In Vitro Anti-Inflammatory and Antioxidant Activities of pH-Responsive Resveratrol-Urocanic Acid Nano-Assemblies
Abstract
1. Introduction
2. Results and Discussion
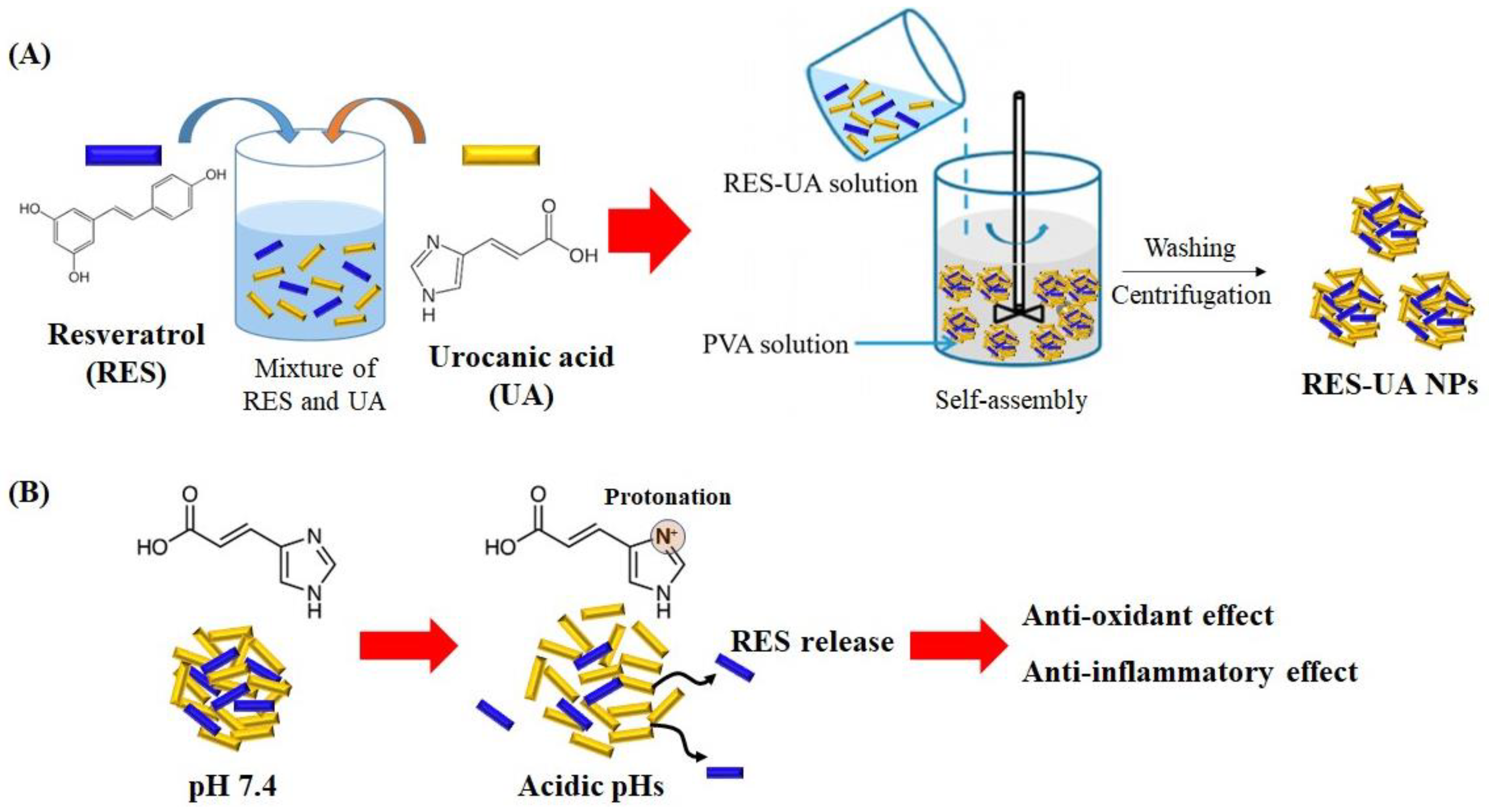
2.1. Characterization
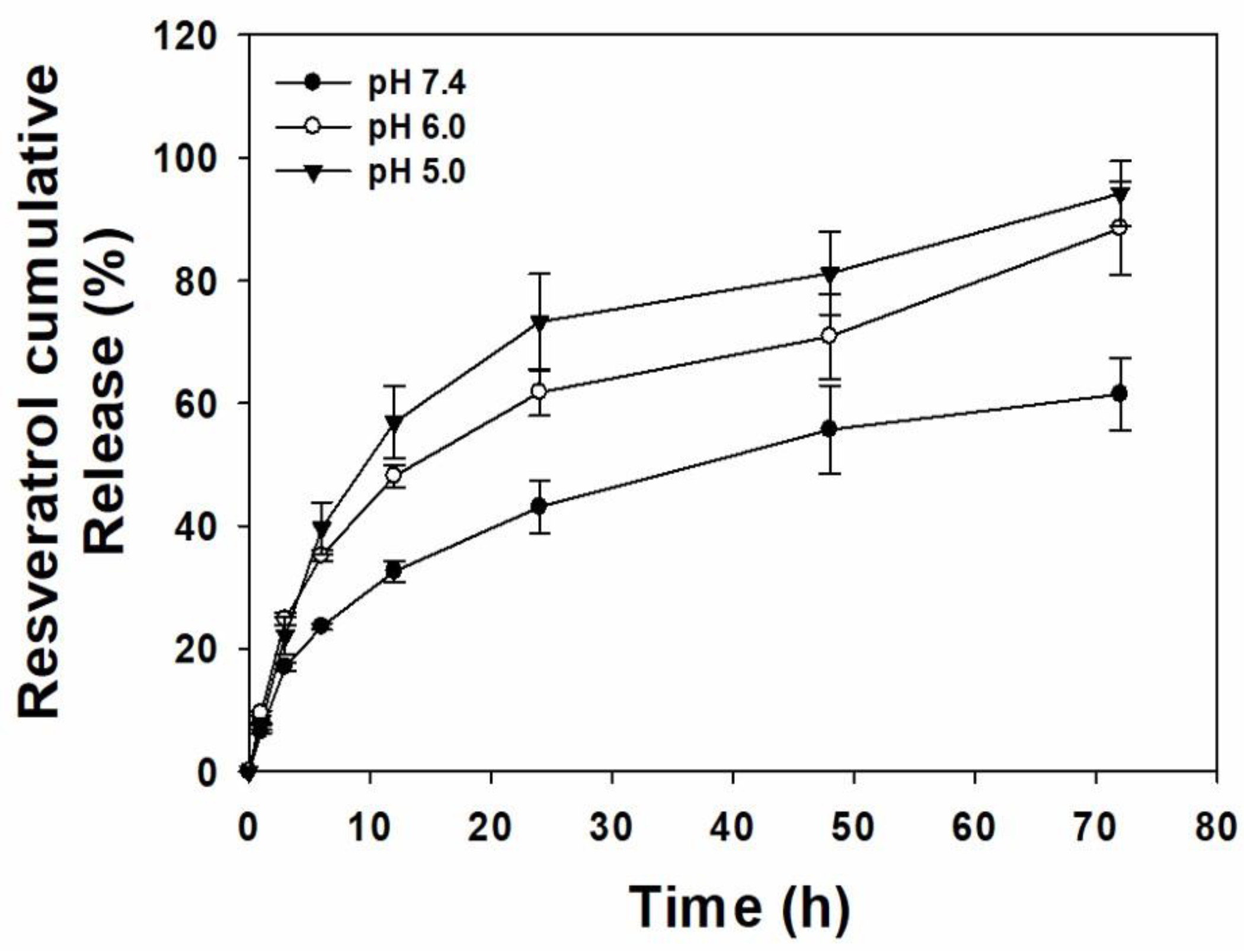
2.2. Resveratrol Release from RES-UA NPs
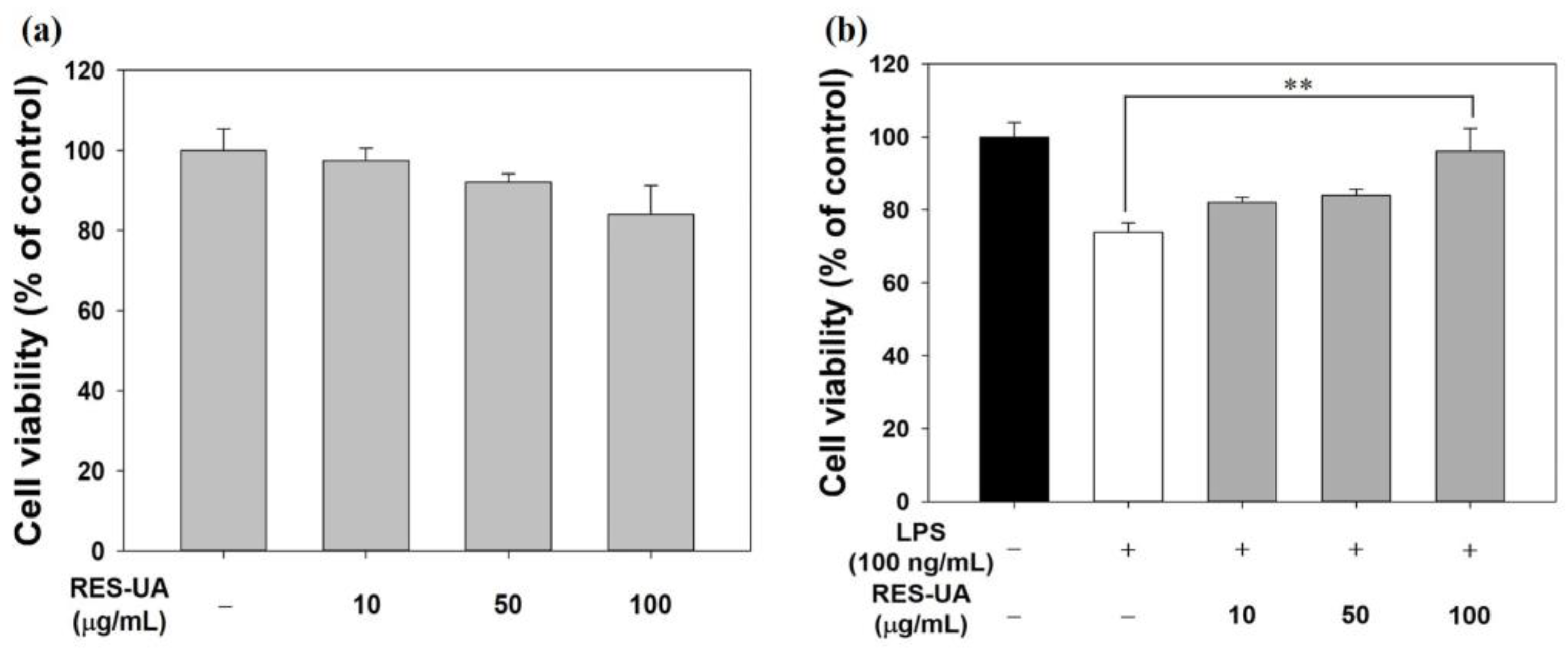
2.3. Cell Viability
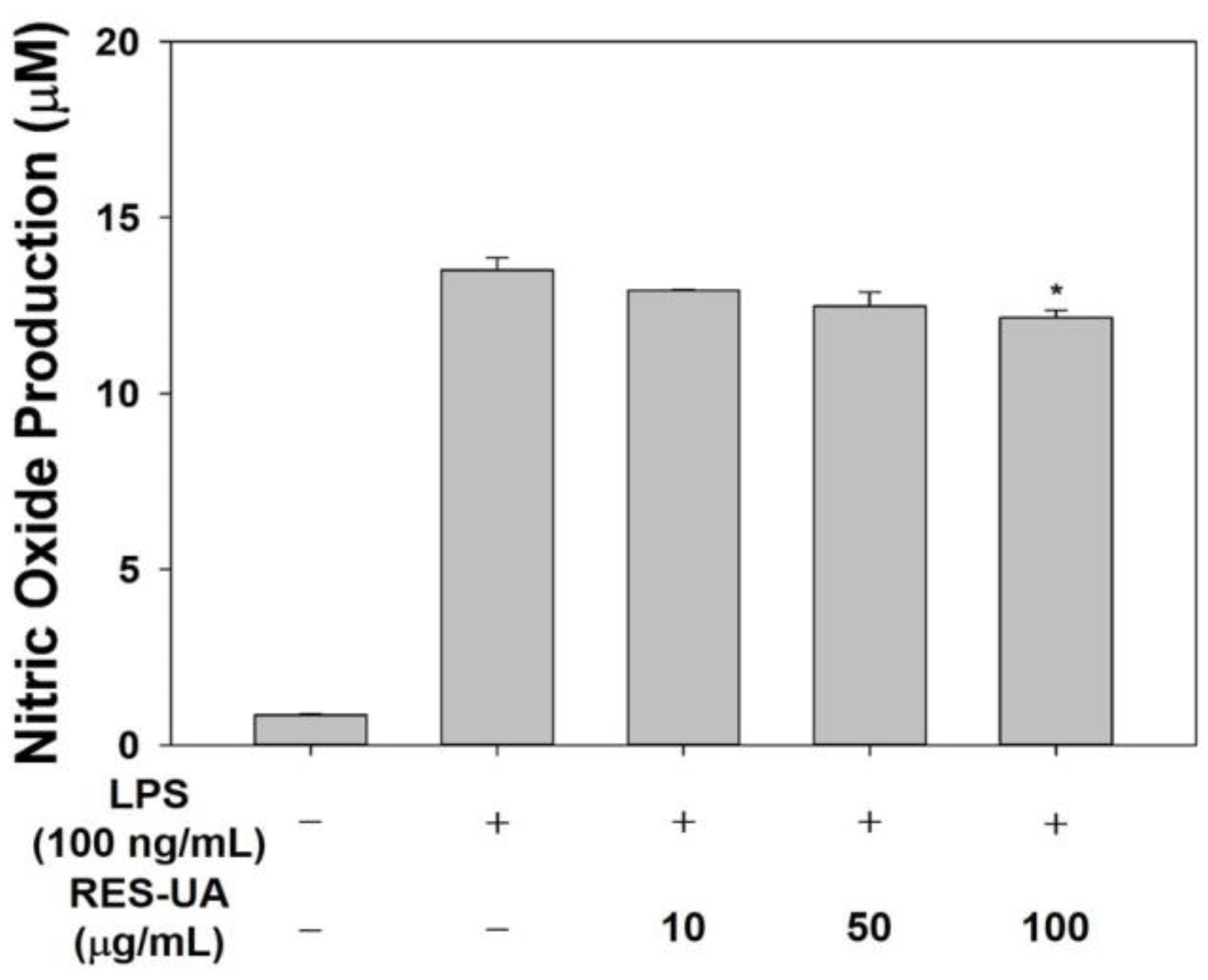
2.4. Effect of RES-UA NPs on Nitric Oxide Production in LPS-Activated RAW 264.7 Macrophages
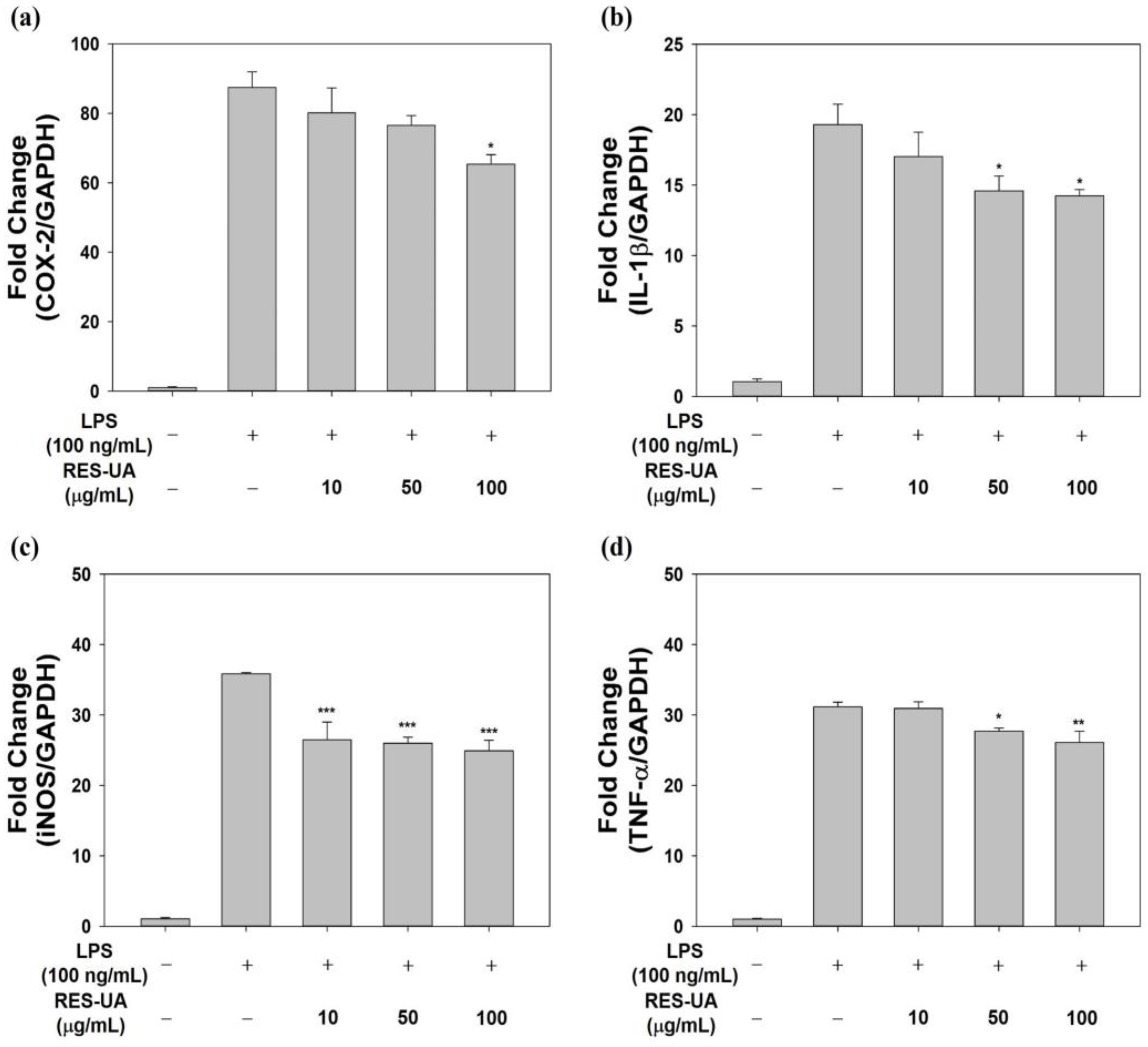
2.5. Effect of RES-UA NPs on Inflammatory Gene Expression in LPS-Treated RAW 264.7 Cells
2.6. Antioxidant Effects of RES-UA NPs and ROS Scavenging Assay at the Cellular Level
3. Materials and Methods
3.1. Preparation of Resveratrol and Urocanic Acid Nanoparticles
3.2. Characterization
3.3. In Vitro pH-Responsive RES Release Behavior of RES-UA NPs
3.4. Cell Viability Assessment
3.5. Anti-Inflammatory Potential
3.5.1. Investigation of Nitric Oxide Production
3.5.2. Real-Time Reverse Transcription-Polymerase Chain Reaction Determination of Transcript Levels of Target Genes
3.6. Antioxidant Effects of RES-UA NPs
3.6.1. In Vitro Antioxidant Capacity
3.6.2. ROS Scavenging Ability of RES-UA NPs
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Zhao, J.M.; Lao, S.; Lu, W.W. Association between bisphosphonate use and risk of undergoing knee replacement in patients with osteoarthritis. Ann. Rheum. Dis. 2019, 78, e13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Moskowitz, R.W.; Nuki, G.; Abramson, S.; Altman, R.D.; Arden, N.; Bierma-Zeinstra, S.; Brandt, K.D.; Croft, P.; Doherty, M.; et al. OARSI recommendations for the management of hip and knee osteoarthritis, Part II: OARSI evidence-based, expert consensus guidelines. Osteoarthr. Cartil. 2008, 16, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Sun, B.H.; Wu, C.W.; Kalunian, K.C. New developments in osteoarthritis. Rheum. Dis. Clin. N. Am. 2007, 33, 135–148. [Google Scholar] [CrossRef]
- van Oostrom, S.H.; Picavet, H.S.; de Bruin, S.R.; Stirbu, I.; Korevaar, J.C.; Schellevis, F.G.; Baan, C.A. Multimorbidity of chronic diseases and health care utilization in general practice. BMC Fam. Pract. 2014, 15, 61. [Google Scholar] [CrossRef]
- Mancipe Castro, L.M.; Garcia, A.J.; Guldberg, R.E. Biomaterial strategies for improved intra-articular drug delivery. J. Biomed. Mater. Res. A 2021, 109, 426–436. [Google Scholar] [CrossRef]
- Cao, P.; Li, Y.; Tang, Y.; Ding, C.; Hunter, D.J. Pharmacotherapy for knee osteoarthritis: Current and emerging therapies. Expert Opin. Pharm. 2020, 21, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.C.; Przkora, R.; Cruz-Almeida, Y. Knee osteoarthritis: Pathophysiology and current treatment modalities. J. Pain Res. 2018, 11, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Wu, W.; Wang, M.; Guo, J.; Li, H.; Zhang, S.; Xu, J.; Zou, J. Targeted treatment for osteoarthritis: Drugs and delivery system. Drug Deliv. 2021, 28, 1861–1876. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.E.; Allen, K.D.; Golightly, Y.M.; Goode, A.P.; Jordan, J.M. A systematic review of recommendations and guidelines for the management of osteoarthritis: The chronic osteoarthritis management initiative of the U.S. bone and joint initiative. Semin. Arthritis Rheum. 2014, 43, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Tian, X.Y.; Zhang, H.X.; Huang, R.; Li, N.; Chen, P.J.; Wang, R. Exercise as a prescription for patients with various diseases. J. Sport Health Sci. 2019, 8, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Bannuru, R.R.; Osani, M.C.; Vaysbrot, E.E.; Arden, N.K.; Bennell, K.; Bierma-Zeinstra, S.M.A.; Kraus, V.B.; Lohmander, L.S.; Abbott, J.H.; Bhandari, M.; et al. OARSI guidelines for the non-surgical management of knee, hip, and polyarticular osteoarthritis. Osteoarthr. Cartil. 2019, 27, 1578–1589. [Google Scholar] [CrossRef]
- Kolasinski, S.L.; Neogi, T.; Hochberg, M.C.; Oatis, C.; Guyatt, G.; Block, J.; Callahan, L.; Copenhaver, C.; Dodge, C.; Felson, D.; et al. 2019 American College of Rheumatology/Arthritis Foundation Guideline for the Management of Osteoarthritis of the Hand, Hip, and Knee. Arthritis Rheumatol. 2020, 72, 220–233. [Google Scholar] [CrossRef]
- Bjarnason, I. Gastrointestinal safety of NSAIDs and over-the-counter analgesics. Int. J. Clin. Pract. Suppl. 2013, 67, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Nissen, S.E.; Yeomans, N.D.; Solomon, D.H.; Luscher, T.F.; Libby, P.; Husni, M.E.; Graham, D.Y.; Borer, J.S.; Wisniewski, L.M.; Wolski, K.E.; et al. Cardiovascular Safety of Celecoxib, Naproxen, or Ibuprofen for Arthritis. N. Engl. J. Med. 2016, 375, 2519–2529. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Wang, K.; Wu, X.; Tu, C.; Gao, C. Stimuli-Sensitive Nanotherapies for the Treatment of Osteoarthritis. Macromol. Biosci. 2021, 21, e2100280. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Lim, T.; Shen, L.Y.; Zheng, G.; Wei, X.J.; Zhang, C.Q.; Zhu, Z.Z. Well-dispersed platelet lysate entrapped nanoparticles incorporate with injectable PDLLA-PEG-PDLLA triblock for preferable cartilage engineering application. Biomaterials 2021, 268, 120605. [Google Scholar] [CrossRef]
- Hoes, J.N.; Jacobs, J.W.; Buttgereit, F.; Bijlsma, J.W. Current view of glucocorticoid co-therapy with DMARDs in rheumatoid arthritis. Nat. Rev. Rheumatol. 2010, 6, 693–702. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Bhardwaj, A.; Aggarwal, R.S.; Seeram, N.P.; Shishodia, S.; Takada, Y. Role of resveratrol in prevention and therapy of cancer: Preclinical and clinical studies. Anticancer Res. 2004, 24, 2783–2840. [Google Scholar]
- Shrestha, A.; Pandey, R.P.; Sohng, J.K. Biosynthesis of resveratrol and piceatannol in engineered microbial strains: Achievements and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2959–2972. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Takami, A.; Trung, L.Q.; Kato, S.; Nakao, S. Resveratrol prevents EBV transformation and inhibits the outgrowth of EBV-immortalized human B cells. PLoS ONE 2012, 7, e51306. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, S.; Zumbrun, E.E.; Guan, H.; Nagarkatti, P.S.; Nagarkatti, M. Resveratrol attenuates lipopolysaccharide-induced acute kidney injury by suppressing inflammation driven by macrophages. Mol. Nutr. Food Res. 2015, 59, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, L.; Zhang, P.; Song, J.; Liu, W. An anti-inflammatory cell-free collagen/resveratrol scaffold for repairing osteochondral defects in rabbits. Acta Biomater. 2014, 10, 4983–4995. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Sun, L.; Liu, B.; Deng, Y.S.; Zhan, D.; Chen, Y.L.; He, Y.; Liu, J.; Zhang, Z.J.; Sun, J.; et al. Resveratrol inhibits LPS-induced MAPKs activation via activation of the phosphatidylinositol 3-kinase pathway in murine RAW 264.7 macrophage cells. PLoS ONE 2012, 7, e44107. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhou, H.; Wang, X.; Mai, K.; He, G. Resveratrol attenuates oxidative stress and inflammatory response in turbot fed with soybean meal based diet. Fish Shellfish Immunol. 2019, 91, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Csaki, C.; Keshishzadeh, N.; Fischer, K.; Shakibaei, M. Regulation of inflammation signalling by resveratrol in human chondrocytes in vitro. Biochem. Pharm. 2008, 75, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Shakibaei, M.; Csaki, C.; Nebrich, S.; Mobasheri, A. Resveratrol suppresses interleukin-1beta-induced inflammatory signaling and apoptosis in human articular chondrocytes: Potential for use as a novel nutraceutical for the treatment of osteoarthritis. Biochem. Pharm. 2008, 76, 1426–1439. [Google Scholar] [CrossRef]
- Elmali, N.; Baysal, O.; Harma, A.; Esenkaya, I.; Mizrak, B. Effects of resveratrol in inflammatory arthritis. Inflammation 2007, 30, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Chen, Y.C.; Wang, D.P. Resveratrol, a natural antioxidant, protects monosodium iodoacetate-induced osteoarthritic pain in rats. Biomed. Pharm. 2016, 83, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.E. Osteoarthritis biomarkers: Year in review. Osteoarthr. Cartil. 2018, 26, 312–318. [Google Scholar] [CrossRef]
- Wu, X.; Ren, G.; Zhou, R.; Ge, J.; Chen, F.H. The role of Ca2+ in acid-sensing ion channel 1a-mediated chondrocyte pyroptosis in rat adjuvant arthritis. Lab. Investig. 2019, 99, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, W.; Cai, C.; Wu, Y.; Li, J.; Dong, S. Advanced application of stimuli-responsive drug delivery system for inflammatory arthritis treatment. Mater. Today Bio 2022, 14, 100223. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.C.; Kuo, C.C.; Chen, B.Y.; Cho, C.J.; Hung, C.C.; Chen, W.C.; Borsali, R. RGB-Switchable Porous Electrospun Nanofiber Chemoprobe-Filter Prepared from Multifunctional Copolymers for Versatile Sensing of pH and Heavy Metals. ACS Appl. Mater. Interfaces 2017, 9, 16381–16396. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Qin, Z.; Chen, H.; Lan, Q.; Wang, Z.; Lan, N.; Yang, Y.; Zheng, L.; Zhao, J.; Kai, D. pH-responsive and hyaluronic acid-functionalized metal-organic frameworks for therapy of osteoarthritis. J. Nanobiotechnol. 2020, 18, 139. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Liu, Y.; Wu, J.; Zhou, P.; Wang, Y.; Li, R.; Yang, X.; Zhang, N. pH-sensitive pullulan-based nanoparticle carrier of methotrexate and combretastatin A4 for the combination therapy against hepatocellular carcinoma. Biomaterials 2013, 34, 7181–7190. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, Y.; Wang, Y.; Wu, J.; Yang, X.; Li, R.; Wang, Y.; Zhang, N. pH-sensitive pullulan-based nanoparticle carrier for adriamycin to overcome drug-resistance of cancer cells. Carbohydr. Polym. 2014, 111, 908–917. [Google Scholar] [CrossRef]
- Wang, Y.S.; Liu, Y.; Liu, Y.Y.; Wang, Y.; Wu, J.; Li, R.S.; Yang, J.R.; Zhang, N. pH-sensitive pullulan-based nanoparticles for intracellular drug delivery. Polym. Chem. 2014, 5, 423–432. [Google Scholar] [CrossRef]
- Luzardo-Alvarez, A.; Lamela-Gomez, I.; Otero-Espinar, F.; Blanco-Mendez, J. Development, Characterization, and In Vitro Evaluation of Resveratrol-Loaded Poly-(epsilon-caprolactone) Microcapsules Prepared by Ultrasonic Atomization for Intra-Articular Administration. Pharmaceutics 2019, 11, 249. [Google Scholar] [CrossRef]
- Porto, I.C.C.M.; Nascimento, T.G.; Oliveira, J.M.S.; Freitas, P.H.; Haimeur, A.; Franca, R. Use of polyphenols as a strategy to prevent bond degradation in the dentin-resin interface. Eur. J. Oral. Sci. 2018, 126, 146–158. [Google Scholar] [CrossRef]
- Pujara, N.; Jambhrunkar, S.; Wong, K.Y.; McGuckin, M.; Popat, A. Enhanced colloidal stability, solubility and rapid dissolution of resveratrol by nanocomplexation with soy protein isolate. J. Colloid Interface Sci. 2017, 488, 303–308. [Google Scholar] [CrossRef]
- Kumar, N.; Thomas, S.; Rao, R.; Maiti, N.; Kshirsagar, R.J. Surface-enhanced Raman scattering based sensing of trans-urocanic acid, an epidermal photoreceptor using silver nanoparticles aided by density functional theoretical calculations. J. Raman Spectrosc. 2019, 50, 837–846. [Google Scholar] [CrossRef]
- Lin, T.L.; Shu, C.C.; Chen, Y.M.; Lu, J.J.; Wu, T.S.; Lai, W.F.; Tzeng, C.M.; Lai, H.C.; Lu, C.C. Like Cures Like: Pharmacological Activity of Anti-Inflammatory Lipopolysaccharides from Gut Microbiome. Front. Pharm. 2020, 11, 554. [Google Scholar] [CrossRef]
- Basso, F.G.; Soares, D.G.; Pansani, T.N.; Turrioni, A.P.; Scheffel, D.L.; de Souza Costa, C.A.; Hebling, J. Effect of LPS treatment on the viability and chemokine synthesis by epithelial cells and gingival fibroblasts. Arch. Oral. Biol. 2015, 60, 1117–1121. [Google Scholar] [CrossRef]
- Tsai, S.H.; Lin-Shiau, S.Y.; Lin, J.K. Suppression of nitric oxide synthase and the down-regulation of the activation of NFkappaB in macrophages by resveratrol. Br. J. Pharm. 1999, 126, 673–680. [Google Scholar] [CrossRef]
- Panaro, M.A.; Carofiglio, V.; Acquafredda, A.; Cavallo, P.; Cianciulli, A. Anti-inflammatory effects of resveratrol occur via inhibition of lipopolysaccharide-induced NF-kappaB activation in Caco-2 and SW480 human colon cancer cells. Br. J. Nutr. 2012, 108, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Muniandy, K.; Gothai, S.; Badran, K.M.H.; Suresh Kumar, S.; Esa, N.M.; Arulselvan, P. Suppression of Proinflammatory Cytokines and Mediators in LPS-Induced RAW 264.7 Macrophages by Stem Extract of Alternanthera sessilis via the Inhibition of the NF-kappaB Pathway. J. Immunol. Res. 2018, 2018, 3430684. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Park, T.; Kang, M.G.; Park, D. Anti-Inflammatory Activity and ROS Regulation Effect of Sinapaldehyde in LPS-Stimulated RAW 264.7 Macrophages. Molecules 2020, 25, 4089. [Google Scholar] [CrossRef]
- Schwager, J.; Richard, N.; Widmer, F.; Raederstorff, D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complement. Altern. Med. 2017, 17, 309. [Google Scholar] [CrossRef]
- Shi, Q.; Wang, X.; Tang, X.; Zhen, N.; Wang, Y.; Luo, Z.; Zhang, H.; Liu, J.; Zhou, D.; Huang, K. In vitro antioxidant and antitumor study of zein/SHA nanoparticles loaded with resveratrol. Food Sci. Nutr. 2021, 9, 3530–3537. [Google Scholar] [CrossRef]
- Tang, L.; Chen, X.; Tong, Q.; Ran, Y.; Ma, L.; Tan, Y.; Yi, Z.; Li, X. Biocompatible, bacteria-targeting resveratrol nanoparticles fabricated by Mannich molecular condensation for accelerating infected wound healing. J. Mater. Chem. B 2022, 10, 9280–9294. [Google Scholar] [CrossRef]
- Hsu, H.Y.; Wen, M.H. Lipopolysaccharide-mediated reactive oxygen species and signal transduction in the regulation of interleukin-1 gene expression. J. Biol. Chem. 2002, 277, 22131–22139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yu, R.; Deng, J.; Zhao, Q.; Li, Y.; Joo, M.; van Breemen, R.B.; Christman, J.W.; Xiao, L. Pivotal role of reactive oxygen species in differential regulation of lipopolysaccharide-induced prostaglandins production in macrophages. Mol. Pharm. 2013, 83, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; You, Q.; Hu, L.; Gao, J.; Meng, Q.; Liu, W.; Wu, X.; Xu, Q. The Antioxidant Procyanidin Reduces Reactive Oxygen Species Signaling in Macrophages and Ameliorates Experimental Colitis in Mice. Front. Immunol. 2017, 8, 1910. [Google Scholar] [CrossRef]
- Canton, M.; Sanchez-Rodriguez, R.; Spera, I.; Venegas, F.C.; Favia, M.; Viola, A.; Castegna, A. Reactive Oxygen Species in Macrophages: Sources and Targets. Front. Immunol. 2021, 12, 734229. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.T.; Tseng, Y.T.; Wong, W.J.; Liu, C.M.; Lo, Y.C. Resveratrol prevents nanoparticles-induced inflammation and oxidative stress via downregulation of PKC-alpha and NADPH oxidase in lung epithelial A549 cells. BMC Complement. Altern. Med. 2018, 18, 211. [Google Scholar] [CrossRef]



| pH | Particle Size (nm) | PDI * | Zeta-Potential (mV) |
|---|---|---|---|
| 7.4 | 106.5 ± 31.0 | 0.545 ± 0.08 | −1.17 ± 0.31 |
| 6.0 | 159.3 ± 36.8 | 0.531 ± 0.05 | −2.63 ± 0.25 |
| 5.0 | 188.7 ± 59.2 | 0.487 ± 0.08 | −2.80 ± 0.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Kang, S.; Yu, Y.; Jung, S.Y.; Park, K.; Kim, S.-M.; Kim, H.-J.; Kim, J.G.; Kim, S.E. In Vitro Anti-Inflammatory and Antioxidant Activities of pH-Responsive Resveratrol-Urocanic Acid Nano-Assemblies. Int. J. Mol. Sci. 2023, 24, 3843. https://doi.org/10.3390/ijms24043843
Song H, Kang S, Yu Y, Jung SY, Park K, Kim S-M, Kim H-J, Kim JG, Kim SE. In Vitro Anti-Inflammatory and Antioxidant Activities of pH-Responsive Resveratrol-Urocanic Acid Nano-Assemblies. International Journal of Molecular Sciences. 2023; 24(4):3843. https://doi.org/10.3390/ijms24043843
Chicago/Turabian StyleSong, Heegyeong, Seok Kang, Ying Yu, Sung Yun Jung, Kyeongsoon Park, Sang-Min Kim, HaK-Jun Kim, Jae Gyoon Kim, and Sung Eun Kim. 2023. "In Vitro Anti-Inflammatory and Antioxidant Activities of pH-Responsive Resveratrol-Urocanic Acid Nano-Assemblies" International Journal of Molecular Sciences 24, no. 4: 3843. https://doi.org/10.3390/ijms24043843
APA StyleSong, H., Kang, S., Yu, Y., Jung, S. Y., Park, K., Kim, S.-M., Kim, H.-J., Kim, J. G., & Kim, S. E. (2023). In Vitro Anti-Inflammatory and Antioxidant Activities of pH-Responsive Resveratrol-Urocanic Acid Nano-Assemblies. International Journal of Molecular Sciences, 24(4), 3843. https://doi.org/10.3390/ijms24043843







