Cherezov, V.; Stevens, R.C. Structure-function of the G protein-coupled receptor superfamily. Annual Rev Pharmacol. Toxicol. 2013, 53, 531–556. [Google Scholar] [CrossRef] [PubMed]
- Audet, N.; Paquin-Gobeil, M.; Landry-Paquet, O.; Schiller, P.W.; Piñeyro, G. Internalization and Src activity regulate the time course of ERK activation by delta opioid receptor ligands. J. Biol. Chem. 2005, 280, 7808–7816. [Google Scholar] [CrossRef]
- Eisinger, D.A.; Schulz, R. Extracellular signal-regulated kinase/mitogen-activated protein kinases block internalization of delta-opioid receptors. J. Pharmacol. Exp. Ther. 2004, 309, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hong, M.H.; Zhang, L.S.; Hou, Y.Y.; Wang, Y.H.; Wang, F.F.; Chen, Y.J.; Xu, X.J.; Chen, J.; Xie, X.; et al. Serine 363 of the {delta}-opioid receptor is crucial for adopting distinct pathways to activate ERK1/2 in response to stimulation with different ligands. J. Cell. Sci. 2010, 123, 4259–4270. [Google Scholar] [CrossRef]
- Heiss, A.; Ammer, H.; Eisinger, D.A. delta-Opioid receptor-stimulated Akt signaling in neuroblastoma x glioma (NG108-15) hybrid cells involves receptor tyrosine kinase-mediated PI3K activation. Exp. Cell. Res. 2009, 315, 2115–2125. [Google Scholar] [CrossRef]
- Wilson, M.A.; Burt, A.R.; Milligan, G.; Anderson, N.G. Mitogenic signalling by delta opioid receptors expressed in rat-1 fibroblasts involves activation of the p70s6k/p85s6k S6 kinase. Biochem. J. 1997, 325, 217–222. [Google Scholar] [CrossRef]
- Berven, L.A.; Willard, F.S.; Crouch, M.F. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell. Res. 2004, 296, 183–195. [Google Scholar] [CrossRef]
- Malamut, G.; El Machhour, R.; Montcuquet, N.; Martin-Lannerée, S.; Dusanter-Fourt, I.; Verkarre, V.; Mention, J.J.; Rahmi, G.; Kiyono, H.; Butz, E.A.; et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease-associated inflammation and lymphomagenesis. J. Clin. Invest. 2010, 120, 2131–2143. [Google Scholar] [CrossRef]
- Fairlie, W.D.; Tran, S.; Lee, E.F. Crosstalk between apoptosis and autophagy signaling pathways. Int. Rev. Cell. Mol. Biol. 2020, 352, 115–158. [Google Scholar] [PubMed]
- Manai, F.; Azzalin, A.; Gabriele, F.; Martinelli, C.; Morandi, M.; Biggiogera, M.; Bozzola, M.; Comincini, S. The in vitro effects of enzymatic digested gliadin on the functionality of the autophagy process. Int. J. Mol. Sci. 2018, 19, 635. [Google Scholar] [CrossRef]
- Manai, F.; Azzalin, A.; Morandi, M.; Riccardi, V.; Zanoletti, L.; Dei Giudici, M.; Gabriele, F.; Martinelli, C.; Bozzola, M.; Comincini, S. Trehalose modulates autophagy process to counteract gliadin cytotoxicity in an in vitro celiac disease model. Cells 2019, 8, 348. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.K.; Tait, S.W.; Lamkanfi, M.; Amer, A.O.; Nunez, G.; Pagès, G.; Pouysségur, J.; McGargill, M.A.; Green, D.R.; Kanneganti, T.D. TLR2 and RIP2 pathways mediate autophagy of Listeria monocytogenes via extracellular signal-regulated kinase (ERK) activation. J. Biol. Chem. 2011, 286, 42981–42991. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell. Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Pickert, G.; Neufert, C.; Leppkes, M.; Zheng, Y.; Wittkopf, N.; Warntjen, M.; Lehr, H.A.; Hirth, S.; Weigmann, B.; Wirtz, S.; et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 2009, 206, 1465–1472. [Google Scholar] [PubMed]
- Neufert, C.; Pickert, G.; Zheng, Y.; Wittkopf, N.; Warntjen, M.; Nikolaev, A.; Ouyang, W.; Neurath, M.F.; Becker, C. Activation of epithelial STAT3 regulates intestinal homeostasis. Cell Cycle 2010, 9, 652–655. [Google Scholar] [CrossRef]
- Wittkopf, N.; Pickert, G.; Billmeier, U.; Mahapatro, M.; Wirtz, S.; Martini, E.; Leppkes, M.; Neurath, M.F.; Becker, C. Activation of intestinal epithelial Stat3 orchestrates tissue defense during gastrointestinal infection. PLoS ONE 2015, 10, e0118401. [Google Scholar] [CrossRef]
- Pang, L.; Huynh, J.; Alorro, M.G.; Li, X.; Ernst, M.; Chand, A.L. STAT3 Signalling via the IL-6ST/gp130 Cytokine Receptor Promotes Epithelial Integrity and Intestinal Barrier Function during DSS-Induced Colitis. Biomedicines 2021, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Shahabi, N.A.; Sharp, B.M. Delta opioid agonists inhibit proliferation of highly purified murine CD4+ and CD8+ T-cells. Adv Exp. Med. Biol. 1995, 373, 29–36. [Google Scholar]
- Janković, B.D.; Radulović, J. Enkephalins, brain and immunity: Modulation of immune responses by methionine-enkephalin injected into the cerebral cavity. Int. J. Neurosci. 1992, 67, 241–270. [Google Scholar] [CrossRef] [PubMed]
- Plotnikoff, N.P.; Miller, G.C. Enkephalins as immunomodulators. Int. J. Immunopharmacol. 1983, 5, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Roscetti, G.; Ausiello, C.M.; Palma, C.; Gulla, P.; Roda, L.G. Enkephalin activity on antigen-induced proliferation of human peripheral blood mononucleate cells. Int. J. Immunopharmacol. 1988, 10, 819–823. [Google Scholar] [CrossRef]
- Li, W.; Chen, W.; Herberman, R.B.; Plotnikoff, N.P.; Youkilis, G.; Griffin, N.; Wang, E.; Lu, C.; Shan, F. Immunotherapy of cancer via mediation of cytotoxic T lymphocytes by methionine enkephalin (MENK). Cancer Lett. 2014, 344, 212–222. [Google Scholar] [CrossRef]
- Moro-García, M.A.; Mayo, J.C.; Sainz, R.M.; Alonso-Arias, R. Influence of Inflammation in the Process of T Lymphocyte Differentiation: Proliferative, Metabolic, and Oxidative Changes. Front. Immunol. 2018, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Shahabi, N.A.; McAllen, K.; Sharp, B.M. delta opioid receptors stimulate Akt-dependent phosphorylation of c-jun in T cells. J. Pharmacol. Exp. Ther. 2006, 316, 933–939. [Google Scholar] [CrossRef]
- Nguyen, K.; Miller, B.C. CD28 costimulation induces delta opioid receptor expression during anti-CD3 activation of T cells. J. Immunol. 2002, 168, 4440–4445. [Google Scholar] [CrossRef] [PubMed]
- Li, M.D.; McAllen, K.; Sharp, B.M. Regulation of delta opioid receptor expression by anti-CD3-epsilon, PMA, and ionomycin in murine splenocytes and T cells. J. Leukoc. Biol. 1999, 65, 707–714. [Google Scholar] [CrossRef]
- Rittié, L.; Fisher, G.J. Isolation and culture of skin fibroblasts. Methods Mol. Med. 2005, 117, 83–98. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Sbalchiero, E.; Azzalin, A.; Palumbo, S.; Barbieri, G.; Arias, A.; Simonelli, L.; Ferretti, L.; Comincini, S. Altered cellular distribution and sub-cellular sorting of doppel (Dpl) protein in human astrocytoma cell lines. Cell. Oncol. 2008, 30, 337–347. [Google Scholar] [CrossRef]
- Ormerod, M.G.; Tribukait, B.; Giaretti, W. Consensus report of the task force on standardisation of DNA flow cytometry in clinical pathology. DNA Flow Cytometry Task Force of the European Society for Analytical Cellular Pathology. Anal. Cell. Pathol. 1998, 17, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Slivinschi, B.; Manai, F.; Martinelli, C.; Carriero, F.; D’Amato, C.; Massarotti, M.; Bresciani, G.; Casali, C.; Milanesi, G.; Artal, L.; et al. Enhanced Delivery of Rose Bengal by Amino Acids Starvation and Exosomes Inhibition in Human Astrocytoma Cells to Potentiate Anticancer Photodynamic Therapy Effects. Cells 2022, 11, 2502. [Google Scholar] [CrossRef]
- Barbieri, G.; Palumbo, S.; Gabrusiewicz, K.; Azzalin, A.; Marchesi, N.; Spedito, A.; Biggiogera, M.; Sbalchiero, E.; Mazzini, G.; Miracco, C.; et al. Silencing of cellular prion protein (PrPC) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy 2011, 7, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Claff, T.; Yu, J.; Blais, V.; Patel, N.; Martin, C.; Wu, L.; Han, G.W.; Holleran, B.J.; Van der Poorten, O.; White, K.L.; et al. Elucidating the active δ-opioid receptor crystal structure with peptide and small-molecule agonists. Sci. Adv. 2019, 5, eaax9115. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shaw, D.E.; Shelley, M.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Physics 2020, 153, 044130. [Google Scholar] [CrossRef]
- Plante, A.; Shore, D.M.; Morra, G.; Khelashvili, G.; Weinstein, H. A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs. Molecules 2019, 24, 2097. [Google Scholar]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comp. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angewandte Chemie Int. 1999, 38, 236–240. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manai, F.; Zanoletti, L.; Morra, G.; Mansoor, S.; Carriero, F.; Bozzola, E.; Muscianisi, S.; Comincini, S. Gluten Exorphins Promote Cell Proliferation through the Activation of Mitogenic and Pro-Survival Pathways. Int. J. Mol. Sci. 2023, 24, 3912. https://doi.org/10.3390/ijms24043912
Manai F, Zanoletti L, Morra G, Mansoor S, Carriero F, Bozzola E, Muscianisi S, Comincini S. Gluten Exorphins Promote Cell Proliferation through the Activation of Mitogenic and Pro-Survival Pathways. International Journal of Molecular Sciences. 2023; 24(4):3912. https://doi.org/10.3390/ijms24043912
Chicago/Turabian StyleManai, Federico, Lisa Zanoletti, Giulia Morra, Samman Mansoor, Francesca Carriero, Elena Bozzola, Stella Muscianisi, and Sergio Comincini. 2023. "Gluten Exorphins Promote Cell Proliferation through the Activation of Mitogenic and Pro-Survival Pathways" International Journal of Molecular Sciences 24, no. 4: 3912. https://doi.org/10.3390/ijms24043912
APA StyleManai, F., Zanoletti, L., Morra, G., Mansoor, S., Carriero, F., Bozzola, E., Muscianisi, S., & Comincini, S. (2023). Gluten Exorphins Promote Cell Proliferation through the Activation of Mitogenic and Pro-Survival Pathways. International Journal of Molecular Sciences, 24(4), 3912. https://doi.org/10.3390/ijms24043912






