Salinity-Induced Cytosolic Alkaline Shifts in Arabidopsis Roots Require the SOS Pathway
Abstract
1. Introduction
2. Results
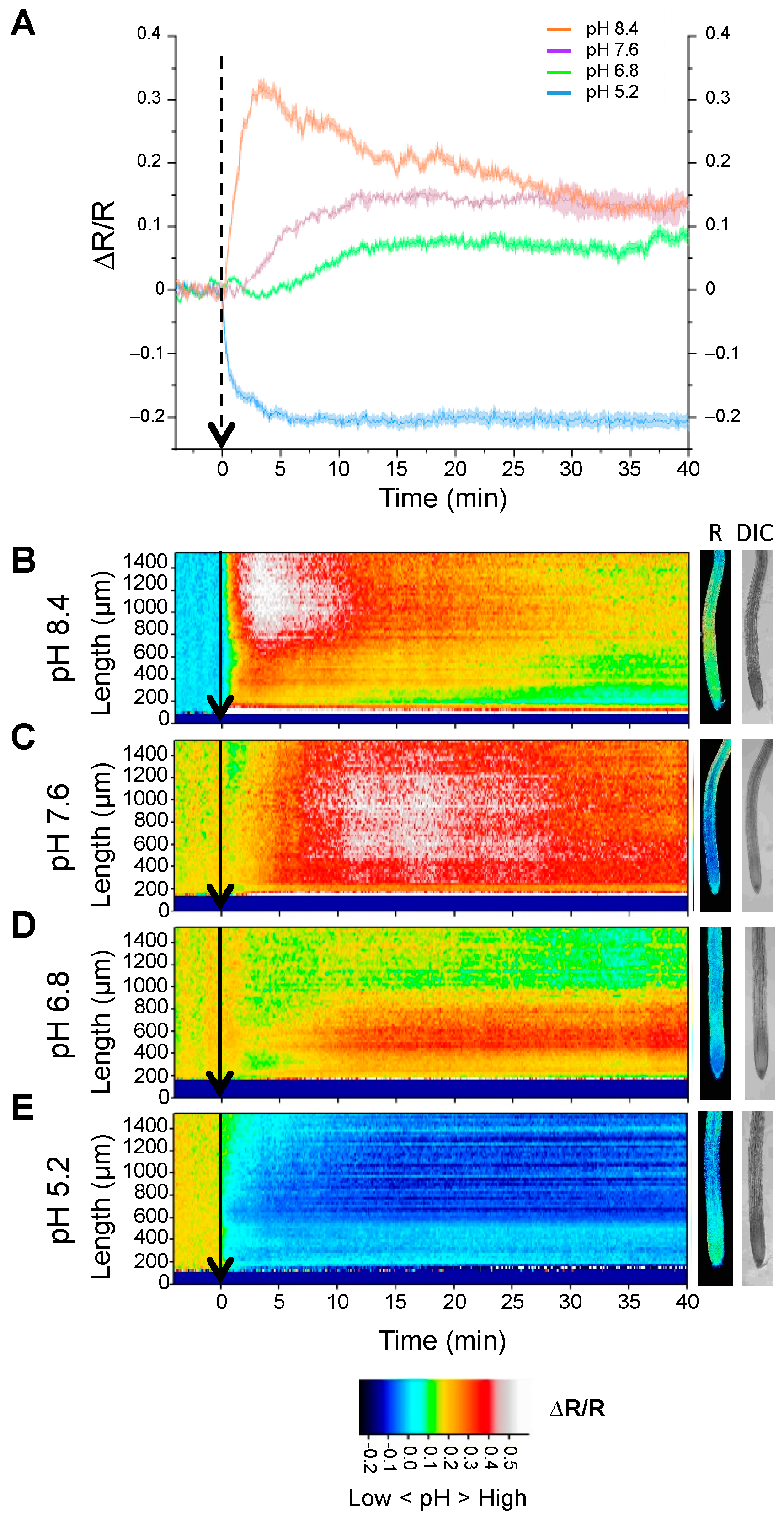
2.1. In Vivo Measurement of Cytosolic pH with Genetically Encoded Sensors
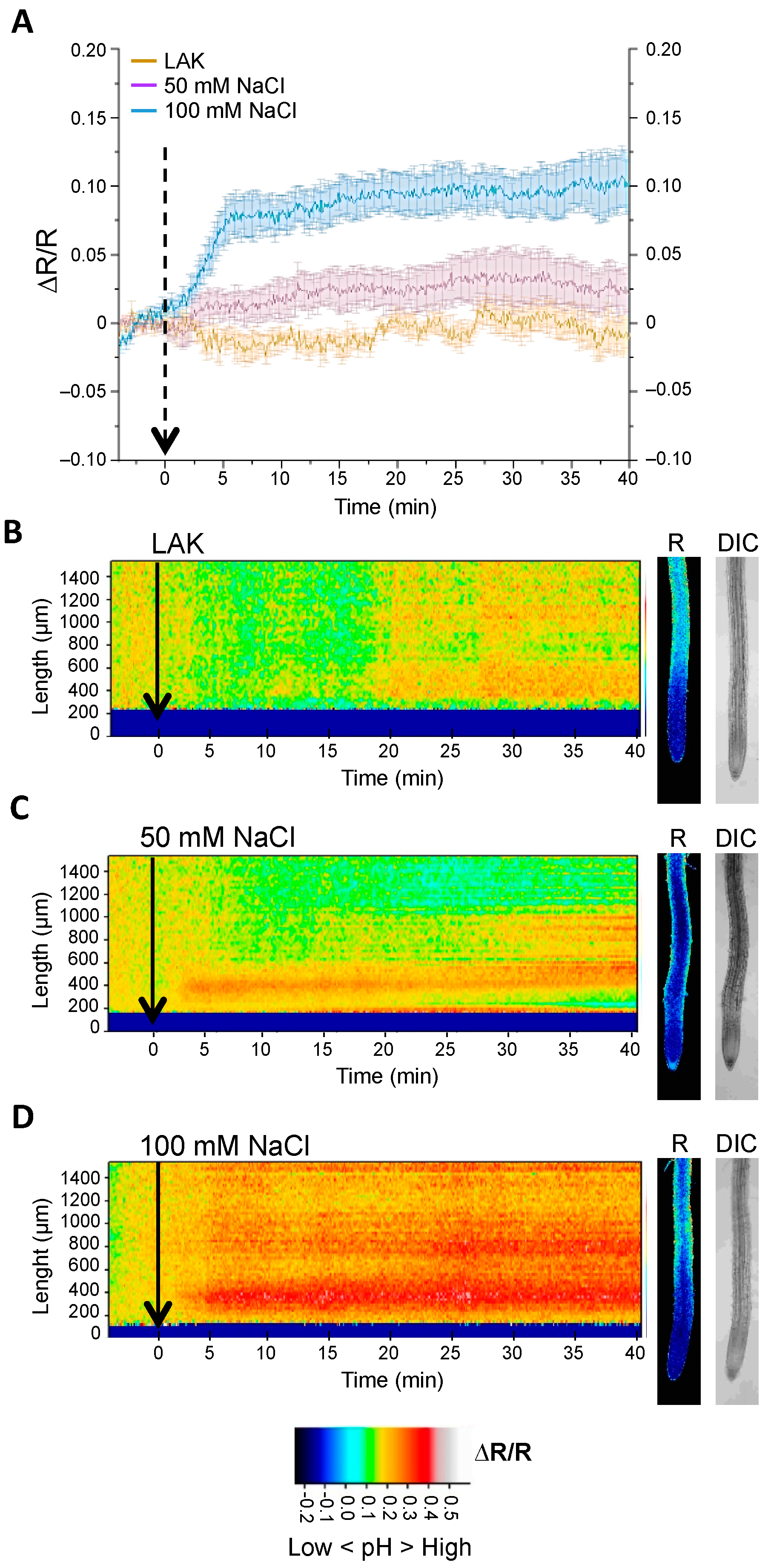
2.2. Cytosolic pH Variations near the Tonoplast and Plasma Membranes under Salt Stress
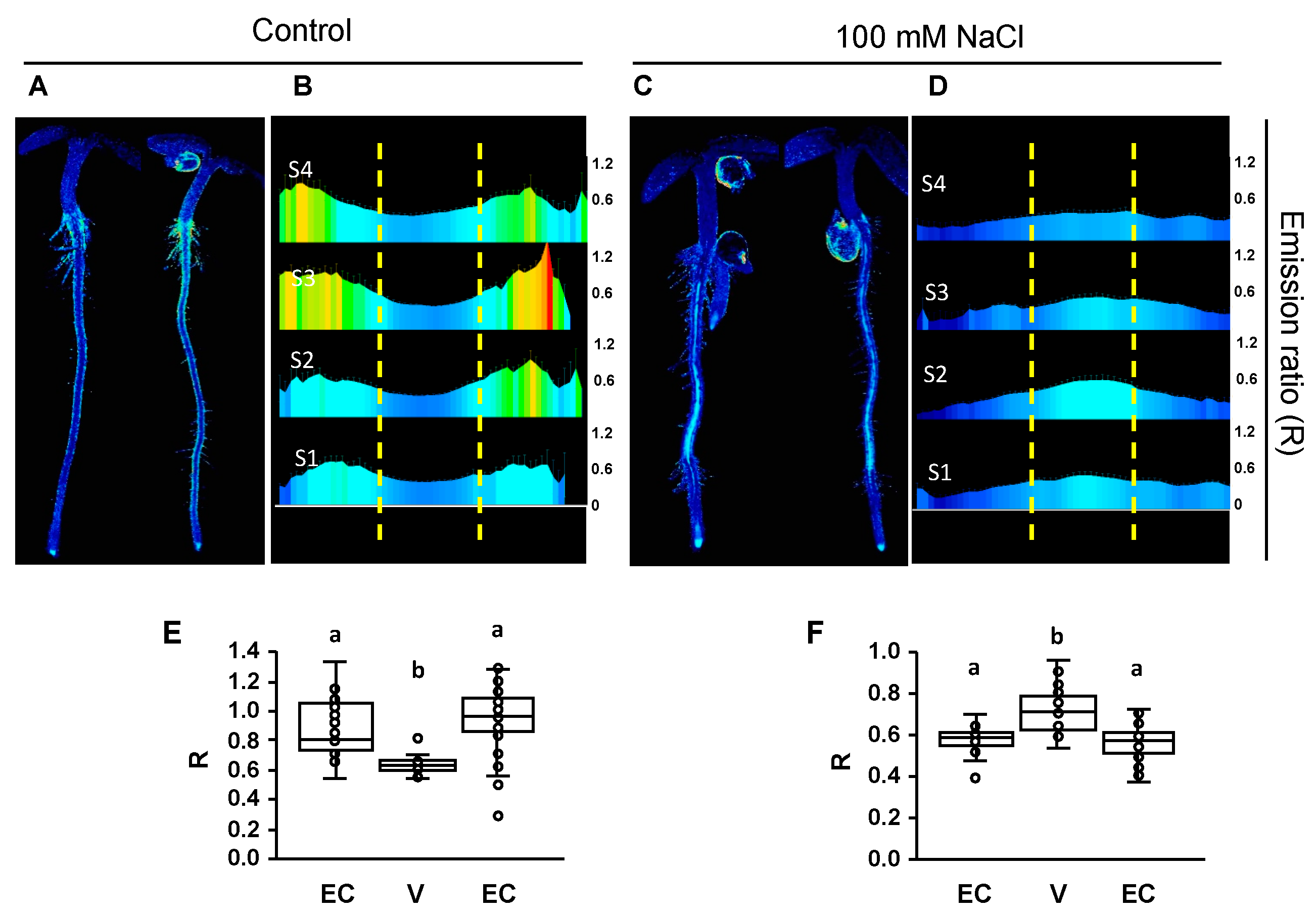
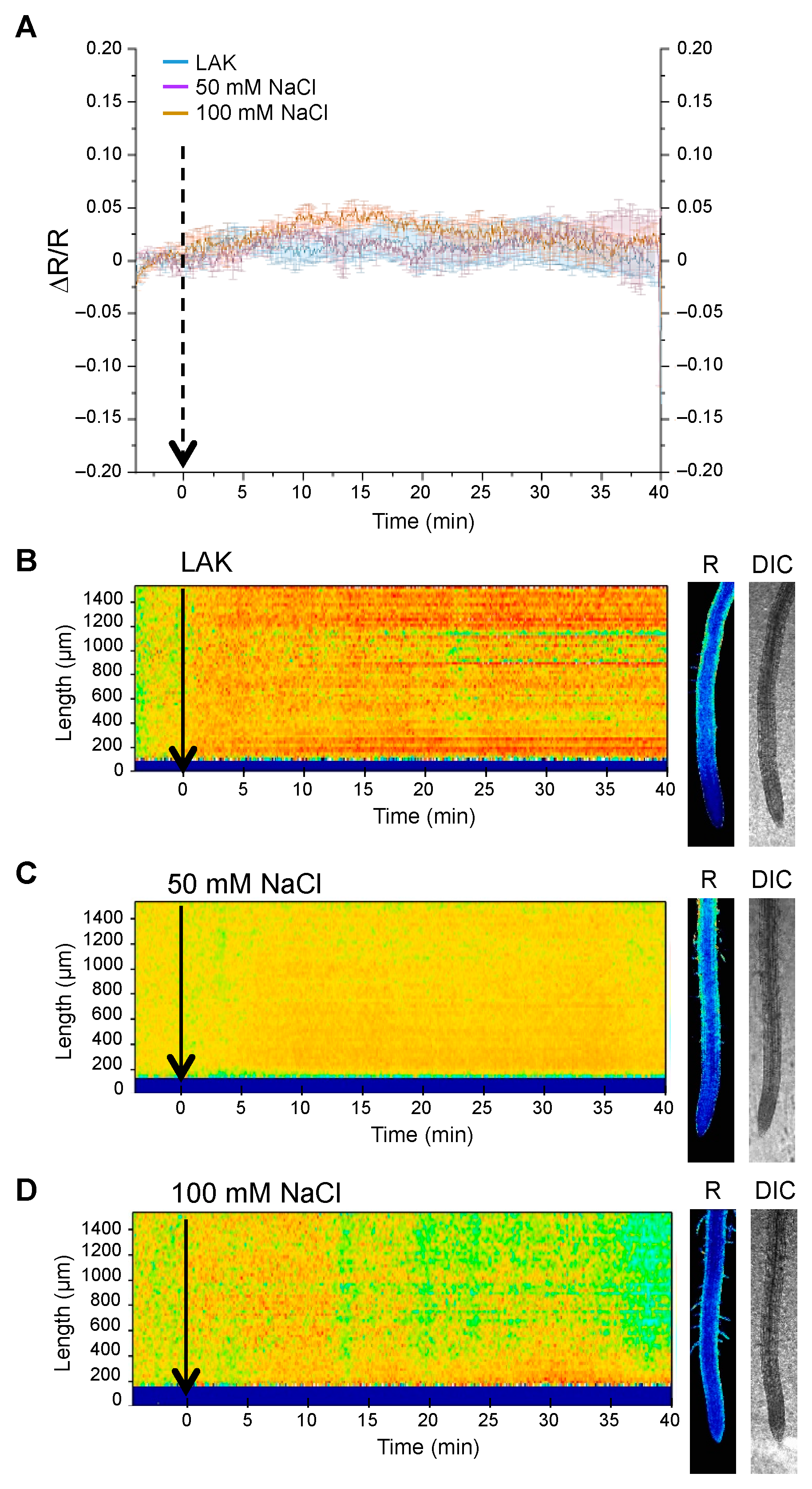
2.3. Changes in Cytosolic pH in Whole Roots under Salt Stress
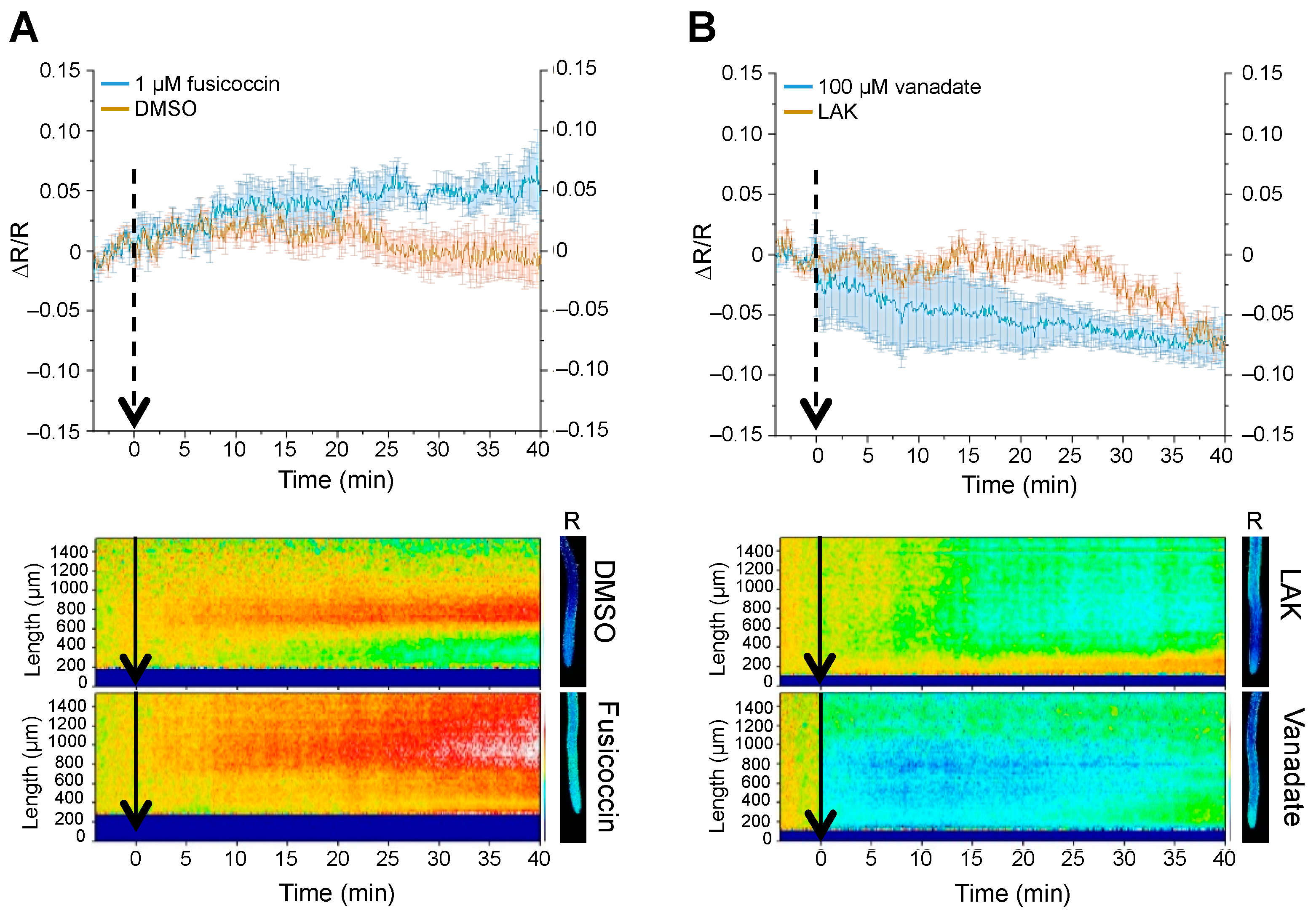
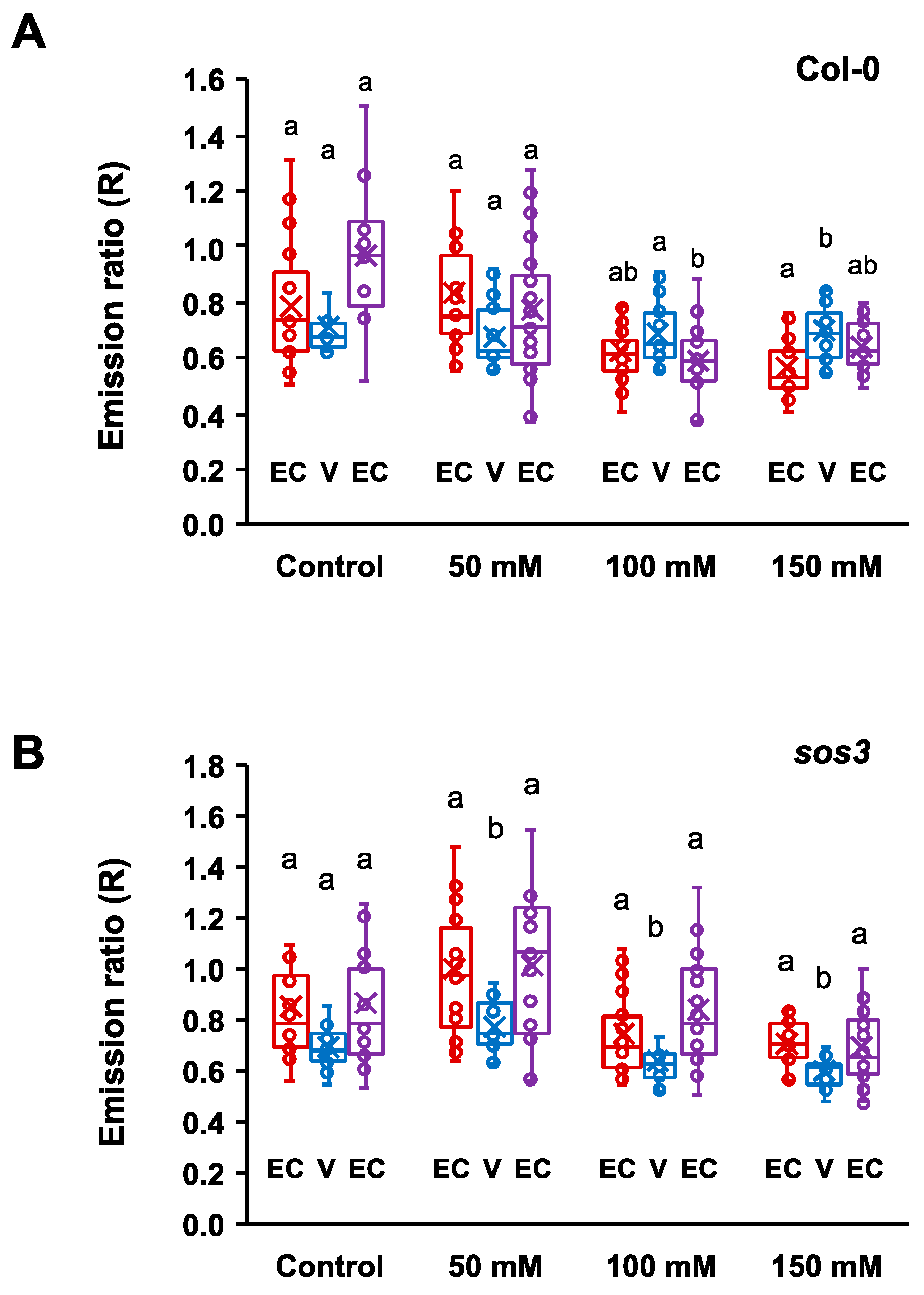
2.4. Cytosolic pH Shifts under Compromised Salt-Stress Sensing
3. Discussion
4. Materials and Methods
4.1. pHGFP Reporters
4.2. Plant Transformation and Reporter Lines Selection
4.3. Fluorescence Imaging of pHGFP Reporter Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Aragon, R.; Haro, R.; Benito, B.; Rodriguez-Navarro, A. Salt intolerance in Arabidopsis: Shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 2016, 243, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Benito, B.; Haro, R.; Amtmann, A.; Cuin, T.A.; Dreyer, I. The twins K+ and Na+ in plants. J. Plant Physiol. 2014, 171, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Aragon, R.; Rodriguez-Navarro, A. Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. Cell Mol. Biol. 2017, 91, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Ragel, P.; Raddatz, N.; Leidi, E.O.; Quintero, F.J.; Pardo, J.M. Regulation of K+ Nutrition in Plants. Front. Plant Sci. 2019, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.G.; Hilleary, R.; Swanson, S.J.; Kim, S.H.; Gilroy, S. Rapid, Long-Distance Electrical and Calcium Signaling in Plants. Annu. Rev. Plant Biol. 2016, 67, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Belfield, E.J.; Mithani, A.; Visscher, A.; Ragoussis, J.; Mott, R.; Smith, J.A.C.; Harberd, N.P. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 2012, 31, 4359–4370. [Google Scholar] [CrossRef]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef]
- Evans, M.J.; Choi, W.G.; Gilroy, S.; Morris, R.J. A ROS-Assisted Calcium Wave Dependent on the AtRBOHD NADPH Oxidase and TPC1 Cation Channel Propagates the Systemic Response to Salt Stress. Plant Physiol. 2016, 171, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Ohta, M.; Shi, H.Z.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Quan, R.D.; Lin, H.X.; Mendoza, I.; Zhang, Y.G.; Cao, W.H.; Yang, Y.Q.; Shang, M.; Chen, S.Y.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 2020, 71, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Steinhorst, L.; He, G.; Moore, L.K.; Schültke, S.; Schmitz-Thom, I.; Cao, Y.; Hashimoto, K.; Andrés, Z.; Piepenburg, K.; Ragel, P.; et al. A Ca(2+)-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev. Cell 2022, 57, 2081–2094.e7. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant 2014, 151, 257–279. [Google Scholar] [CrossRef]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef]
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci 2015, 241, 109–119. [Google Scholar] [CrossRef]
- Koster, P.; Wallrad, L.; Edel, K.H.; Faisal, M.; Alatar, A.A.; Kudla, J. The battle of two ions: Ca(2+) signalling against Na(+) stress. Plant Biol. 2019, 21 (Suppl. S1), 39–48. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Jin, L.; Peng, R. Crosstalk between Ca(2+) and Other Regulators Assists Plants in Responding to Abiotic Stress. Plants 2022, 11, 1351. [Google Scholar] [CrossRef]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. Cell Mol. Biol. 2000, 23, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Laohavisit, A.; Richards, S.L.; Shabala, L.; Chen, C.; Colaco, R.D.; Swarbreck, S.M.; Shaw, E.; Dark, A.; Shabala, S.; Shang, Z.; et al. Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 2013, 163, 253–262. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef]
- Wilkins, K.A.; Matthus, E.; Swarbreck, S.M.; Davies, J.M. Calcium-Mediated Abiotic Stress Signaling in Roots. Front. Plant Sci. 2016, 7, 1296. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal 2009, 2, ra45. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na(+)/K(+)homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012, 63, 305–317. [Google Scholar] [CrossRef]
- Drerup, M.M.; Schlucking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Kader, M.A.; Lindberg, S. Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 2010, 5, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.; Xu, Z.; Luoni, L.; Bonza, M.C.; Doccula, F.G.; De Michelis, M.I.; Morris, R.J.; Schwarzländer, M.; Costa, A. Cellular Ca2+ Signals Generate Defined pH Signatures in Plants. Plant Cell 2018, 30, 2704. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Köster, P.; Andrés, Z.; Waadt, C.; Bradamante, G.; Lampou, K.; Kudla, J.; Schumacher, K. Dual-Reporting Transcriptionally Linked Genetically Encoded Fluorescent Indicators Resolve the Spatiotemporal Coordination of Cytosolic Abscisic Acid and Second Messenger Dynamics in Arabidopsis. Plant Cell 2020, 32, 2582–2601. [Google Scholar] [CrossRef]
- Li, K.; Prada, J.; Damineli, D.S.C.; Liese, A.; Romeis, T.; Dandekar, T.; Feijó, J.A.; Hedrich, R.; Konrad, K.R. An optimized genetically encoded dual reporter for simultaneous ratio imaging of Ca(2+) and H(+) reveals new insights into ion signaling in plants. New Phytol. 2021, 230, 2292–2310. [Google Scholar] [CrossRef]
- Waadt, R.; Kudla, J.; Kollist, H. Multiparameter in vivo imaging in plants using genetically encoded fluorescent indicator multiplexing. Plant Physiol. 2021, 187, 537–549. [Google Scholar] [CrossRef]
- Moseyko, N.; Feldman, L.J. Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ. 2001, 24, 557–563. [Google Scholar] [CrossRef]
- Takemoto, K.; Ebine, K.; Askani, J.C.; Krüger, F.; Gonzalez, Z.A.; Ito, E.; Goh, T.; Schumacher, K.; Nakano, A.; Ueda, T. Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E2457–E2466. [Google Scholar] [CrossRef]
- Kesten, C.; Gámez-Arjona, F.M.; Menna, A.; Scholl, S.; Dora, S.; Huerta, A.I.; Huang, H.-Y.; Tintor, N.; Kinoshita, T.; Rep, M.; et al. Pathogen-induced pH changes regulate the growth-defense balance in plants. EMBO J. 2019, 38, e101822. [Google Scholar] [CrossRef]
- Zheng, J.; Han, S.W.; Rodriguez-Welsh, M.F.; Rojas-Pierce, M. Homotypic Vacuole Fusion Requires VTI11 and Is Regulated by Phosphoinositides. Mol. Plant 2014, 7, 1026–1040. [Google Scholar] [CrossRef]
- Brillada, C.; Zheng, J.; Krüger, F.; Rovira-Diaz, E.; Askani, J.C.; Schumacher, K.; Rojas-Pierce, M. Phosphoinositides control the localization of HOPS subunit VPS41, which together with VPS33 mediates vacuole fusion in plants. Proc. Natl. Acad. Sci. USA 2018, 115, E8305–E8314. [Google Scholar] [CrossRef] [PubMed]
- Würtele, M.; Jelich-Ottmann, C.; Wittinghofer, A.; Oecking, C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003, 22, 987–994. [Google Scholar] [CrossRef]
- Ottmann, C.; Marco, S.; Jaspert, N.; Marcon, C.; Schauer, N.; Weyand, M.; Vandermeeren, C.; Duby, G.; Boutry, M.; Wittinghofer, A.; et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell 2007, 25, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Lew, R.R. Electrogenic Transport Properties of Growing Arabidopsis Root Hairs1 The Plasma Membrane Proton Pump and Potassium Channels. Plant Physiol. 1991, 97, 1527–1534. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Meyer, S.; De Angeli, A.; Nagy, R. Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 2012, 63, 183–213. [Google Scholar] [CrossRef]
- Seidel, T.; Siek, M.; Marg, B.; Dietz, K.J. Energization of vacuolar transport in plant cells and its significance under stress. Int. Rev. Cell Mol. Biol. 2013, 304, 57–131. [Google Scholar]
- Peiter, E. The plant vacuole: Emitter and receiver of calcium signals. Cell Calcium 2011, 50, 120–128. [Google Scholar] [CrossRef]
- Tan, X.; Li, K.; Wang, Z.; Zhu, K.; Tan, X.; Cao, J. A Review of Plant Vacuoles: Formation, Located Proteins, and Functions. Plants 2019, 8, 327. [Google Scholar] [CrossRef]
- Shabala, S.; Chen, G.; Chen, Z.H.; Pottosin, I. The energy cost of the tonoplast futile sodium leak. New Phytol. 2020, 225, 1105–1110. [Google Scholar] [CrossRef]
- Waadt, R.; Krebs, M.; Kudla, J.; Schumacher, K. Multiparameter imaging of calcium and abscisic acid and high-resolution quantitative calcium measurements using R-GECO1-mTurquoise in Arabidopsis. New Phytol. 2017, 216, 303–320. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Z.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- El Mahi, H.; Perez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gamez-Arjona, F.; Fernandez, J.L.; Bundo, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na(+)/H(+) Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, Q.S.; Quintero, F.J.; Pardo, J.M.; Ohta, M.; Zhang, C.Q.; Schumaker, K.S.; Zhu, J.K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 2004, 16, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Villalta, I.; García, E.; Hornero-Mendez, D.; Carranco, R.; Tello, C.; Mendoza, I.; De Luca, A.; Andrés, Z.; Schumacher, K.; Pardo, J.M.; et al. Distinct Roles of N-Terminal Fatty Acid Acylation of the Salinity-Sensor Protein SOS3. Front. Plant Sci. 2021, 12, 691124. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Kronzucker, H.J.; Coskun, D.; Schulze, L.M.; Wong, J.R.; Britto, D.T. Sodium as nutrient and toxicant. Plant Soil 2013, 369, 1–23. [Google Scholar] [CrossRef]
- Xie, Q.; Zhou, Y.; Jiang, X. Structure, Function, and Regulation of the Plasma Membrane Na(+)/H(+) Antiporter Salt Overly Sensitive 1 in Plants. Front. Plant Sci. 2022, 13, 866265. [Google Scholar] [CrossRef]
- Martiniere, A.; Bassil, E.; Jublanc, E.; Alcon, C.; Reguera, M.; Sentenac, H.; Blumwald, E.; Paris, N. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell 2013, 25, 4028–4043. [Google Scholar] [CrossRef]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis Endomembrane System. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Pan, T.; Zhang, X.; Fan, L.; Quintero, F.J.; Zhao, H.; Su, X.; Li, X.; Villalta, I.; Mendoza, I.; et al. K(+) Efflux Antiporters 4, 5, and 6 Mediate pH and K(+) Homeostasis in Endomembrane Compartments. Plant Physiol. 2018, 178, 1657–1678. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, C.; Lindberg, S. Sodium induces simultaneous changes in cytosolic calcium and pH in salt-tolerant quince protoplasts. J. Plant Physiol. 2009, 166, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.H.; Maity, P.J.; Geilfus, C.-M.; Lindberg, S.; Mühling, K.H. Leaf ion homeostasis and plasma Membr. H+-ATPase Act. Vicia Faba Change After Extra Calcium Potassium Supply Under Salinity. Plant Physiol Biochem. 2014, 82, 244–253. [Google Scholar] [CrossRef]
- Pecherina, A.; Grinberg, M.; Ageyeva, M.; Zanegina, D.; Akinchits, E.; Brilkina, A.; Vodeneev, V. Salt-Induced Changes in Cytosolic pH and Photosynthesis in Tobacco and Potato Leaves. Int. J. Mol. Sci. 2023, 24, 491. [Google Scholar] [CrossRef]
- Gao, D.; Knight, M.R.; Trewavas, A.J.; Sattelmacher, B.; Plieth, C. Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol. 2004, 134, 898–908. [Google Scholar] [CrossRef]
- Schulte, A.; Lorenzen, I.; Böttcher, M.; Plieth, C. A novel fluorescent pH probe for expression in plants. Plant Methods 2006, 2, 7. [Google Scholar] [CrossRef]
- Kader, M.A.; Lindberg, S.; Seidel, T.; Golldack, D.; Yemelyanov, V. Sodium sensing induces different changes in free cytosolic calcium concentration and pH in salt-tolerant and -sensitive rice (Oryza sativa) cultivars. Physiol. Plant. 2007, 130, 99–111. [Google Scholar] [CrossRef]
- Sun, Y.; Lindberg, S.; Shabala, L.; Morgan, S.; Shabala, S.; Jacobsen, S.E. A comparative analysis of cytosolic Na+ changes under salinity between halophyte quinoa (Chenopodium quinoa) and glycophyte pea (Pisum sativum). Environ. Exp. Bot. 2017, 141, 154–160. [Google Scholar] [CrossRef]
- Zhou, X.; Li, J.; Wang, Y.; Liang, X.; Zhang, M.; Lu, M.; Guo, Y.; Qin, F.; Jiang, C. The classical SOS pathway confers natural variation of salt tolerance in maize. New Phytol. 2022, 236, 479–494. [Google Scholar] [CrossRef]
- Demidchik, V. Mechanisms and physiological roles of K+ efflux from root cells. J. Plant Physiol. 2014, 171, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Jayakannan, M.; Bose, J.; Babourina, O.; Rengel, Z.; Shabala, S. Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 2013, 64, 2255–2268. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2015, 115, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-H.; Leidi, E.; Zhang, Q.; Hwang, S.-M.; Li, Y.; Quintero, F.J.; Jiang, X.; D’Urzo, M.P.; Lee, S.Y.; Zhao, Y.; et al. Loss of Halophytism by Interference with SOS1 Expression. Plant Physiol. 2009, 151, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.H.; Lee, S.Y.; Bressan, R.A.; Yun, D.J.; Bohnert, H.J. Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 2010, 61, 1205–1213. [Google Scholar] [CrossRef]
- Barragan, V.; Leidi, E.; Andres, Z.; Rubio, L.; De Luca, A.; Fernandez, J.; Cubero, B.; Pardo, J. Ion Exchangers NHX1 and NHX2 Mediate Active Potassium Uptake into Vacuoles to Regulate Cell Turgor and Stomatal Function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef]
- Bassil, E.; Tajima, H.; Liang, Y.C.; Ohto, M.A.; Ushijima, K.; Nakano, R.; Esumi, T.; Coku, A.; Belmonte, M.; Blumwald, E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 2011, 23, 3482–3497. [Google Scholar] [CrossRef]
- Park, H.J.; Gámez-Arjona, F.M.; Lindahl, M.; Aman, R.; Villalta, I.; Cha, J.Y.; Carranco, R.; Lim, C.J.; García, E.; Bressan, R.A.; et al. S-acylated and nucleus-localized SALT OVERLY SENSITIVE3/CALCINEURIN B-LIKE4 stabilizes GIGANTEA to regulate Arabidopsis flowering time under salt stress. Plant Cell 2023, 35, 298–317. [Google Scholar] [CrossRef]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef]
- Shabala, L.; Cuin, T.; Newman, I.; Shabala, S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 2005, 222, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Song, T.; Wallrad, L.; Kudla, J.; Wang, X.; Zhang, W. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 2019, 5, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Olias, R.; Eljakaoui, Z.; Li, J.; Alvarez De Morales, P.; Marin-Manzano, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, P.; Ma, Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. The Effect of AtHKT1;1 or AtSOS1 Mutation on the Expressions of Na+ or K+ Transporter Genes and Ion Homeostasis in Arabidopsis thaliana under Salt Stress. Int. J. Mol. Sci. 2019, 20, 1085. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.S.; Shukla, P.S.; Jha, A.; Agarwal, P.K.; Jha, B. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na(+) loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012, 12, 188. [Google Scholar] [CrossRef]
- Zhu, J.K.; Liu, J.P.; Xiong, L.M. Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 1998, 10, 1181–1191. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef]
- Kardash, E.; Bandemer, J.; Raz, E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nat. Protoc. 2011, 6, 1835–1846. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rombolá-Caldentey, B.; Andrés, Z.; Waadt, R.; Quintero, F.J.; Schumacher, K.; Pardo, J.M. Salinity-Induced Cytosolic Alkaline Shifts in Arabidopsis Roots Require the SOS Pathway. Int. J. Mol. Sci. 2023, 24, 3549. https://doi.org/10.3390/ijms24043549
Rombolá-Caldentey B, Andrés Z, Waadt R, Quintero FJ, Schumacher K, Pardo JM. Salinity-Induced Cytosolic Alkaline Shifts in Arabidopsis Roots Require the SOS Pathway. International Journal of Molecular Sciences. 2023; 24(4):3549. https://doi.org/10.3390/ijms24043549
Chicago/Turabian StyleRombolá-Caldentey, Belén, Zaida Andrés, Rainer Waadt, Francisco J. Quintero, Karin Schumacher, and José M. Pardo. 2023. "Salinity-Induced Cytosolic Alkaline Shifts in Arabidopsis Roots Require the SOS Pathway" International Journal of Molecular Sciences 24, no. 4: 3549. https://doi.org/10.3390/ijms24043549
APA StyleRombolá-Caldentey, B., Andrés, Z., Waadt, R., Quintero, F. J., Schumacher, K., & Pardo, J. M. (2023). Salinity-Induced Cytosolic Alkaline Shifts in Arabidopsis Roots Require the SOS Pathway. International Journal of Molecular Sciences, 24(4), 3549. https://doi.org/10.3390/ijms24043549





