Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum)
Abstract
1. Introduction
2. Results
2.1. Clean Reads and De Novo Assembly of Transcripts
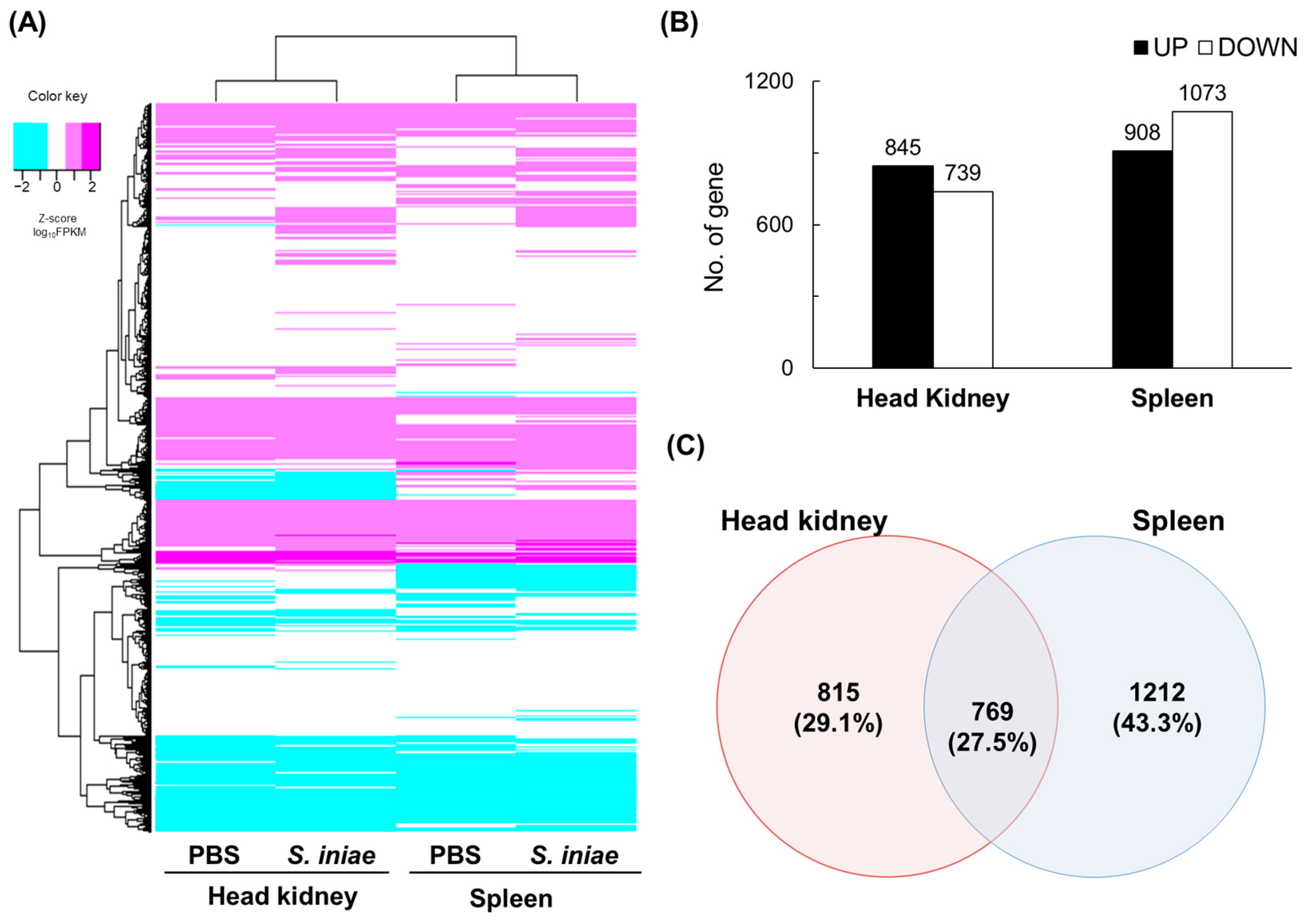
2.2. Differentially Expressed Genes after S. iniae Infection and their Validation Using RT-qPCR
2.3. Functional Enrichment Analysis of DEGs in the KEGG Pathways
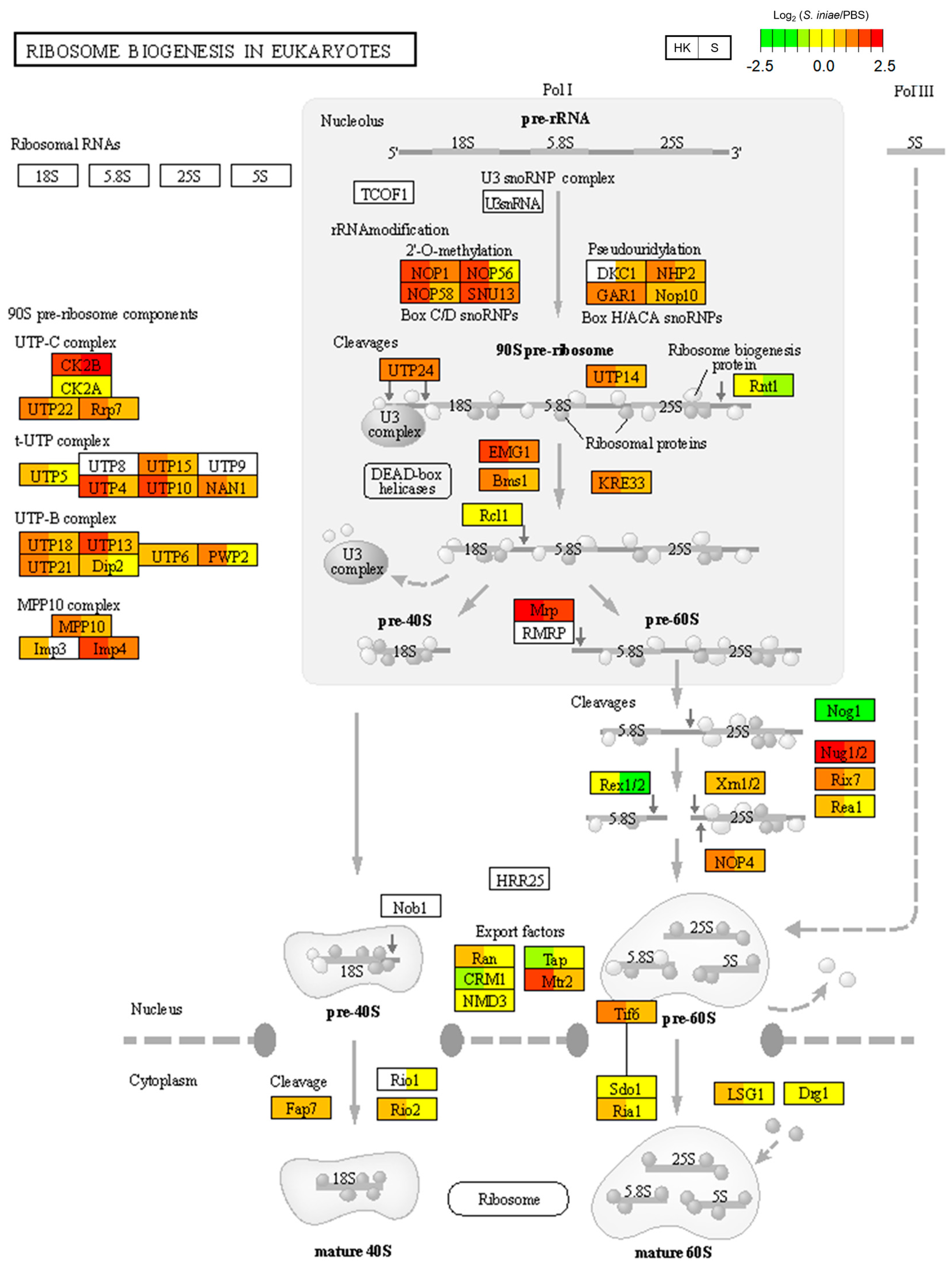
2.4. Specific Induction of Ribosome-Biogenesis-Related Genes in the Head Kidney
2.5. Splenic DEGs Assigned to the KEGG Pathways
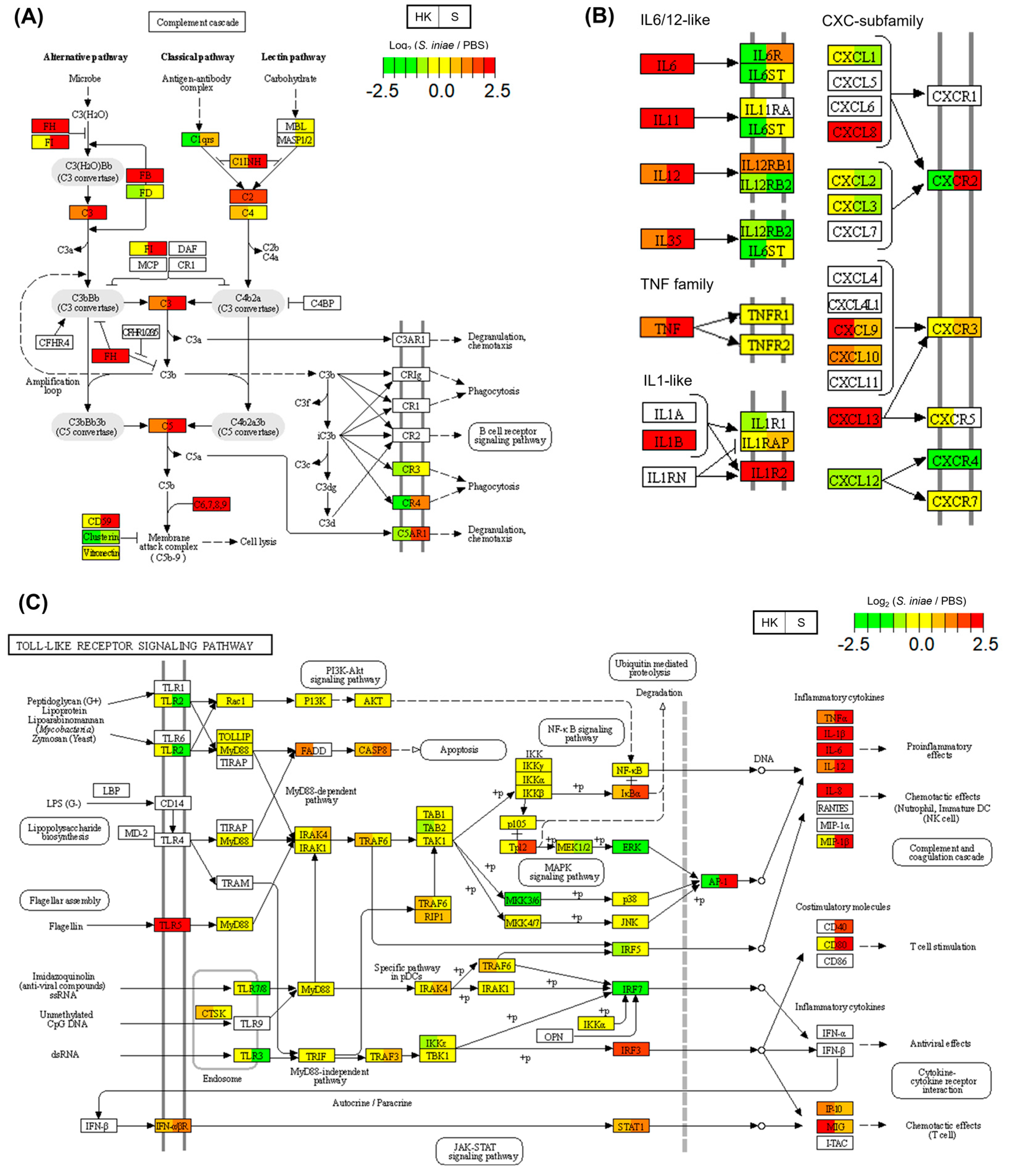
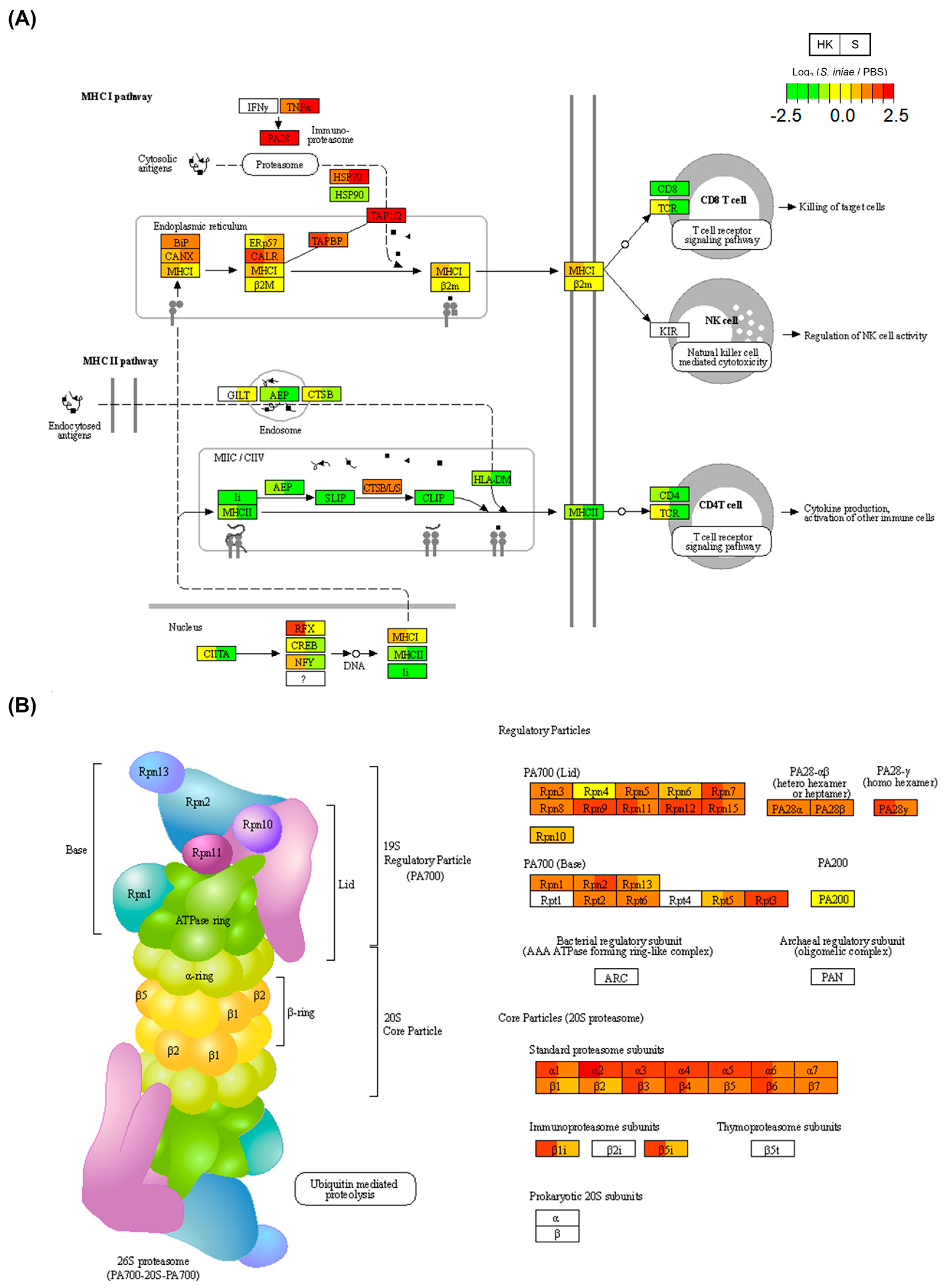
2.6. Common DEGs Enriched in Immune-Related KEGG Pathways
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Streptococcus Iniae Challenge
4.3. Total RNA Extraction, Sequence Library Preparation, and Sequencing
4.4. Reads Data Filtration and De Novo Transcriptome Assembly
4.5. Gene Function Annotation
4.6. Differentially Expressed Genes and Functional Enrichment Analysis
4.7. Real-Time Reverse Transcription Polymerase Chain Reaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agnew, W.; Barnes, A.C. Streptococcus iniae: An aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 2007, 122, 1–15. [Google Scholar] [CrossRef]
- Rowe, H.M.; Withey, J.H.; Neely, M.N. Zebrafish as a model for zoonotic aquatic pathogens. Dev. Comp. Immunol. 2014, 46, 96–107. [Google Scholar] [CrossRef]
- Weinstein, M.R.; Litt, M.; Kertesz, D.A.; Wyper, P.; Rose, D.; Coulter, M.; McGeer, A.; Facklam, R.; Ostach, C.; Willey, B.M.; et al. Invasive Infections Due to a Fish Pathogen, Streptococcus iniae. N. Engl. J. Med. 1997, 337, 589–594. [Google Scholar] [CrossRef]
- Bromage, E.; Owens, L. Infection of barramundi Lates calcarifer with Streptococcus iniae: Effects of different routes of exposure. Dis. Aquat. Org. 2002, 52, 199–205. [Google Scholar] [CrossRef]
- Shoemaker, C.A.; Klesius, P.H.; Evans, J.J. Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am. J. Vet. Res. 2001, 62, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, M.; Li, C.; Li, L.-P.; Gan, X.; Huang, J.; Lei, A.-Y.; Xu, Z.-H.; Liang, W.-W. Identification of multiple genes and their expression profiles in four strains of Oreochromis spp. in response to Streptococcus iniae. J. Fish Biol. 2013, 82, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Madhuranga, W.; Tharuka, M.N.; Harasgama, J.; Kwon, H.; Wan, Q.; Lee, J. Immune responses, subcellular localization, and antiviral activity of interferon-induced protein 35 (IFP35) in rock bream (Oplegnathus fasciatus). Dev. Comp. Immunol. 2021, 123, 104142. [Google Scholar] [CrossRef] [PubMed]
- Bathige, S.; Umasuthan, N.; Godahewa, G.; Thulasitha, W.S.; Jayasinghe, J.; Wan, Q.; Lee, J. Molecular insights of two STAT1 variants from rock bream (Oplegnathus fasciatus) and their transcriptional regulation in response to pathogenic stress, interleukin-10, and tissue injury. Fish Shellfish. Immunol. 2017, 69, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Tharuka, M.N.; Yang, H.; Lee, J. Expression, subcellular localization, and potential antiviral function of three interferon regulatory factors in the big-belly seahorse (Hippocampus abdominalis). Fish Shellfish. Immunol. 2020, 96, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Cao, M.; Zhang, L.; Fu, Q.; Yang, N.; Tan, F.; Song, L.; Su, B.; Li, C. Characterization and initial functional analysis of cathepsin K in turbot (Scophthalmus maximus L.). Fish Shellfish. Immunol. 2019, 93, 153–160. [Google Scholar] [CrossRef]
- Choi, K.-M.; Hwang, S.D.; Joo, M.S.; Hwang, J.Y.; Kwon, M.-G.; Jeong, J.-M.; Seo, J.S.; Jee, B.-Y.; Park, C.-I. The atypical chemokine receptor 4 in red sea bream (Pagrus major): Molecular characterization and gene expression analysis. Fish Shellfish. Immunol. 2019, 93, 50–54. [Google Scholar] [CrossRef]
- Harvie, E.A.; Green, J.M.; Neely, M.N.; Huttenlocher, A. Innate Immune Response to Streptococcus iniae Infection in Zebrafish Larvae. Infect. Immun. 2013, 81, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Wang, P.-C.; Chen, S.-C. Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis. Front. Immunol. 2019, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, C.; Ao, Q.; Tan, Y.; Luo, Y.; Guo, Y.; Lan, G.; Jiang, H.; Gan, X. Trancriptomic profiling revealed the signatures of acute immune response in tilapia (Oreochromis niloticus) following Streptococcus iniae challenge. Fish Shellfish. Immunol. 2015, 46, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Eldar, A.; Bejerano, Y.; Bercovier, H. Streptococcus shiloi andStreptococcus difficile: Two new streptococcal species causing a meningoencephalitis in fish. Curr. Microbiol. 1994, 28, 139–143. [Google Scholar] [CrossRef]
- Shihab, I.; Gopalakrishnan, A.; Vineesh, N.; Muktha, M.; Akhilesh, K.V.; Vijayagopal, P. Histological profiling of gonads depicting protandrous hermaphroditism in Eleutheronema tetradactylum. J. Fish Biol. 2017, 90, 2402–2411. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Iadevaia, V.; Liu, R.; Proud, C.G. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin. Cell Dev. Biol. 2014, 36, 113–120. [Google Scholar] [CrossRef]
- Bianco, C.; Mohr, I. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife 2019, 8, e49551. [Google Scholar] [CrossRef]
- Fuhrman, L.E.; Goel, A.K.; Smith, J.; Shianna, K.V.; Aballay, A. Nucleolar Proteins Suppress Caenorhabditis elegans Innate Immunity by Inhibiting p53/CEP-1. PLoS Genet. 2009, 5, e1000657. [Google Scholar] [CrossRef] [PubMed]
- Harvie, E.A.; Huttenlocher, A. Neutrophils in host defense: New insights from zebrafish. J. Leukoc. Biol. 2015, 98, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Oates, A.C.; Crowhurst, M.O.; Ward, A.C.; Layton, J.E. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 2001, 98, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.C.; Quiceno, D.G.; Ochoa, A.C. l-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 2007, 109, 1568–1573. [Google Scholar] [CrossRef] [PubMed]
- Highfill, S.L.; Rodriguez, P.C.; Zhou, Q.; Goetz, C.A.; Koehn, B.H.; Veenstra, R.; Taylor, P.A.; Panoskaltsis-Mortari, A.; Serody, J.; Munn, D.; et al. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1–dependent mechanism that is up-regulated by interleukin-13. Blood 2010, 116, 5738–5747. [Google Scholar] [CrossRef]
- Makarenkova, V.P.; Bansal, V.; Matta, B.M.; Perez, L.A.; Ochoa, J.B. CD11b+/Gr-1+ Myeloid Suppressor Cells Cause T Cell Dysfunction after Traumatic Stress. J. Immunol. 2006, 176, 2085–2094. [Google Scholar] [CrossRef]
- Maekawa, S.; Wang, P.-C.; Chen, S.-C. Differential expression of immune-related genes in head kidney and spleen of cobia (Rachycentron canadum) having Streptococcus dysgalactiae infection. Fish Shellfish. Immunol. 2019, 92, 842–850. [Google Scholar] [CrossRef]
- Nagaishi, T.; Pao, L.; Lin, S.-H.; Iijima, H.; Kaser, A.; Qiao, S.-W.; Chen, Z.; Glickman, J.; Najjar, S.M.; Nakajima, A.; et al. SHP1 Phosphatase-Dependent T Cell Inhibition by CEACAM1 Adhesion Molecule Isoforms. Immunity 2006, 25, 769–781. [Google Scholar] [CrossRef]
- Boshra, H.; Li, J.; Sunyer, J. Recent advances on the complement system of teleost fish. Fish Shellfish. Immunol. 2006, 20, 239–262. [Google Scholar] [CrossRef]
- Nakao, M.; Tsujikura, M.; Ichiki, S.; Vo, T.K.; Somamoto, T. The complement system in teleost fish: Progress of post-homolog-hunting researches. Dev. Comp. Immunol. 2011, 35, 1296–1308. [Google Scholar] [CrossRef]
- Walport, M.J. Complement. First of two parts. N. Engl. J. Med. 2001, 344, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Miyata, M.; Chan, C.; Ngoh, S.Y.; Liew, W.C.; Saju, J.M.; Ng, K.S.; Wong, F.S.; Lee, Y.S.; Chang, S.F.; et al. Differential Transcriptomic Response in the Spleen and Head Kidney Following Vaccination and Infection of Asian Seabass with Streptococcus iniae. PLoS ONE 2014, 9, e99128. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Su, B.; Fu, Q.; Shang, M.; Gao, C.; Tan, F.; Li, C. Identification, characterization and expression analysis of TLR5 in the mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish Shellfish. Immunol. 2017, 68, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Wearsch, P.A.; Cresswell, P. The quality control of MHC class I peptide loading. Curr. Opin. Cell Biol. 2008, 20, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Kerimoglu, B.; Yaparla, A.; Hodgkinson, J.W.; Xie, J.; Belosevic, M. Mechanisms of Fish Macrophage Antimicrobial Immunity. Front. Immunol. 2018, 9, 1105. [Google Scholar] [CrossRef]
- Sever, L.; Vo, N.T.; Lumsden, J.; Bols, N.C.; Dixon, B. Induction of rainbow trout MH class I and accessory proteins by viral haemorrhagic septicaemia virus. Mol. Immunol. 2014, 59, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Pulpipat, T.; Wang, P.-C.; Chen, S.-C. Transcriptome analysis of immune- and iron-related genes after Francisella noatunensis subsp. orientalis infection in Nile tilapia (Oreochromis niloticus). Fish Shellfish. Immunol. 2021, 111, 36–48. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]


| ID | Pathway Name | Count | (%) | Q-Value |
|---|---|---|---|---|
| Head kidney-specific DEGs | ||||
| ko03008 | Ribosome biogenesis in eukaryotes | 23 | (2.82) | 3.82 × 10−5 |
| ko03020 | RNA polymerase | 11 | (1.35) | 2.16 × 10−2 |
| Spleen-specific DEGs | ||||
| ko04080 | Neuroactive ligand-receptor interaction | 53 | (4.37) | 8.22 × 10−7 |
| ko04721 | Synaptic vesicle cycle | 21 | (1.73) | 1.03 × 10−5 |
| ko05171 | Coronavirus disease—COVID-19 | 53 | (4.37) | 2.53 × 10−5 |
| ko05323 | Rheumatoid arthritis | 23 | (1.90) | 4.48 × 10−5 |
| ko04145 | Phagosome | 29 | (2.39) | 2.76 × 10−4 |
| ko04514 | Cell adhesion molecules | 27 | (2.23) | 5.01 × 10−4 |
| ko04966 | Collecting duct acid secretion | 10 | (0.83) | 5.71 × 10−4 |
| ko05340 | Primary immunodeficiency | 14 | (1.16) | 2.97 × 10−3 |
| ko04658 | Th1 and Th2 cell differentiation | 19 | (1.57) | 5.44 × 10−3 |
| ko04911 | Insulin secretion | 18 | (1.49) | 6.12 × 10−3 |
| ko04610 | Complement and coagulation cascades | 21 | (1.73) | 7.12 × 10−3 |
| ko03010 | Ribosome | 31 | (2.56) | 1.66 × 10−2 |
| ko04961 | Endocrine and other factor-regulated calcium reabsorption | 12 | (0.99) | 1.57 × 10−2 |
| ko04924 | Renin secretion | 16 | (1.32) | 2.54 × 10−2 |
| ko04659 | Th17 cell differentiation | 20 | (1.65) | 2.56 × 10−2 |
| ko04640 | Hematopoietic cell lineage | 17 | (1.40) | 3.07 × 10−2 |
| ko05200 | Pathways in cancer | 77 | (6.35) | 3.07 × 10−2 |
| ko05110 | Vibrio cholerae infection | 13 | (1.07) | 2.93 × 10−2 |
| ko04921 | Oxytocin signaling pathway | 25 | (2.06) | 3.20 × 10−2 |
| ko05120 | Epithelial cell signaling in Helicobacter pylori infection | 17 | (1.40) | 3.21 × 10−2 |
| ko04217 | Necroptosis | 23 | (1.90) | 3.23 × 10−2 |
| ko04630 | JAK-STAT signaling pathway | 23 | (1.90) | 3.08 × 10−2 |
| ko00010 | Glycolysis/Gluconeogenesis | 13 | (1.07) | 3.20 × 10−2 |
| ko04071 | Sphingolipid signaling pathway | 22 | (1.82) | 4.44 × 10−2 |
| ko04725 | Cholinergic synapse | 19 | (1.57) | 4.98 × 10−2 |
| Common DEGs | ||||
| ko03050 | Proteasome | 29 | (3.77) | 3.55 × 10−17 |
| ko05144 | Malaria | 15 | (1.95) | 3.82 × 10−6 |
| ko05146 | Amoebiasis | 20 | (2.60) | 3.22 × 10−5 |
| ko05020 | Prion disease | 42 | (5.46) | 5.65 × 10−5 |
| ko05017 | Spinocerebellar ataxia | 27 | (3.51) | 6.15 × 10−5 |
| ko04610 | Complement and coagulation cascades | 18 | (2.34) | 1.40 × 10−4 |
| ko05322 | Systemic lupus erythematosus | 13 | (1.69) | 2.32 × 10−4 |
| ko05133 | Pertussis | 16 | (2.08) | 2.20 × 10−4 |
| ko00970 | Aminoacyl-tRNA biosynthesis | 12 | (1.56) | 4.71 × 10−4 |
| ko04060 | Cytokine–cytokine receptor interaction | 29 | (3.77) | 5.66 × 10−4 |
| ko04080 | Neuroactive ligand-receptor interaction | 27 | (3.51) | 1.18 × 10−3 |
| ko04612 | Antigen processing and presentation | 12 | (1.56) | 5.30 × 10−3 |
| ko01110 | Biosynthesis of secondary metabolites | 47 | (6.11) | 1.03 × 10−2 |
| ko05169 | Epstein–Barr virus infection | 28 | (3.64) | 1.05 × 10−2 |
| ko05012 | Parkinson disease | 33 | (4.29) | 1.17 × 10−2 |
| ko05142 | Chagas disease | 16 | (2.08) | 1.16 × 10−2 |
| ko05150 | Staphylococcus aureus infection | 9 | (1.17) | 2.04 × 10−2 |
| ko00260 | Glycine, serine and threonine metabolism | 10 | (1.30) | 2.03 × 10−2 |
| ko05010 | Alzheimer disease | 41 | (5.33) | 2.10 × 10−2 |
| ko05134 | Legionellosis | 11 | (1.43) | 2.35 × 10−2 |
| ko04640 | Hematopoietic cell lineage | 13 | (1.69) | 3.57 × 10−2 |
| ko00590 | Arachidonic acid metabolism | 8 | (1.04) | 3.87 × 10−2 |
| ko04061 | Viral protein interaction with cytokine and cytokine receptor | 12 | (1.56) | 3.71 × 10−2 |
| ko05323 | Rheumatoid arthritis | 12 | (1.56) | 3.56 × 10−2 |
| ko05022 | Pathways of neurodegeneration—multiple diseases | 46 | (5.98) | 3.88 × 10−2 |
| ko04657 | IL-17 signaling pathway | 14 | (1.82) | 4.34 × 10−2 |
| ko04974 | Protein digestion and absorption | 11 | (1.43) | 4.41 × 10−2 |
| ko05016 | Huntington disease | 34 | (4.42) | 4.25 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maekawa, S.; Wang, P.-C.; Chen, S.-C. Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum). Int. J. Mol. Sci. 2023, 24, 3832. https://doi.org/10.3390/ijms24043832
Maekawa S, Wang P-C, Chen S-C. Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum). International Journal of Molecular Sciences. 2023; 24(4):3832. https://doi.org/10.3390/ijms24043832
Chicago/Turabian StyleMaekawa, Shun, Pei-Chi Wang, and Shih-Chu Chen. 2023. "Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum)" International Journal of Molecular Sciences 24, no. 4: 3832. https://doi.org/10.3390/ijms24043832
APA StyleMaekawa, S., Wang, P.-C., & Chen, S.-C. (2023). Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum). International Journal of Molecular Sciences, 24(4), 3832. https://doi.org/10.3390/ijms24043832





