Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas
Abstract
1. Introduction
2. Results
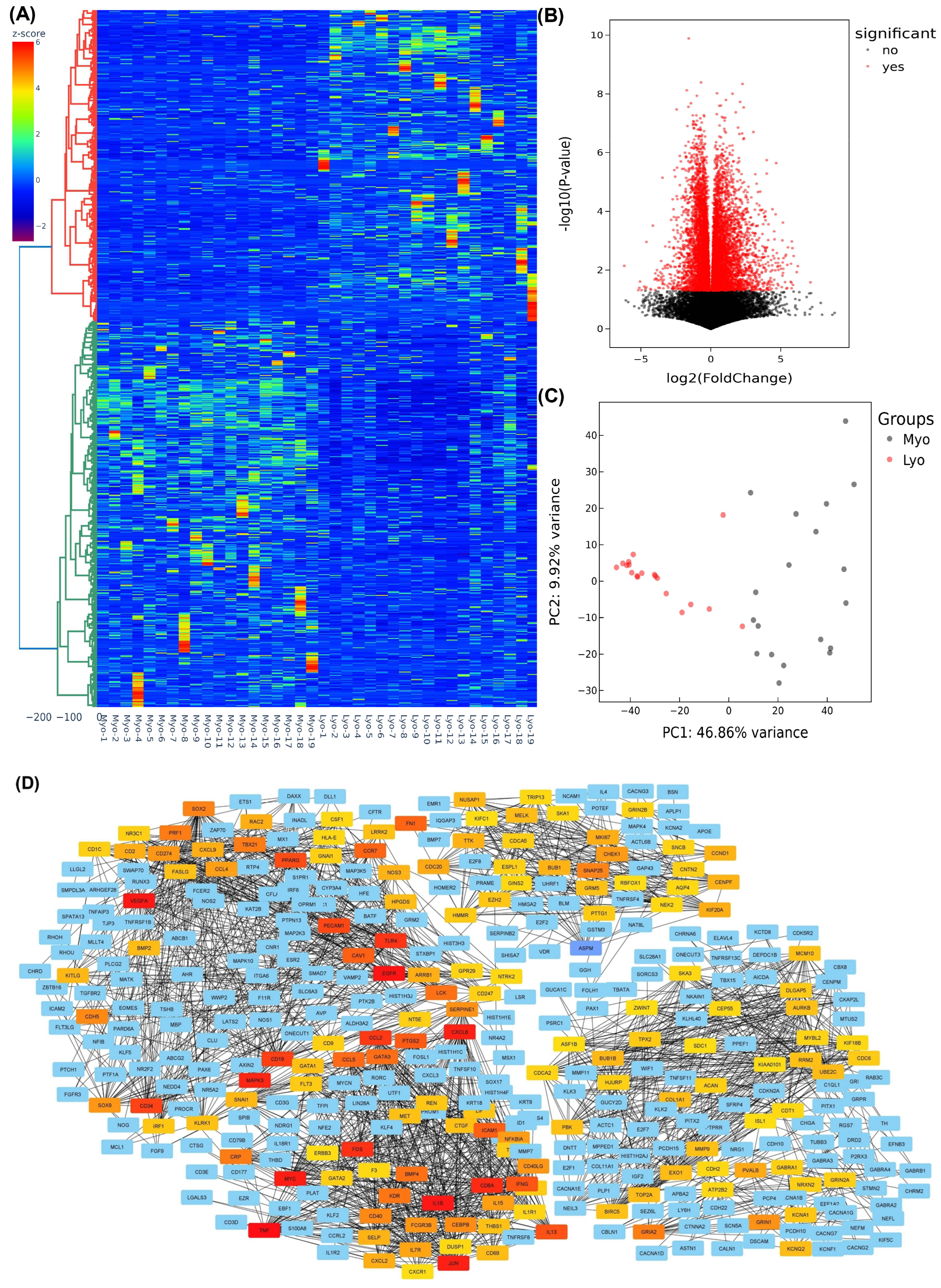
2.1. High-Throughput Sequencing of Coding RNA Transcripts in Leiomyoma and Matched Myometrium
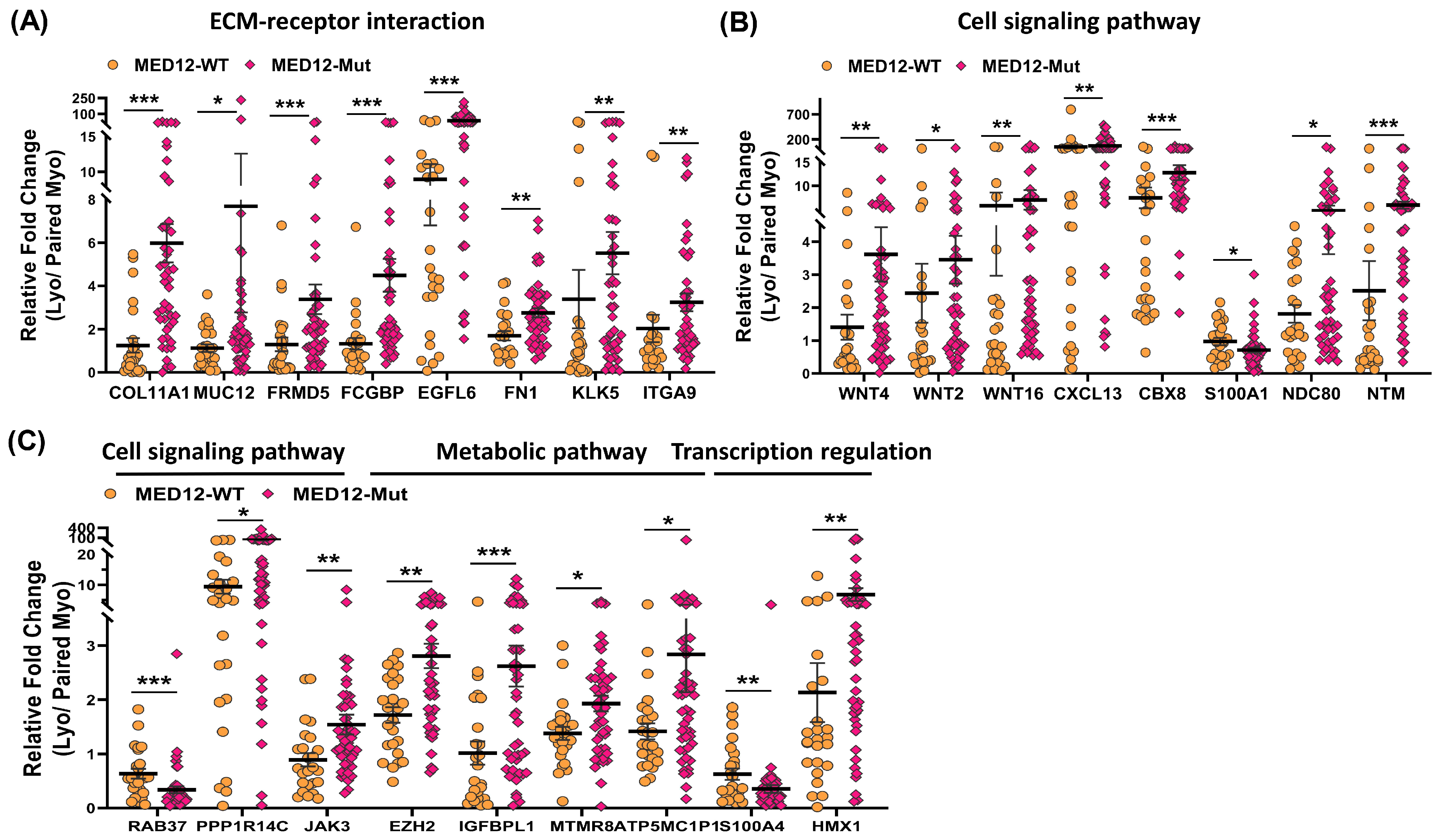
2.2. Differential Expression of MED12-Associated Coding RNA Transcripts in Leiomyoma and Matched Myometrium
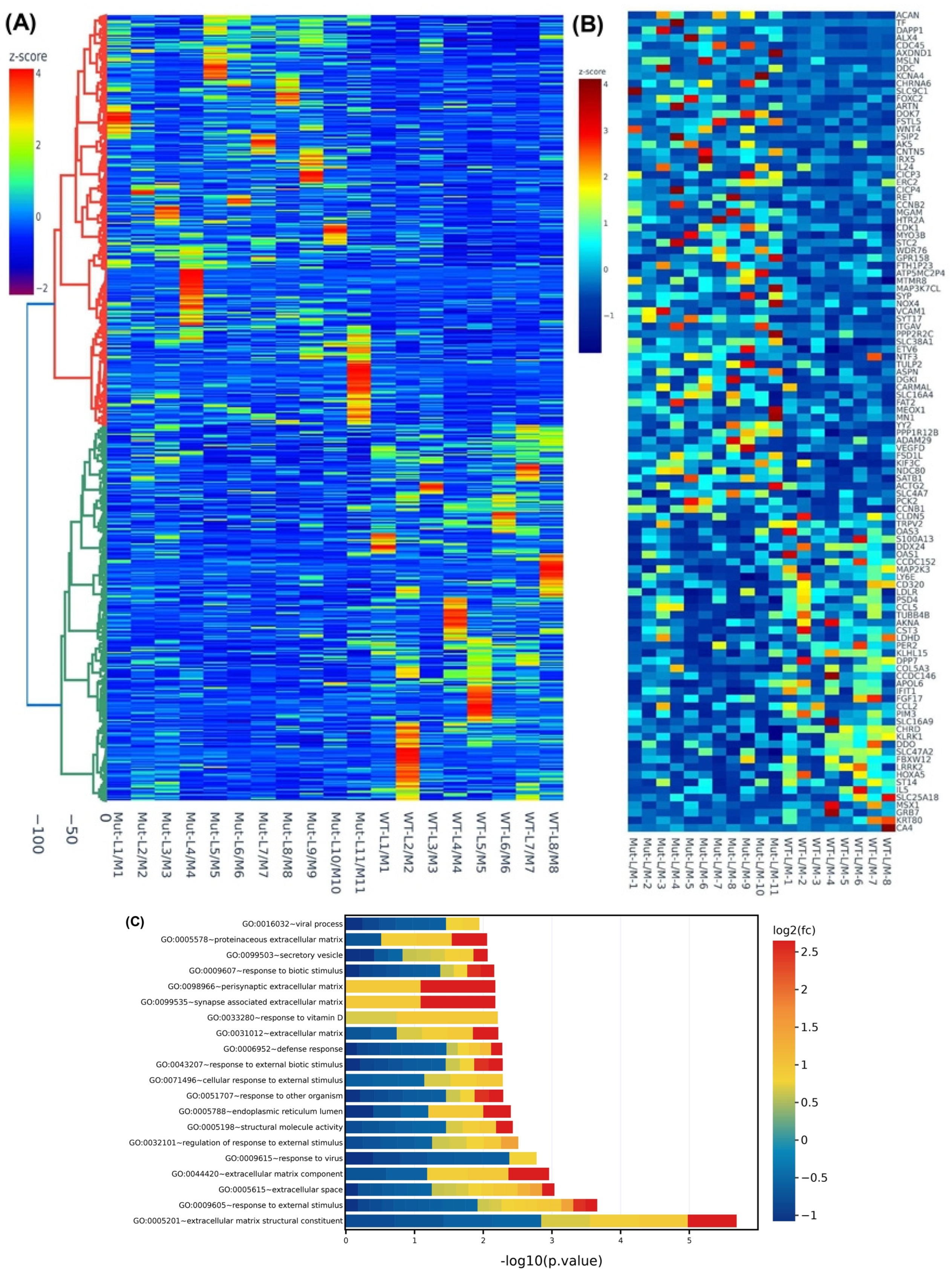
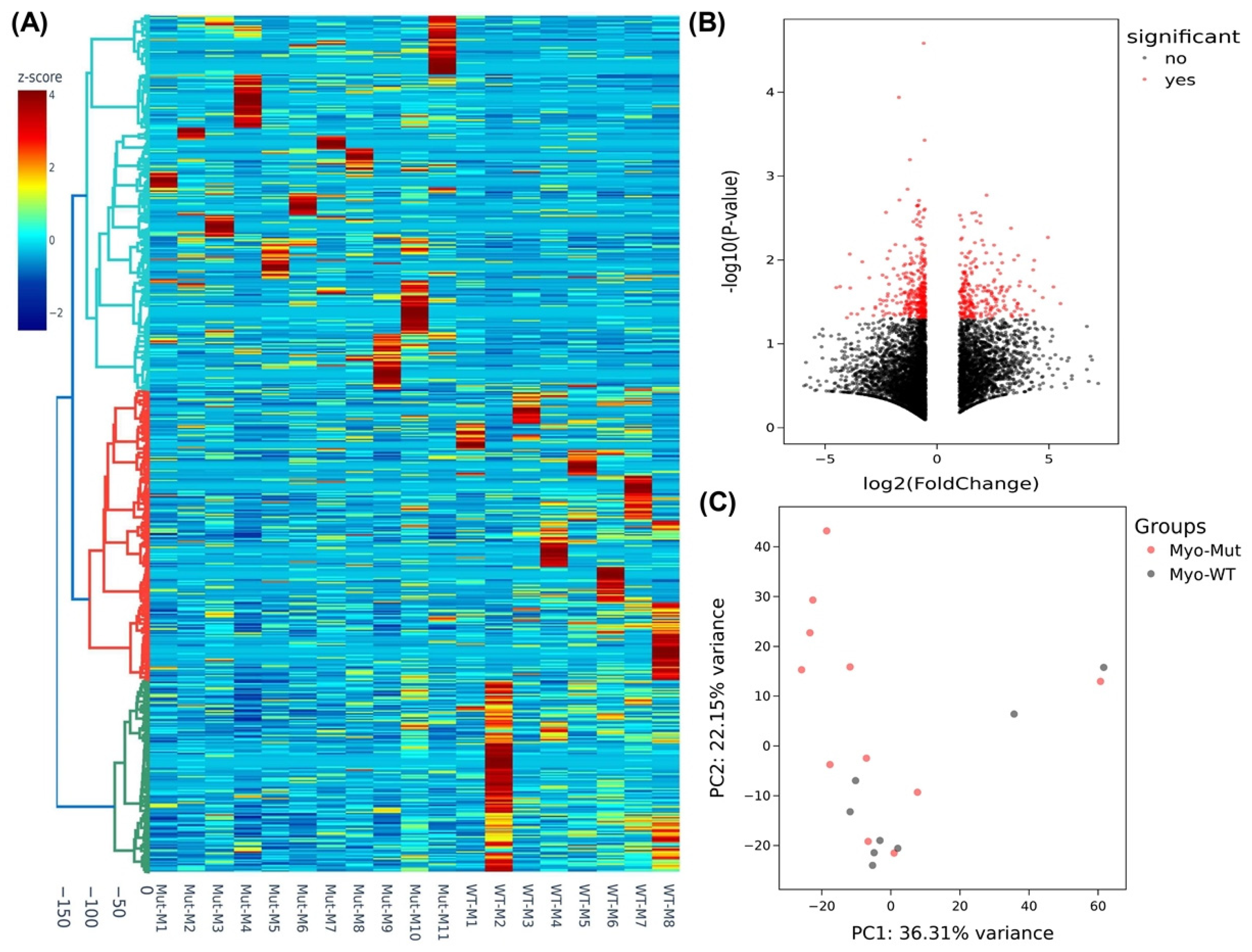
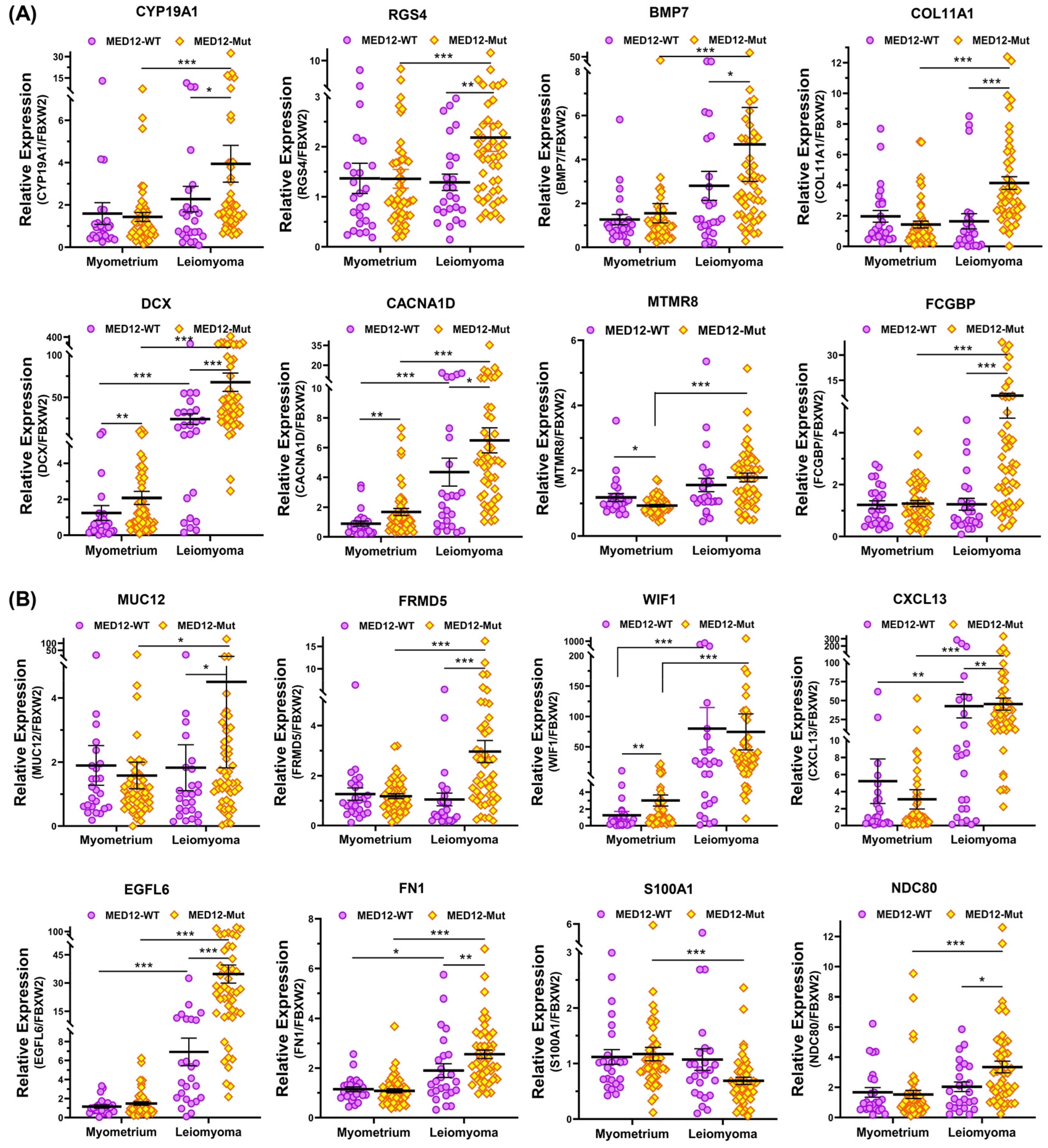
2.3. Differential Expression of MED12-Associated Coding RNA Transcripts in Myometrium
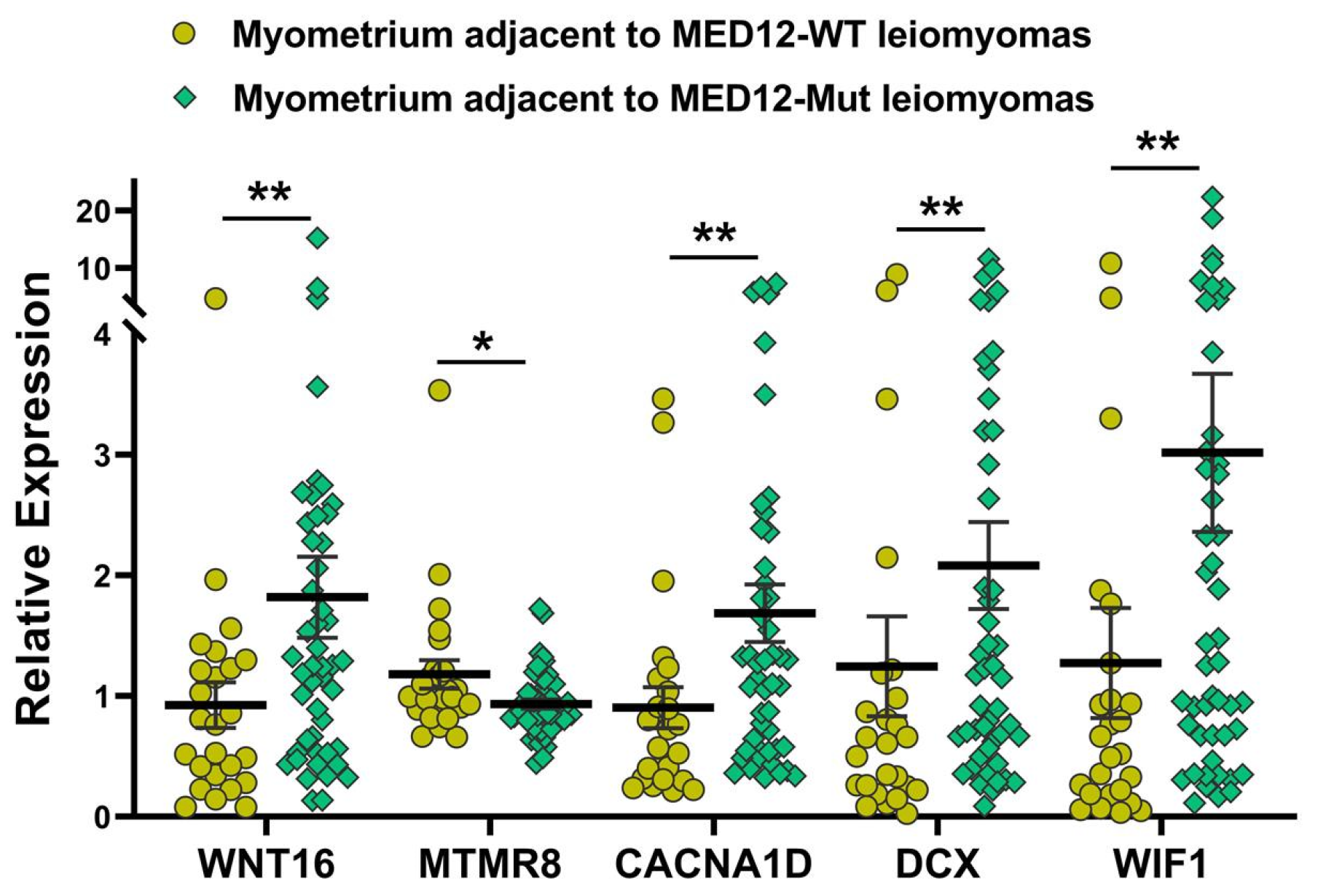
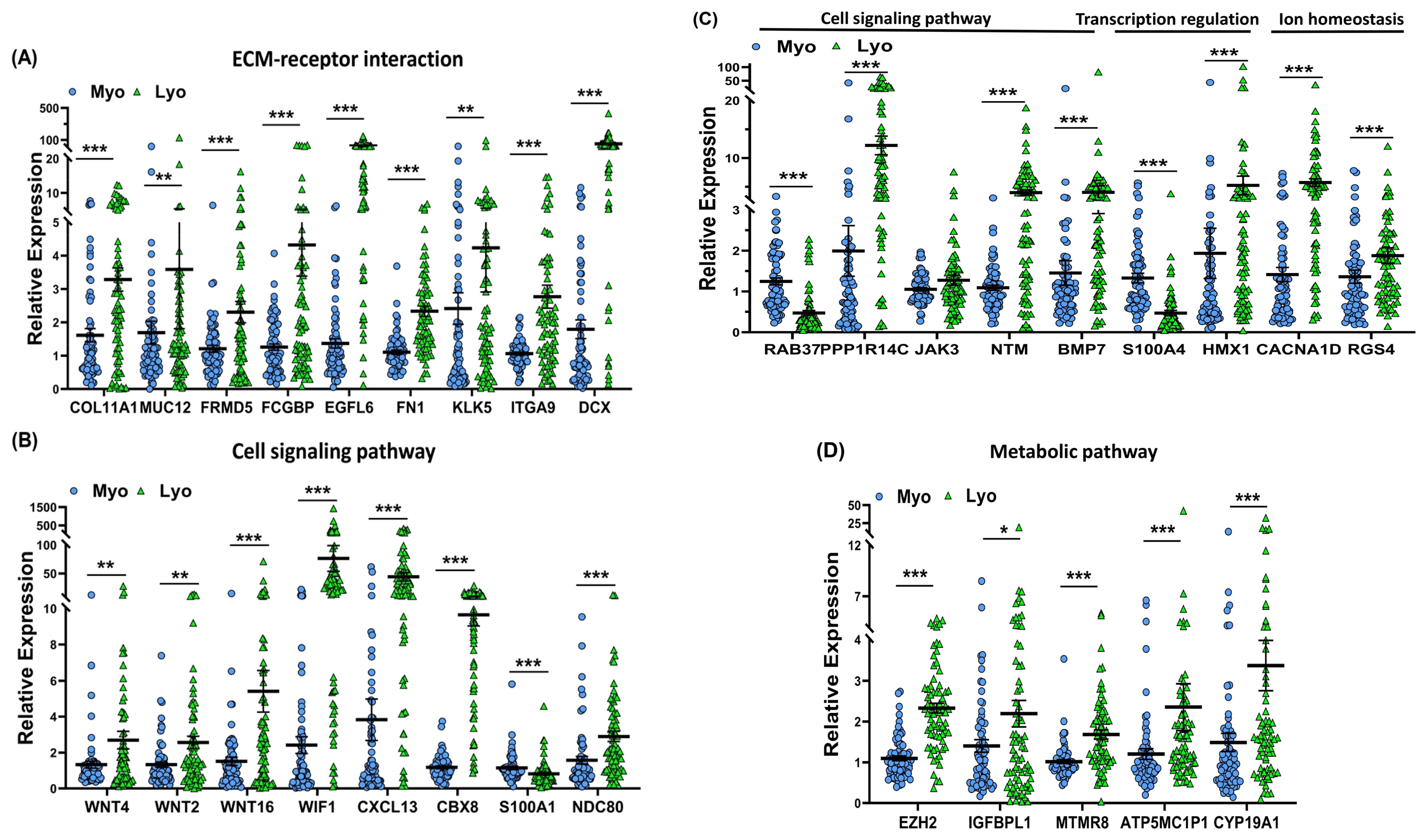
2.4. Validation of MED12-Associated Coding RNA Transcripts in Leiomyoma and Matched Myometrium
3. Discussion
4. Materials and Methods
4.1. Myometrium and Leiomyoma Tissue Collection
4.2. MED12-Mutation Analysis
4.3. RNA Sequencing and Bioinformatic Analysis
4.4. Quantitative RT-PCR
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stewart, E.A.; Cookson, C.L.; Gandolfo, R.A.; Schulze-Rath, R. Epidemiology of uterine fibroids: A systematic review. Bjog 2017, 124, 1501–1512. [Google Scholar] [CrossRef]
- Islam, M.S.; Ciavattini, A.; Petraglia, F.; Castellucci, M.; Ciarmela, P. Extracellular matrix in uterine leiomyoma pathogenesis: A potential target for future therapeutics. Hum. Reprod. Update 2018, 24, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Chegini, N. Proinflammatory and profibrotic mediators: Principal effectors of leiomyoma development as a fibrotic disorder. Semin. Reprod. Med. 2010, 28, 180–203. [Google Scholar] [CrossRef] [PubMed]
- Segars, J.H.; Parrott, E.C.; Nagel, J.D.; Guo, X.C.; Gao, X.; Birnbaum, L.S.; Pinn, V.W.; Dixon, D. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: Comprehensive review, conference summary and future recommendations. Hum. Reprod. Update 2014, 20, 309–333. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.K.; Holthouser, K.; Segars, J.H.; Leppert, P.C. Recent scientific advances in leiomyoma (uterine fibroids) research facilitates better understanding and management. F1000Res 2015, 4, 183. [Google Scholar] [CrossRef]
- Chuang, T.D.; Khorram, O. Expression Profiling of lncRNAs, miRNAs, and mRNAs and Their Differential Expression in Leiomyoma Using Next-Generation RNA Sequencing. Reprod. Sci. 2018, 25, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, X.; Dong, R.; Liu, X.; Li, Y.; Lu, S.; Xu, L.; Wang, Y.; Wang, X.; Hou, D.; et al. Integrated analysis of long noncoding RNAs and mRNAs reveals their potential roles in the pathogenesis of uterine leiomyomas. Oncotarget 2014, 5, 8625–8636. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Rehan, A.; Khorram, O. Functional role of the long noncoding RNA X-inactive specific transcript in leiomyoma pathogenesis. Fertil. Steril. 2021, 115, 238–247. [Google Scholar] [CrossRef]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Long Noncoding RNA MIAT Modulates the Extracellular Matrix Deposition in Leiomyomas by Sponging MiR-29 Family. Endocrinology 2021, 162, bqab186. [Google Scholar] [CrossRef]
- Falahati, Z.; Mohseni-Dargah, M.; Mirfakhraie, R. Emerging Roles of Long Non-coding RNAs in Uterine Leiomyoma Pathogenesis: A Review. Reprod. Sci. 2022, 29, 1086–1101. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Differential Expression of Super-Enhancer-Associated Long Non-coding RNAs in Uterine Leiomyomas. Reprod. Sci. 2022, 29, 2960–2976. [Google Scholar] [CrossRef] [PubMed]
- Lagana, A.S.; Vergara, D.; Favilli, A.; La Rosa, V.L.; Tinelli, A.; Gerli, S.; Noventa, M.; Vitagliano, A.; Triolo, O.; Rapisarda, A.M.C.; et al. Epigenetic and genetic landscape of uterine leiomyomas: A current view over a common gynecological disease. Arch. Gynecol. Obstet. 2017, 296, 855–867. [Google Scholar] [CrossRef]
- Mehine, M.; Makinen, N.; Heinonen, H.R.; Aaltonen, L.A.; Vahteristo, P. Genomics of uterine leiomyomas: Insights from high-throughput sequencing. Fertil. Steril. 2014, 102, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.C.; Kim, T.M.; Dreyfuss, J.M.; Somasundaram, P.; Christacos, N.C.; Rousselle, M.; Quade, B.J.; Park, P.J.; Stewart, E.A.; Morton, C.C. Expression profiling of uterine leiomyomata cytogenetic subgroups reveals distinct signatures in matched myometrium: Transcriptional profilingof the t(12;14) and evidence in support of predisposing genetic heterogeneity. Hum. Mol. Genet. 2012, 21, 2312–2329. [Google Scholar] [CrossRef] [PubMed]
- Liegl-Atzwanger, B.; Heitzer, E.; Flicker, K.; Muller, S.; Ulz, P.; Saglam, O.; Tavassoli, F.; Devouassoux-Shisheboran, M.; Geigl, J.; Moinfar, F. Exploring chromosomal abnormalities and genetic changes in uterine smooth muscle tumors. Mod. Pathol. 2016, 29, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Makinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Markowski, D.N.; Bartnitzke, S.; Löning, T.; Drieschner, N.; Helmke, B.M.; Bullerdiek, J. MED12 mutations in uterine fibroids--their relationship to cytogenetic subgroups. Int. J. Cancer 2012, 131, 1528–1536. [Google Scholar] [CrossRef] [PubMed]
- Al-Hendy, A.; Laknaur, A.; Diamond, M.P.; Ismail, N.; Boyer, T.G.; Halder, S.K. Silencing Med12 Gene Reduces Proliferation of Human Leiomyoma Cells Mediated via Wnt/beta-Catenin Signaling Pathway. Endocrinology 2017, 158, 592–603. [Google Scholar]
- El Andaloussi, A.; Al-Hendy, A.; Ismail, N.; Boyer, T.G.; Halder, S.K. Introduction of Somatic Mutation in MED12 Induces Wnt4/β-Catenin and Disrupts Autophagy in Human Uterine Myometrial Cell. Reprod. Sci. 2020, 27, 823–832. [Google Scholar] [CrossRef]
- Verger, A.; Monté, D.; Villeret, V. Twenty years of Mediator complex structural studies. Biochem. Soc. Trans. 2019, 47, 399–410. [Google Scholar] [CrossRef]
- Zhang, S.; O’Regan, R.; Xu, W. The emerging role of mediator complex subunit 12 in tumorigenesis and response to chemotherapeutics. Cancer 2020, 126, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, H.R.; Pasanen, A.; Heikinheimo, O.; Tanskanen, T.; Palin, K.; Tolvanen, J.; Vahteristo, P.; Sjoberg, J.; Pitkanen, E.; Butzow, R.; et al. Multiple clinical characteristics separate MED12-mutation-positive and -negative uterine leiomyomas. Sci. Rep. 2017, 7, 1015. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Shin, Y.H.; Yatsenko, S.A.; Castro, C.A.; Surti, U.; Rajkovic, A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Investig. 2015, 125, 3280–3284. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Makinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef]
- Park, M.J.; Shen, H.; Spaeth, J.M.; Tolvanen, J.H.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Yang, Q.; Bulun, S.E.; et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 2018, 293, 4870–4882. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Kim, S.; Ishii, S.; Boyer, T.G. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol. Cell. Biol. 2006, 26, 8667–8682. [Google Scholar] [CrossRef]
- Huang, S.; Holzel, M.; Knijnenburg, T.; Schlicker, A.; Roepman, P.; McDermott, U.; Garnett, M.; Grernrum, W.; Sun, C.; Prahallad, A.; et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell 2012, 151, 937–950. [Google Scholar] [CrossRef]
- Chuang, T.D.; Khorram, O. Regulation of Cell Cycle Regulatory Proteins by MicroRNAs in Uterine Leiomyoma. Reprod. Sci. 2019, 26, 250–258. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Khorram, O. Cross-talk between miR-29c and transforming growth factor-β3 is mediated by an epigenetic mechanism in leiomyoma. Fertil. Steril. 2019, 112, 1180–1189. [Google Scholar] [CrossRef]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Tryptophan catabolism is dysregulated in leiomyomas. Fertil. Steril. 2021, 116, 1160–1171. [Google Scholar] [CrossRef]
- Mas, A.; Cervello, I.; Fernandez-Alvarez, A.; Faus, A.; Diaz, A.; Burgues, O.; Casado, M.; Simon, C. Overexpression of the truncated form of High Mobility Group A proteins (HMGA2) in human myometrial cells induces leiomyoma-like tissue formation. Mol. Hum. Reprod. 2015, 21, 330–338. [Google Scholar] [CrossRef]
- Yang, Q.; Ciebiera, M.; Victoria Bariani, M.; Ali, M.; Elkafas, H.; Boyer, T.G.; Al-Hendy, A. Comprehensive Review of Uterine Fibroids: Developmental Origin, Pathogenesis, and Treatment. Endocr. Rev. 2022, 43, 678–719. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Luo, X.; Panda, H.; Chegini, N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Mol. Endocrinol. 2012, 26, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Bariani, M.V.; Yang, Q.; Al-Hendy, A. Understanding the Impact of Uterine Fibroids on Human Endometrium Function. Front. Cell Dev. Biol. 2021, 9, 633180. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Panda, H.; Luo, X.; Chegini, N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr. Relat. Cancer 2012, 19, 541–556. [Google Scholar] [CrossRef]
- George, J.W.; Fan, H.; Johnson, B.; Carpenter, T.J.; Foy, K.K.; Chatterjee, A.; Patterson, A.L.; Koeman, J.; Adams, M.; Madaj, Z.B.; et al. Integrated Epigenome, Exome, and Transcriptome Analyses Reveal Molecular Subtypes and Homeotic Transformation in Uterine Fibroids. Cell Rep. 2019, 29, 4069–4085.e6. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Luo, E.Y.; Adams, S.M.; Adams, T.; Ye, Y.; Shetye, S.S.; Soslowsky, L.J.; Birk, D.E. Collagen XI regulates the acquisition of collagen fibril structure, organization and functional properties in tendon. Matrix Biol. 2020, 94, 77–94. [Google Scholar] [CrossRef]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Gaweł, A.M.; Ratajczak, M.; Gajda, E.; Grzanka, M.; Paziewska, A.; Cieślicka, M.; Kulecka, M.; Oczko-Wojciechowska, M.; Godlewska, M. Analysis of the Role of FRMD5 in the Biology of Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2021, 22, 6726. [Google Scholar] [CrossRef]
- Hu, J.; Niu, M.; Li, X.; Lu, D.; Cui, J.; Xu, W.; Li, G.; Zhan, J.; Zhang, H. FERM domain-containing protein FRMD5 regulates cell motility via binding to integrin β5 subunit and ROCK1. FEBS Lett. 2014, 588, 4348–4356. [Google Scholar] [CrossRef]
- Yan, T.; Tian, D.; Chen, J.; Tan, Y.; Cheng, Y.; Ye, L.; Deng, G.; Liu, B.; Yuan, F.; Zhang, S.; et al. FCGBP Is a Prognostic Biomarker and Associated With Immune Infiltration in Glioma. Front. Oncol. 2021, 11, 769033. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Niu, X.; Li, Y.; Zhang, J.R.; Zhu, S.J.; Yang, Q.Y.; Zhang, W.; Gong, L. Role of the mucin-like glycoprotein FCGBP in mucosal immunity and cancer. Front. Immunol. 2022, 13, 863317. [Google Scholar] [CrossRef]
- Kang, J.; Wang, J.; Tian, J.; Shi, R.; Jia, H.; Wang, Y. The emerging role of EGFL6 in angiogenesis and tumor progression. Int. J. Med. Sci. 2020, 17, 1320. [Google Scholar] [CrossRef] [PubMed]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115 Pt 20, 3861–3863. [Google Scholar] [CrossRef] [PubMed]
- Yue, B. Biology of the extracellular matrix: An overview. J. Glaucoma 2014, 23 (Suppl. S1), S20–S23. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Lai, J.; Clements, J.A. Kallikreins on steroids: Structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr. Rev. 2010, 31, 407–446. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Orikawa, H.; Baba, T.; Fukushima, T.; Kataoka, H. Hepatocyte growth factor activator is a serum activator of single-chain precursor macrophage-stimulating protein. FEBS J. 2009, 276, 3481–3490. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, J.; Tan, F.; Wang, B.; Xu, W.; Yuan, C. ITGA9: Potential Biomarkers and Therapeutic Targets in Different Tumors. Curr. Pharm. Des. 2022, 28, 1412–1418. [Google Scholar] [CrossRef]
- Ayanlaja, A.A.; Xiong, Y.; Gao, Y.; Ji, G.; Tang, C.; Abdikani Abdullah, Z.; Gao, D. Distinct Features of Doublecortin as a Marker of Neuronal Migration and Its Implications in Cancer Cell Mobility. Front. Mol. Neurosci. 2017, 10, 199. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, T.; Zhang, S.; Wang, J.; Chen, Y.; Zhao, H.; Yang, Y.; Shi, S.; Chen, Q.; Liu, K. The Wnt signaling pathway in tumorigenesis, pharmacological targets, and drug development for cancer therapy. Biomark. Res. 2021, 9, 68. [Google Scholar] [CrossRef]
- Kitaya, K.; Yasuo, T. Aberrant expression of selectin E, CXCL1, and CXCL13 in chronic endometritis. Mod. Pathol. 2010, 23, 1136–1146. [Google Scholar] [CrossRef]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef]
- Völkers, M.; Rohde, D.; Goodman, C.; Most, P. S100A1: A regulator of striated muscle sarcoplasmic reticulum Ca2+ handling, sarcomeric, and mitochondrial function. J. Biomed. Biotechnol. 2010, 2010, 178614. [Google Scholar] [CrossRef]
- Sun, B.; Kekenes-Huskey, P.M. Molecular Basis of S100A1 Activation and Target Regulation Within Physiological Cytosolic Ca(2+) Levels. Front. Mol. Biosci. 2020, 7, 77. [Google Scholar] [CrossRef]
- Tooley, J.; Stukenberg, P.T. The Ndc80 complex: Integrating the kinetochore’s many movements. Chromosome Res. 2011, 19, 377–391. [Google Scholar] [CrossRef]
- Gil, O.D.; Zhang, L.; Chen, S.; Ren, Y.Q.; Pimenta, A.; Zanazzi, G.; Hillman, D.; Levitt, P.; Salzer, J.L. Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J. Neurobiol. 2002, 51, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Cheng, H.C.; Wang, Y.S.; Lin, P.; Jen, J.; Kuo, I.Y.; Chang, Y.H.; Liao, P.C.; Chen, R.H.; Yuan, W.C.; et al. Small GTPase Rab37 targets tissue inhibitor of metalloproteinase 1 for exocytosis and thus suppresses tumour metastasis. Nat. Commun. 2014, 5, 4804. [Google Scholar] [CrossRef] [PubMed]
- Drgonova, J.; Zimonjic, D.B.; Hall, F.S.; Uhl, G.R. Effect of KEPI (Ppp1r14c) deletion on morphine analgesia and tolerance in mice of different genetic backgrounds: When a knockout is near a relevant quantitative trait locus. Neuroscience 2010, 165, 882–895. [Google Scholar] [CrossRef]
- Xiong, L.; Xia, W.F.; Tang, F.L.; Pan, J.X.; Mei, L.; Xiong, W.C. Retromer in Osteoblasts Interacts With Protein Phosphatase 1 Regulator Subunit 14C, Terminates Parathyroid Hormone’s Signaling, and Promotes Its Catabolic Response. EBioMedicine 2016, 9, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Mengie Ayele, T.; Tilahun Muche, Z.; Behaile Teklemariam, A.; Bogale Kassie, A.; Chekol Abebe, E. Role of JAK2/STAT3 Signaling Pathway in the Tumorigenesis, Chemotherapy Resistance, and Treatment of Solid Tumors: A Systemic Review. J. Inflamm. Res. 2022, 15, 1349–1364. [Google Scholar] [CrossRef]
- Wissmann, C.; Wild, P.J.; Kaiser, S.; Roepcke, S.; Stoehr, R.; Woenckhaus, M.; Kristiansen, G.; Hsieh, J.C.; Hofstaedter, F.; Hartmann, A.; et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J. Pathol. 2003, 201, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.; Tao, Q.; Zhong, S.; Fields, C.R.; Kim, W.J.; Lee, M.W.; Cui, Y.; Brown, K.D.; Robertson, K.D. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis 2006, 27, 1341–1348. [Google Scholar] [CrossRef]
- Dituri, F.; Cossu, C.; Mancarella, S.; Giannelli, G. The Interactivity between TGFβ and BMP Signaling in Organogenesis, Fibrosis, and Cancer. Cells 2019, 8, 1130. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, E.; Chen, D.F. Emerging roles for insulin-like growth factor binding protein like protein 1. Neural Regen. Res. 2019, 14, 258–259. [Google Scholar]
- Yoo, K.Y.; Son, J.Y.; Lee, J.U.; Shin, W.; Im, D.W.; Kim, S.J.; Ryu, S.E.; Heo, Y.S. Structure of the catalytic phosphatase domain of MTMR8: Implications for dimerization, membrane association and reversible oxidation. Acta Crystallogr. D Biol. Crystallogr. 2015, 71 Pt 7, 1528–1539. [Google Scholar] [CrossRef]
- Vives-Bauza, C.; Magrané, J.; Andreu, A.L.; Manfredi, G. Novel role of ATPase subunit C targeting peptides beyond mitochondrial protein import. Mol. Biol. Cell 2010, 21, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Y.; Lin, H. Estrogen disorders: Interpreting the abnormal regulation of aromatase in granulosa cells (Review). Int. J. Mol. Med. 2021, 47, 73. [Google Scholar] [CrossRef] [PubMed]
- Ambartsumian, N.; Klingelhöfer, J.; Grigorian, M. The Multifaceted S100A4 Protein in Cancer and Inflammation. Methods Mol. Biol. 2019, 1929, 339–365. [Google Scholar]
- Boulling, A.; Wicht, L.; Schorderet, D.F. Identification of HMX1 target genes: A predictive promoter model approach. Mol. Vis. 2013, 19, 1779–1794. [Google Scholar] [PubMed]
- Banciu, A.; Banciu, D.D.; Mustaciosu, C.C.; Radu, M.; Cretoiu, D.; Xiao, J.; Cretoiu, S.M.; Suciu, N.; Radu, B.M. Beta-Estradiol Regulates Voltage-Gated Calcium Channels and Estrogen Receptors in Telocytes from Human Myometrium. Int. J. Mol. Sci. 2018, 19, 1413. [Google Scholar] [CrossRef]
- Georgoussi, Z.; Leontiadis, L.; Mazarakou, G.; Merkouris, M.; Hyde, K.; Hamm, H. Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell. Signal. 2006, 18, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Moyo, M.B.; Parker, J.B.; Chakravarti, D. Altered chromatin landscape and enhancer engagement underlie transcriptional dysregulation in MED12 mutant uterine leiomyomas. Nat. Commun. 2020, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Xie, Y.; Yan, W.; Khorram, O. Next-generation sequencing reveals differentially expressed small noncoding RNAs in uterine leiomyoma. Fertil. Steril. 2018, 109, 919–929. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Asif, H.; Feng, Y.; Kohrn, B.F.; Kennedy, S.R.; Kim, J.J.; Wei, J.J. Myometrial oxidative stress drives MED12 mutations in leiomyoma. Cell Biosci. 2022, 12, 111. [Google Scholar] [CrossRef]
- Paul, E.N.; Burns, G.W.; Carpenter, T.J.; Grey, J.A.; Fazleabas, A.T.; Teixeira, J.M. Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium. Int. J. Mol. Sci. 2021, 22, 3618. [Google Scholar] [CrossRef]
- Mayer, A.; Höckel, M.; Wree, A.; Leo, C.; Horn, L.C.; Vaupel, P. Lack of hypoxic response in uterine leiomyomas despite severe tissue hypoxia. Cancer Res. 2008, 68, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chen, X.; He, B.; Liu, S.; Li, Y.; Wang, Q.; Gao, H.; Wang, S.; Liu, J.; Zhang, S.; et al. ROS are critical for endometrial breakdown via NF-κB-COX-2 signaling in a female mouse menstrual-like model. Endocrinology 2014, 155, 3638–3648. [Google Scholar] [CrossRef]
- Tanwar, P.S.; Lee, H.J.; Zhang, L.; Zukerberg, L.R.; Taketo, M.M.; Rueda, B.R.; Teixeira, J.M. Constitutive activation of Beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol. Reprod. 2009, 81, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yin, P.; Dotts, A.J.; Kujawa, S.A.; Coon, V.J.; Wei, J.J.; Chakravarti, D.; Bulun, S.E. Activation of protein kinase B by WNT4 as a regulator of uterine leiomyoma stem cell function. Fertil. Steril. 2020, 114, 1339–1349. [Google Scholar] [CrossRef]
- Ono, M.; Yin, P.; Navarro, A.; Moravek, M.B.; Coon, J.S.t.; Druschitz, S.A.; Serna, V.A.; Qiang, W.; Brooks, D.C.; Malpani, S.S.; et al. Paracrine activation of WNT/beta-catenin pathway in uterine leiomyoma stem cells promotes tumor growth. Proc. Natl. Acad. Sci. USA 2013, 110, 17053–17058. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.A.; Jamaluddin, M.F.B.; Adebayo, M.; Bajwa, P.; Scott, R.J.; Dharmarajan, A.M.; Nahar, P.; Tanwar, P.S. Extracellular matrix (ECM) activates beta-catenin signaling in uterine fibroids. Reproduction 2018, 155, 61–71. [Google Scholar] [PubMed]
- Malinauskas, T.; Aricescu, A.R.; Lu, W.; Siebold, C.; Jones, E.Y. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nat. Struct. Mol. Biol. 2011, 18, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Litovkin, K.V.; Ivanova, O.V.; Bauer, A.; Hoheisel, J.D.; Bubnov, V.V.; Zaporozhan, V.N. Microarray study of gene expression in uterine leiomyoma. Exp. Oncol. 2008, 30, 106–111. [Google Scholar]
- Maekawa, R.; Sato, S.; Tamehisa, T.; Sakai, T.; Kajimura, T.; Sueoka, K.; Sugino, N. Different DNA methylome, transcriptome and histological features in uterine fibroids with and without MED12 mutations. Sci. Rep. 2022, 12, 8912. [Google Scholar] [CrossRef]
- Arslan, A.A.; Gold, L.I.; Mittal, K.; Suen, T.C.; Belitskaya-Levy, I.; Tang, M.S.; Toniolo, P. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: New evidence and a systematic review. Hum. Reprod. 2005, 20, 852–863. [Google Scholar] [CrossRef]
- Bae, S.M.; Kim, Y.W.; Lee, J.M.; Namkoong, S.E.; Kim, C.K.; Ahn, W.S. Expression profiling of the cellular processes in uterine leiomyomas: Omic approaches and IGF-2 association with leiomyosarcomas. Cancer Res. Treat. 2004, 36, 31–42. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Li, Q.; Gong, Y.; Liu, X.; Bi, L.; Hu, C. Upregulation of kallikrein-related peptidase 5 is associated with the malignant behavior of colorectal cancer. Mol. Med. Rep. 2016, 14, 2164–2170. [Google Scholar] [CrossRef]
- Johnson, J.J.; Miller, D.L.; Jiang, R.; Liu, Y.; Shi, Z.; Tarwater, L.; Williams, R.; Balsara, R.; Sauter, E.R.; Stack, M.S. Protease-activated Receptor-2 (PAR-2)-mediated Nf-κB Activation Suppresses Inflammation-associated Tumor Suppressor MicroRNAs in Oral Squamous Cell Carcinoma. J. Biol. Chem. 2016, 291, 6936–6945. [Google Scholar] [CrossRef]
- Abuduhadeer, X.; Xu, X.; Aihesan, K.; Yilihamu, M.; Zhao, Y.; Zhang, W. Clinical significance of kallikrein 5 as a novel prognostic biomarker in gastric adenocarcinoma. J. Clin. Lab. Anal. 2021, 35, e23958. [Google Scholar] [CrossRef]
- Kryza, T.; Silva, M.L.; Loessner, D.; Heuzé-Vourc’h, N.; Clements, J.A. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie 2016, 122, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. miR-200c Regulates IL8 Expression by Targeting IKBKB: A Potential Mediator of Inflammation in Leiomyoma Pathogenesis. PLoS ONE 2014, 9, e95370. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil. Steril. 2016, 105, 236–245.e1. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.T.; Cannon, B.R.; Zimmer, D.B.; Weber, D.J. S100A1: Structure, Function, and Therapeutic Potential. Curr. Chem. Biol. 2009, 3, 138–145. [Google Scholar]
- Diaz-Romero, J.; Nesic, D. S100A1 and S100B: Calcium Sensors at the Cross-Roads of Multiple Chondrogenic Pathways. J. Cell. Physiol. 2017, 232, 1979–1987. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Liu, S.; Qin, Z. Extracellular S100A4 as a key player in fibrotic diseases. J. Cell Mol. Med. 2020, 24, 5973–5983. [Google Scholar] [CrossRef]
- Schneider, M.; Hansen, J.L.; Sheikh, S.P. S100A4: A common mediator of epithelial-mesenchymal transition, fibrosis and regeneration in diseases? J. Mol. Med. 2008, 86, 507–522. [Google Scholar] [CrossRef]
- Dahlmann, M.; Kobelt, D.; Walther, W.; Mudduluru, G.; Stein, U. S100A4 in Cancer Metastasis: Wnt Signaling-Driven Interventions for Metastasis Restriction. Cancers 2016, 8, 59. [Google Scholar] [CrossRef]
- Bulun, S.E.; Imir, G.; Utsunomiya, H.; Thung, S.; Gurates, B.; Tamura, M.; Lin, Z. Aromatase in endometriosis and uterine leiomyomata. J. Steroid Biochem. Mol. Biol. 2005, 95, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Reierstad, S.; Demura, M.; Rademaker, A.W.; Kasai, T.; Inoue, M.; Usui, H.; Shozu, M.; Bulun, S.E. High aromatase expression in uterine leiomyoma tissues of African-American women. J. Clin. Endocrinol. Metab. 2009, 94, 1752–1756. [Google Scholar] [CrossRef]
- Mlodawska, O.W.; Saini, P.; Parker, J.B.; Wei, J.J.; Bulun, S.E.; Simon, M.A.; Chakravarti, D. Epigenomic and enhancer dysregulation in uterine leiomyomas. Hum. Reprod. Update 2022, 28, 518–547. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Nair, S.; Laknaur, A.; Ismail, N.; Diamond, M.P.; Al-Hendy, A. The Polycomb Group Protein EZH2 Impairs DNA Damage Repair Gene Expression in Human Uterine Fibroids. Biol. Reprod. 2016, 94, 69. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.D.; Khorram, O. Tranilast Inhibits Genes Functionally Involved in Cell Proliferation, Fibrosis, and Epigenetic Regulation and Epigenetically Induces miR-29c Expression in Leiomyoma Cells. Reprod. Sci. 2017, 24, 1253–1263. [Google Scholar] [CrossRef]
- Chuang, T.D.; Quintanilla, D.; Boos, D.; Khorram, O. Further characterization of tryptophan metabolism and its dysregulation in fibroids. F&S Sci. 2022, 3, 392–400. [Google Scholar]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Res 2019, 8, 1874. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Delhomme, N.; Padioleau, I.; Furlong, E.E.; Steinmetz, L.M. easyRNASeq: A bioconductor package for processing RNA-Seq data. Bioinformatics 2012, 28, 2532–2533. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Duitama, C.; Metge, F.; Rosskopp, D.; Boucas, J. Flaski (2.0.3). Zenodo. 2021. Available online: https://zenodo.org/record/4890981#.Y-bRDXbMK3A (accessed on 28 December 2022).
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Almeida, T.A.; Quispe-Ricalde, A.; Montes de Oca, F.; Foronda, P.; Hernandez, M.M. A high-throughput open-array qPCR gene panel to identify housekeeping genes suitable for myometrium and leiomyoma expression analysis. Gynecol. Oncol. 2014, 134, 138–143. [Google Scholar] [CrossRef] [PubMed]


| GO/KEGG Pathway Enrichment | Symbol | Lyo vs Myo | Lyo/Myo(+) vs Lyo/Myo(−) | Lyo(+) vs Lyo(−) | Myo(+) vs Myo(−) | Function |
|---|---|---|---|---|---|---|
| ECM-receptor interaction | COL11A1 | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Minor fibrillar collagen; expression has been associated with advanced tumorigenic disease and epithelial-mesenchymal transition (EMT) [37]. |
| ECM-receptor interaction | MUC12 | Up (p < 0.05) | Up (p < 0.05) | Up (p < 0.05) | No Significance | An O-glycosylated protein of the mucin family; mucous barrier on epithelial surfaces; involves in adhesion modulation, epithelial renewal, differentiation and intracellular signaling via EGF-like domains in its extracellular region [38]. |
| ECM-receptor interaction | FRMD5 | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Part of adherens junction and involved in regulation of cell migration, cellular metabolism, and signal transduction [39,40]. |
| ECM-receptor interaction | FCGBP | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Contains multiple von Willebrand D (VWD) domains that form complexes through disulfide-linked heterodimers with members of the mucin and trefoil factor family, which affect the attachment and motility of pathogens on mucosal surfaces [41,42]. |
| ECM-receptor interaction | EGFL6 | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Member of EGF superfamily, is expressed at significant levels during developmental processes and various malignant cancers; involved in the regulation of cell cycle, tumor proliferation, invasion, and metastasis through activation of multiple signaling pathways including PI3K/AKT, ERK/MAPK, Wnt/β-catenin and integrin-mediated signaling pathway [43]. |
| ECM-receptor interaction | FN1 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.01) | No Significance | A major component of the extracellular matrix, exists as a dimeric or multimeric form with other extracellular matrix proteins such as integrins, collagen, fibrin, and heparan sulfate proteoglycans linked by disulfide bonds [44,45]. |
| ECM-receptor interaction | KLK5 | Up (p < 0.01) | Up (p < 0.01) | Up (p < 0.001) | No Significance | KLK5, a member of the kallikrein subfamily, is involved in collagen formation and MSP-RON signaling [46,47]. |
| ECM-receptor interaction | ITGA9 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.01) | No Significance | An integrin subunit that mediates cell-cell and cell-matrix adhesion and accelerates cell migration and regulates various biological functions including cancer cell proliferation, angiogenesis, adhesion and invasion [48]. |
| ECM-receptor interaction | DCX | Up (p < 0.001) | No Significance | Up (p < 0.001) | Up (p < 0.01) | A microtubule-associated protein, contains two internal tandem repeats and stabilizes microtubules through bundling to the microtubule cytoskeleton [49]. |
| Cell signaling pathway | WNT4 | Up (p < 0.01) | Up (p < 0.01) | Up (p < 0.001) | No Significance | Ligand for members of the frizzled family of seven transmembrane receptors; works as a biphasic initiator for activating the canonical and non-canonical Wnt signaling [50]. |
| Cell signaling pathway | WNT2 | Up (p < 0.01) | Up (p < 0.05) | Up (p < 0.01) | No Significance | Enriched in cancer-associated fibroblasts; has the potential to enhance the growth and invasion of colorectal cancer [50]. |
| Cell signaling pathway | WNT16 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.001) | Up (p < 0.01) | Has no homology to any other Wnts signaling molecule; implicated in tumorigenesis and in skeletal development and postnatal bone homeostasis [50]. |
| Cell signaling pathway | CXCL13 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.01) | No Significance | Potent B lymphocyte chemoattractant, which promotes the migration of B lymphocytes, and is one of the most abundant chemokines in endometrial epithelial cells [51]. |
| Cell signaling pathway | CBX8 | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Component of a PcG PRC1-like complex, is involved in the RNA polymerase II-mediated transcription repression of genes [52]. |
| Cell signaling pathway | S100A1 | Down (p < 0.001) | Down (p < 0.05) | No Significance | No Significance | Member of the S100 family of calcium-binding proteins; interacts with specific target proteins, resulting in the modulation of their activity [53,54]. |
| Cell signaling pathway | NDC80 | Up (p < 0.001) | Up (p < 0.05) | Up (p < 0.05) | No Significance | Component of the essential kinetochore-related NDC80 complex, which is involved in the organization and stabilization of microtubule-kinetochore interactions, spindle checkpoint signaling and chromosome segregation [55]. |
| Cell signaling pathway | NTM | Up (p < 0.001) | Up (p < 0.001) | Up (p < 0.001) | No Significance | Neural cell adhesion molecule, promotes adhesion and neurite outgrowth via a homophilic mechanism [56]. |
| Cell signaling pathway | RAB37 | Down (p < 0.001) | Down (p < 0.001) | Down (p < 0.001) | No Significance | Rab small GTPase protein, through switching its guanine nucleotide binding status between GDP-bound (inactive) and GTP-bound (active) functions as a critical regulator in exocytotic pathway [57]. |
| Cell signaling pathway | PPP1R14C | Up (p < 0.001) | Up (p < 0.05) | Up (p < 0.01) | No Significance | Inhibitor of the PP1 serine/threonine phosphatase, which regulates the activation of PTH(1–34)-induced catabolic response and the non-canonical PTH1R signaling pathway [58,59]. |
| Cell signaling pathway | JAK3 | No Significance | Up (p < 0.01) | Up (p < 0.01) | No Significance | Member of the JAK family of non-receptor tyrosine kinases, plays a pivotal role in cytokine and growth factor-mediated intracellular signal transduction via the JAK/STAT pathway [60]. |
| Cell signaling pathway | WIF1 | Up (p < 0.001) | No Significance | No Significance | Up (p < 0.01) | Secreted protein that binds and inhibits the activity of Wnt proteins.Downregulated in numerous cancers via epigenetic transcriptional silencing mechanism [61,62]. |
| Cell signaling pathway | BMP7 | Up (p < 0.001) | No Significance | Up (p < 0.05) | No Significance | Secreted ligand of the TGF-β superfamily, activates TGF-β signaling via receptor-mediated activation of Smad transcription factors leading to target genes regulation [63]. |
| Metabolic pathways | EZH2 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.01) | No Significance | Catalytic core protein in PRC2 and facilitated PRC2-mediated H3K27me3 reaction. The dysregulation of EZH2 is acting as an important driver of tumorigenesis and progression [64]. |
| Metabolic pathways | IGFBPL1 | Up (p < 0.05) | Up (p < 0.001) | Up (p < 0.01) | No Significance | Located in extracellular space, is an IGF-binding protein that could prolong the half-life of IGF proteins and either promotes or inhibits the effects of IGF proteins on cell growth [65]. |
| Metabolic pathways | MTMR8 | Up (p < 0.001) | Up (p < 0.05) | No Significance | Down (p < 0.05) | Member of the myotubularin-related family; has phosphatase activity towards lipids containing phosphoinositol headgroup and acts on phosphatidylinositol 3-phosphate and phosphatidylinositol 3,5-bisphosphate; functions in membrane trafficking, cytoskeletal regulation, and receptor signaling [66]. |
| Metabolic pathways | ATP5MC1P1 | Up (p < 0.001) | Up (p < 0.05) | Up (p < 0.01) | No Significance | Pseudogene of ATP5MC1, which is encoded by the mitochondrial DNA and is subunit c of mitochondrial ATP synthase (F1F0 ATP synthase or Complex V) [67]. |
| Metabolic pathways | CYP19A1 | Up (p < 0.001) | No Significance | Up (p < 0.05) | No Significance | Member of the cytochrome P450 superfamily, is a monooxygenase involved in many reactions such as synthesis of steroids, lipids, cholesterol, and drug metabolism [68]. |
| Transcription regulation | S100A4 | Down (p < 0.001) | Down (p < 0.01) | Down (p < 0.01) | No Significance | Member of the S100 calcium-binding protein family, through its interaction with other proteins plays essential roles in many cellular processes and the development of cancers including metastasis, differentiation, inflammation, metastasis, and cell cycle progression [69]. |
| Transcription regulation | HMX1 | Up (p < 0.001) | Up (p < 0.01) | Up (p < 0.01) | No Significance | Transcription factor that belongs to the homeobox proteins (H6 family), and it recognizes and binds to the 5′-CAAG-3′ core DNA sequence. HMX1 may act as a transcriptional repressor and involved in the development of craniofacial structures [70]. |
| Ion homeostasis | CACNA1D | Up (p < 0.001) | No Significance | Up (p < 0.05) | Up (p < 0.01) | Member of the high-voltage-activated Ca2+ channels (HVA), is expressed in uterus and involved in a variety of calcium signaling related processes [71]. |
| Ion homeostasis | RGS4 | Up (p < 0.001) | No Significance | Up (p < 0.01) | No Significance | Regulates numerous G protein-coupled receptors (GPCRs) associated post-receptor signaling cascades [72]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, T.-D.; Gao, J.; Quintanilla, D.; McSwiggin, H.; Boos, D.; Yan, W.; Khorram, O. Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas. Int. J. Mol. Sci. 2023, 24, 3742. https://doi.org/10.3390/ijms24043742
Chuang T-D, Gao J, Quintanilla D, McSwiggin H, Boos D, Yan W, Khorram O. Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas. International Journal of Molecular Sciences. 2023; 24(4):3742. https://doi.org/10.3390/ijms24043742
Chicago/Turabian StyleChuang, Tsai-Der, Jianjun Gao, Derek Quintanilla, Hayden McSwiggin, Drake Boos, Wei Yan, and Omid Khorram. 2023. "Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas" International Journal of Molecular Sciences 24, no. 4: 3742. https://doi.org/10.3390/ijms24043742
APA StyleChuang, T.-D., Gao, J., Quintanilla, D., McSwiggin, H., Boos, D., Yan, W., & Khorram, O. (2023). Differential Expression of MED12-Associated Coding RNA Transcripts in Uterine Leiomyomas. International Journal of Molecular Sciences, 24(4), 3742. https://doi.org/10.3390/ijms24043742






