Positive Linear Relationship between Nucleophosmin Protein Expression and the Viral Load in HPV-Associated Oropharyngeal Squamous Cell Carcinoma: A Possible Tool for Stratification of Patients
Abstract
1. Introduction
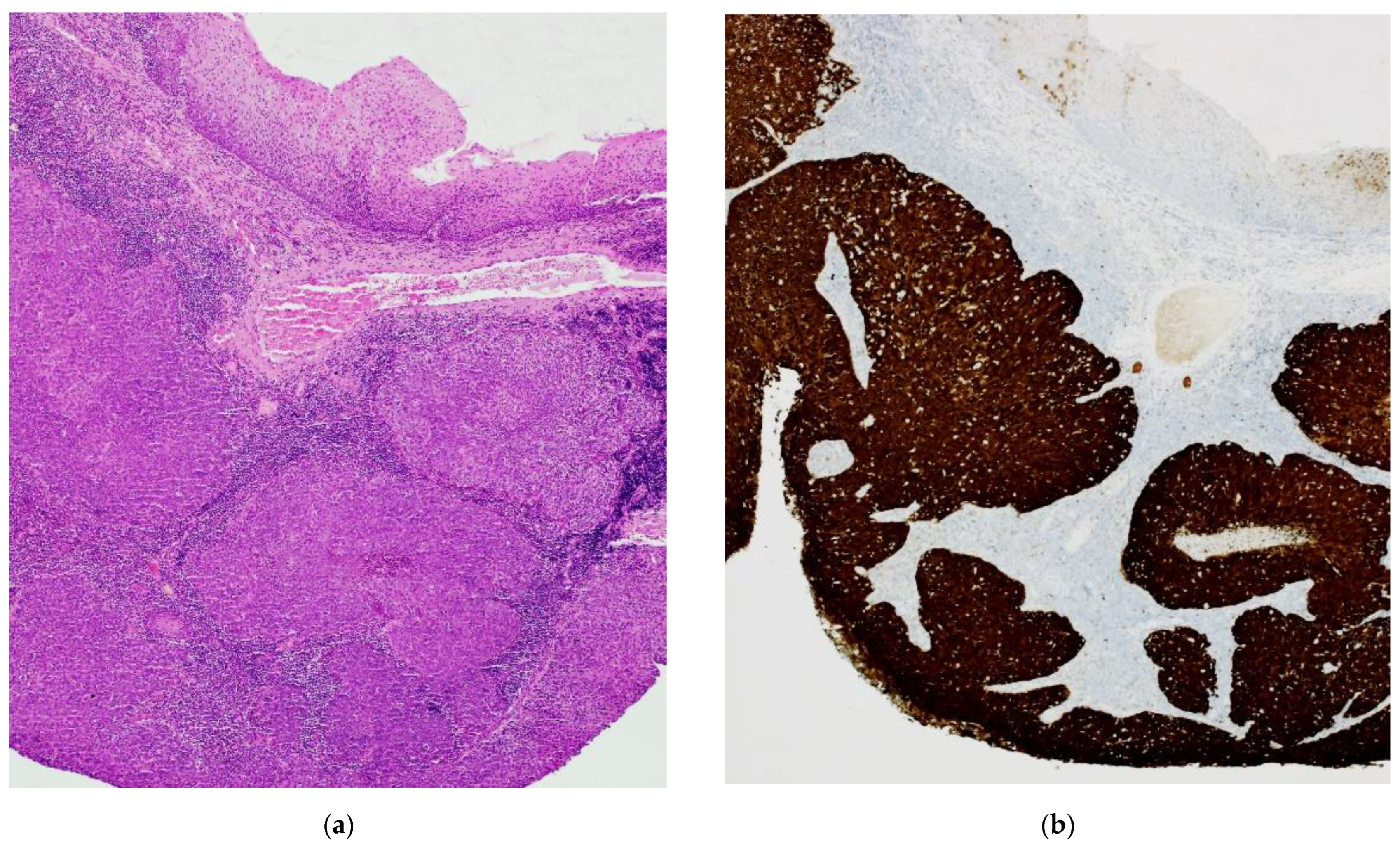
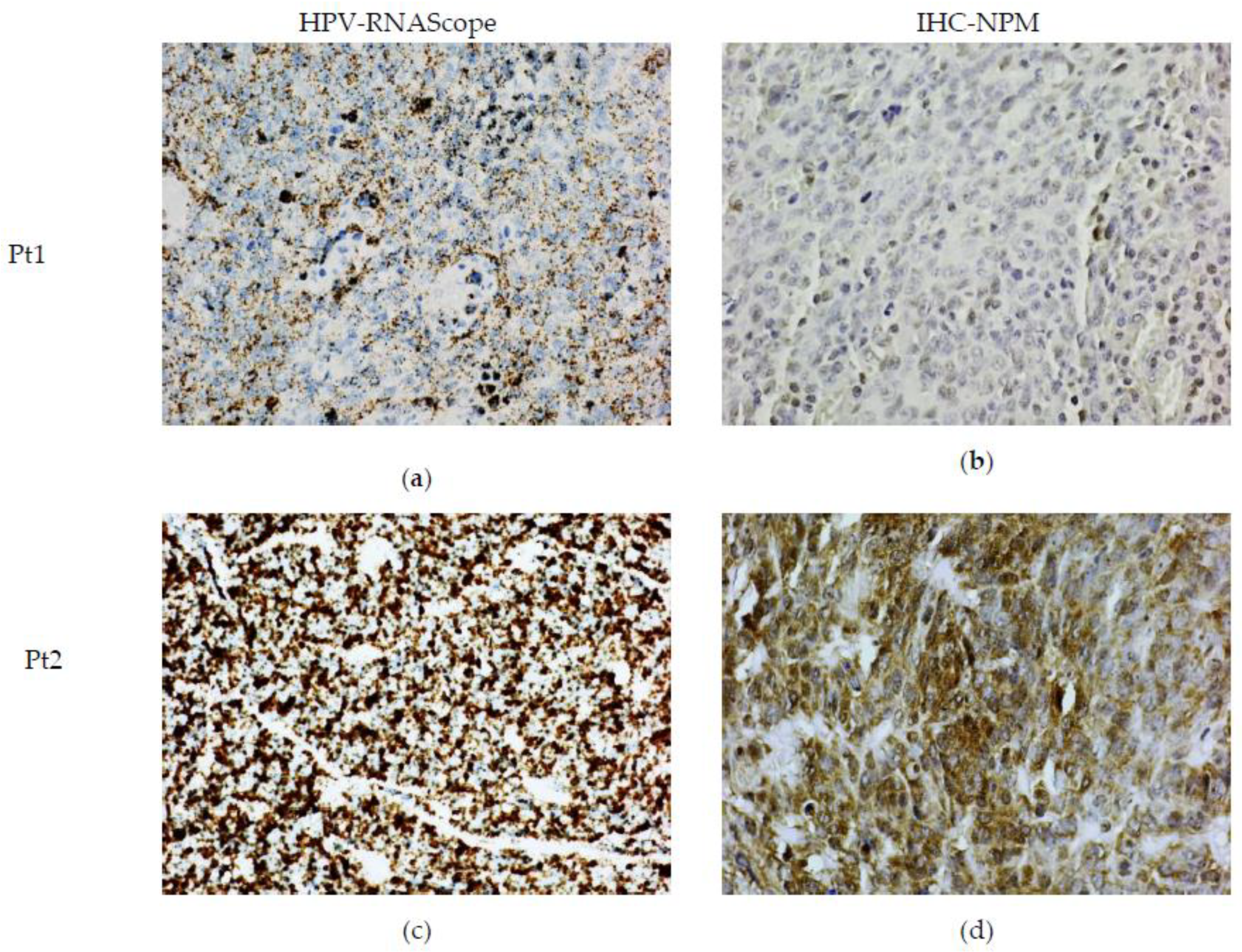
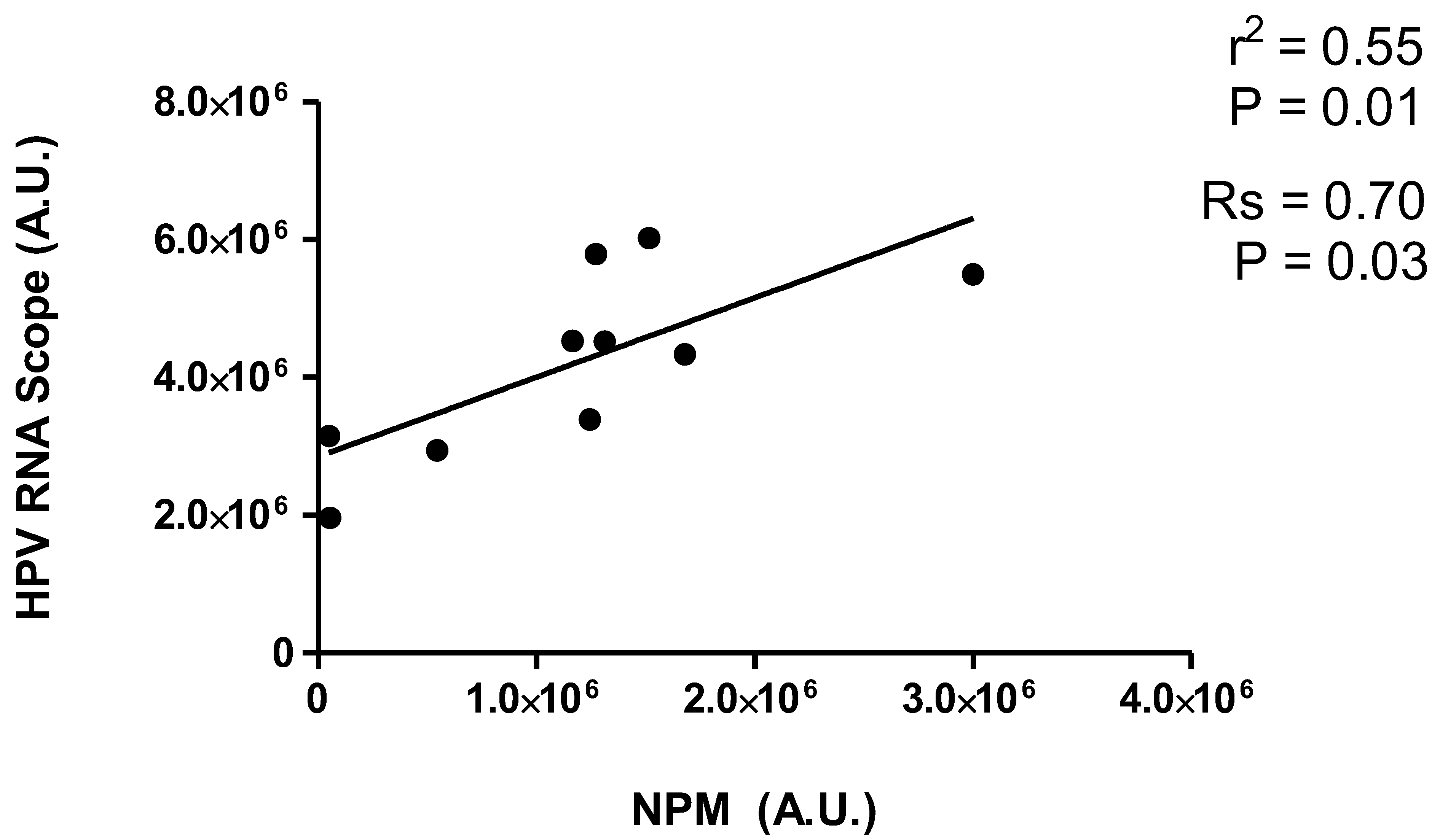
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Immunohistochemistry (IHC)
4.3. RNAscope-In Situ Hybridization (ISH)
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global Cancer Statistics. CA. Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Habib Awan, K.; Arakeri, G.; Jayampath Seneviratne, C.; Muddur, N.; Malik, S.; Ferrari, M.; Rahimi, S.; Brennan, P.A. Machine Learning and Its Potential Applications to the Genomic Study of Head and Neck Cancer—A Systematic Review. J. Oral Pathol. Med. 2019, 48, 773–779. [Google Scholar] [CrossRef]
- Green, B.; Elhamshary, A.; Gomez, R.; Rahimi, S.; Brennan, P.A. An Update on the Current Management of Head and Neck Mucosal Melanoma. J. Oral Pathol. Med. 2017, 46, 475–479. [Google Scholar] [CrossRef]
- Sturgis, E.M.; Cinciripini, P.M. Trends in Head and Neck Cancer Incidence in Relation to Smoking Prevalence: An Emerging Epidemic of Human Papillomavirus-Associated Cancers? Cancer 2007, 110, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Hashibe, M.; Brennan, P.; Benhamou, S.; Castellsague, X.; Chen, C.; Curado, M.P.; Dal Maso, L.; Daudt, A.W.; Fabianova, E.; Fernandez, L.; et al. Alcohol Drinking in Never Users of Tobacco, Cigarette Smoking in Never Drinkers, and the Risk of Head and Neck Cancer: Pooled Analysis in the International Head and Neck Cancer Epidemiology Consortium. J. Natl. Cancer Inst. 2007, 99, 777–789. [Google Scholar] [CrossRef]
- Chai, R.C.; Lambie, D.; Verma, M.; Punyadeera, C. Current Trends in the Etiology and Diagnosis of HPV-Related Head and Neck Cancers. Cancer Med. 2015, 4, 596–607. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Marur, S.; D’Souza, G.; Westra, W.H.; Forastiere, A.A. HPV-Associated Head and Neck Cancer: A Virus-Related Cancer Epidemic. Lancet. Oncol. 2010, 11, 781–789. [Google Scholar] [CrossRef]
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide Burden of Cancer Attributable to HPV by Site, Country and HPV Type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human Papillomavirus Types in Head and Neck Squamous Cell Carcinomas Worldwide: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2005, 14, 467–475. [Google Scholar] [CrossRef]
- Castle, P.E.; Shields, T.; Kirnbauer, R.; Manos, M.M.; Burk, R.D.; Glass, A.G.; Scott, D.R.; Sherman, M.E.; Schiffman, M. Sexual Behavior, Human Papillomavirus Type 16 (HPV 16) Infection, and HPV 16 Seropositivity. Sex. Transm. Dis. 2002, 29, 182–187. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved Survival of Patients with Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef]
- Albers, A.E.; Qian, X.; Kaufmann, A.M.; Coordes, A. Meta Analysis: HPV and P16 Pattern Determines Survival in Patients with HNSCC and Identifies Potential New Biologic Subtype. Sci. Rep. 2017, 7, 16715. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 Oncoprotein Encoded by Human Papillomavirus Types 16 and 18 Promotes the Degradation of P53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Ramón, A.C.; Basukala, O.; Massimi, P.; Thomas, M.; Perera, Y.; Banks, L.; Perea, S.E. CIGB-300 Peptide Targets the CK2 Phospho-Acceptor Domain on Human Papillomavirus E7 and Disrupts the Retinoblastoma (RB) Complex in Cervical Cancer Cells. Viruses 2022, 14, 1681. [Google Scholar] [CrossRef]
- Rampias, T.; Sasaki, C.; Weinberger, P.; Psyrri, A. E6 and E7 Gene Silencing and Transformed Phenotype of Human Papillomavirus 16-Positive Oropharyngeal Cancer Cells. J. Natl. Cancer Inst. 2009, 101, 412–423. [Google Scholar] [CrossRef]
- Iarovaia, O.V.; Ioudinkova, E.S.; Velichko, A.K.; Razin, S. V Manipulation of Cellular Processes via Nucleolus Hijaking in the Course of Viral Infection in Mammals. Cells 2021, 10, 1597. [Google Scholar] [CrossRef]
- Zatsepina, O.; Braspenning, J.; Robberson, D.; Hajibagheri, M.A.; Blight, K.J.; Ely, S.; Hibma, M.; Spitkovsky, D.; Trendelenburg, M.; Crawford, L.; et al. The Human Papillomavirus Type 16 E7 Protein Is Associated with the Nucleolus in Mammalian and Yeast Cells. Oncogene 1997, 14, 1137–1145. [Google Scholar] [CrossRef]
- Dichamp, I.; Séité, P.; Agius, G.; Barbarin, A.; Beby-Defaux, A. Human Papillomavirus 16 Oncoprotein E7 Stimulates UBF1-Mediated RDNA Gene Transcription, Inhibiting a P53-Independent Activity of P14ARF. PLoS ONE 2014, 9, e96136. [Google Scholar] [CrossRef]
- Yoshimoto, M.; Tokuda, A.; Nishiwaki, K.; Sengoku, K.; Yaginuma, Y. The Protein Interacting with Carboxyl Terminus-1 Codon 389 Polymorphism Impairs Protein Interacting with Carboxyl Terminus-1 Function and Is a Risk Factor for Uterine Cervical Cancer. Mol. Carcinog. 2017, 56, 1484–1492. [Google Scholar] [CrossRef]
- Grinstein, E.; Wernet, P.; Snijders, P.J.F.; Rösl, F.; Weinert, I.; Jia, W.; Kraft, R.; Schewe, C.; Schwabe, M.; Hauptmann, S.; et al. Nucleolin as Activator of Human Papillomavirus Type 18 Oncogene Transcription in Cervical Cancer. J. Exp. Med. 2002, 196, 1067–1078. [Google Scholar] [CrossRef]
- Tosi, P.; Cintorino, M.; Santopietro, R.; Lio, R.; Barbini, P.; Ji, H.; Chang, F.; Kataja, V.; Syrjänen, S.; Syrjänen, K. Prognostic Factors in Invasive Cervical Carcinomas Associated with Human Papillomavirus (HPV). Quantitative Data and Cytokeratin Expression. Pathol. Res. Pract. 1992, 188, 866–873. [Google Scholar] [CrossRef]
- Del Carmen Alarcón-Romero, L.; Illades-Aguiar, B.; Flores-Alfaro, E.; Terán-Porcayo, M.A.; Antonio-Véjar, V.; Reyes-Maldonado, E. AgNOR Polymorphism Association with Squamous Intraepithelial Lesions and Invasive Carcinoma with HPV Infection. Salud Publica Mex. 2009, 51, 134–140. [Google Scholar] [CrossRef]
- Donofrio, V.; Lo Muzio, L.; Mignogna, M.D.; Troncone, G.; Staibano, S.; Boscaino, A.; De Rosa, G. Prognostic Evaluation of HPV-Associated Precancerous and Microinvasive Carcinoma of the Oral Cavity: Combined Use of Nucleolar Organiser Regions (AgNOR) and Proliferating Cell Nuclear Antigen (PCNA). Eur. J. Cancer. B. Oral Oncol. 1995, 31, 174–180. [Google Scholar] [CrossRef]
- Lo Muzio, L.; Mignogna, M.D.; Staibano, S.; de Vico, G.; Salvatore, G.; Damiano, S.; Bucci, E.; Procaccini, M.; Mezza, E.; De Rosa, G. Morphometric Study of Nucleolar Organiser Regions (AgNOR) in HPV-Associated Precancerous Lesions and Microinvasive Carcinoma of the Oral Cavity. Oral Oncol. 1997, 33, 247–259. [Google Scholar] [CrossRef]
- Hata, H.; Okayama, K.; Iijima, J.; Teruya, K.; Shiina, N.; Caniz, T.; Ishii, Y.; Fujii, M.; Oda, M.; Okodo, M. A Comparison of Cytomorphological Features of ASC-H Cells Based on Histopathological Results Obtained from a Colposcopic Target Biopsy Immediately after Pap Smear Sampling. Asian Pac. J. Cancer Prev. 2019, 20, 2139–2143. [Google Scholar] [CrossRef]
- Hidalgo, J.V.; Rocher, A.E.; López, J.L.; Gamboni, M.; Vighi, S.; Canessa, O.E.; Peressini, S.; Guerra, F.; di Carlo, M.B.; Palaoro, L.A.; et al. AgNOR, P16 and Human Papillomavirus in Low-Grade Squamous Intra-Epithelial Lesions of the Uterine Cervix. Preliminary Report. Biotech. Histochem. 2012, 87, 257–264. [Google Scholar] [CrossRef]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From Structure and Function to Disease Development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef]
- Lobaina, Y.; Perera, Y. Implication of B23/NPM1 in Viral Infections, Potential Uses of B23/NPM1 Inhibitors as Antiviral Therapy. Infect. Disord. Drug Targets 2019, 19, 2–16. [Google Scholar] [CrossRef]
- McCloskey, R.; Menges, C.; Friedman, A.; Patel, D.; McCance, D.J. Human Papillomavirus Type 16 E6/E7 Upregulation of Nucleophosmin Is Important for Proliferation and Inhibition of Differentiation. J. Virol. 2010, 84, 5131–5139. [Google Scholar] [CrossRef]
- Day, P.M.; Thompson, C.D.; Pang, Y.Y.; Lowy, D.R.; Schiller, J.T. Involvement of Nucleophosmin (NPM1/B23) in Assembly of Infectious HPV16 Capsids. Papillomavirus Res. 2015, 1, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, S.; Akaev, I.; Brennan, P.A.; Virgo, A.; Marani, C.; Gomez, R.S.; Yeoh, C.C. A Proposal for Classification of Oropharyngeal Squamous Cell Carcinoma: Morphology and Status of HPV by Immunohistochemistry and Molecular Biology. J. Oral Pathol. Med. 2020, 49, 110–116. [Google Scholar] [CrossRef]
- Lewis, J.S.; Chernock, R.D.; Ma, X.-J.; Flanagan, J.J.; Luo, Y.; Gao, G.; Wang, X.; El-Mofty, S.K. Partial P16 Staining in Oropharyngeal Squamous Cell Carcinoma: Extent and Pattern Correlate with Human Papillomavirus RNA Status. Mod. Pathol. 2012, 25, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, S. HPV-Related Squamous Cell Carcinoma of Oropharynx: A Review. J. Clin. Pathol. 2020, 73, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Beadle, B.; Bishop, J.A.; Chernock, R.D.; Colasacco, C.; Lacchetti, C.; Moncur, J.T.; Rocco, J.W.; Schwartz, M.R.; Seethala, R.R.; et al. Human Papillomavirus Testing in Head and Neck Carcinomas: Guideline from the College of American Pathologists. Arch. Pathol. Lab. Med. 2018, 142, 559–597. [Google Scholar] [CrossRef] [PubMed]
- Lydiatt, W.M.; Patel, S.G.; O’Sullivan, B.; Brandwein, M.S.; Ridge, J.A.; Migliacci, J.C.; Loomis, A.M.; Shah, J.P. Head and Neck Cancers-Major Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA. Cancer J. Clin. 2017, 67, 122–137. [Google Scholar] [CrossRef]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; de Sanjosé, S.; Trottier, H. HPV DNA, E6/E7 MRNA, and P16INK4a Detection in Head and Neck Cancers: A Systematic Review and Meta-Analysis. Lancet. Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- Mirghani, H.; Casiraghi, O.; Guerlain, J.; Amen, F.; He, M.-X.; Ma, X.-J.; Luo, Y.; Mourareau, C.; Drusch, F.; Lakdhar, A.B.; et al. Diagnosis of HPV Driven Oropharyngeal Cancers: Comparing P16 Based Algorithms with the RNAscope HPV-Test. Oral Oncol. 2016, 62, 101–108. [Google Scholar] [CrossRef]
- Smith, E.M.; Wang, D.; Kim, Y.; Rubenstein, L.M.; Lee, J.H.; Haugen, T.H.; Turek, L.P. P16INK4a Expression, Human Papillomavirus, and Survival in Head and Neck Cancer. Oral Oncol. 2008, 44, 133–142. [Google Scholar] [CrossRef]
- Rietbergen, M.M.; Brakenhoff, R.H.; Bloemena, E.; Witte, B.I.; Snijders, P.J.F.; Heideman, D.A.M.; Boon, D.; Koljenovic, S.; Baatenburg-de Jong, R.J.; Leemans, C.R. Human Papillomavirus Detection and Comorbidity: Critical Issues in Selection of Patients with Oropharyngeal Cancer for Treatment De-Escalation Trials. Ann. Oncol. 2013, 24, 2740–2745. [Google Scholar] [CrossRef]
- Schache, A.G.; Liloglou, T.; Risk, J.M.; Jones, T.M.; Ma, X.-J.; Wang, H.; Bui, S.; Luo, Y.; Sloan, P.; Shaw, R.J.; et al. Validation of a Novel Diagnostic Standard in HPV-Positive Oropharyngeal Squamous Cell Carcinoma. Br. J. Cancer 2013, 108, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Ukpo, O.C.; Flanagan, J.J.; Ma, X.-J.; Luo, Y.; Thorstad, W.L.; Lewis, J.S. High-Risk Human Papillomavirus E6/E7 MRNA Detection by a Novel in Situ Hybridization Assay Strongly Correlates with P16 Expression and Patient Outcomes in Oropharyngeal Squamous Cell Carcinoma. Am. J. Surg. Pathol. 2011, 35, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Ukpo, O.C.; Ma, X.-J.; Flanagan, J.J.; Luo, Y.; Thorstad, W.L.; Chernock, R.D. Transcriptionally-Active High-Risk Human Papillomavirus Is Rare in Oral Cavity and Laryngeal/Hypopharyngeal Squamous Cell Carcinomas—A Tissue Microarray Study Utilizing E6/E7 MRNA in Situ Hybridization. Histopathology 2012, 60, 982–991. [Google Scholar] [CrossRef]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.-C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.-T.; Ma, X.-J.; Luo, Y. RNAscope: A Novel in Situ RNA Analysis Platform for Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.J.; Wang, X.W. Nucleophosmin and Human Cancer. Cancer Detect. Prev. 2006, 30, 481–490. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Beji, S.; Sileno, S.; Lulli, D.; Mercurio, L.; Madonna, S.; Cirielli, C.; Pallotta, S.; Albanesi, C.; Capogrossi, M.C.; et al. Extracellular Nucleophosmin Is Increased in Psoriasis and Correlates with the Determinants of Cardiovascular Diseases. Front. Cardiovasc. Med. 2022, 9, 867813. [Google Scholar] [CrossRef]
- Beji, S.; D’Agostino, M.; Gambini, E.; Sileno, S.; Scopece, A.; Vinci, M.C.; Milano, G.; Melillo, G.; Napolitano, M.; Pompilio, G.; et al. Doxorubicin Induces an Alarmin-like TLR4-Dependent Autocrine/Paracrine Action of Nucleophosmin in Human Cardiac Mesenchymal Progenitor Cells. BMC Biol. 2021, 19, 124. [Google Scholar] [CrossRef]
- Williams, V.M.; Filippova, M.; Soto, U.; Duerksen-Hughes, P.J. HPV-DNA Integration and Carcinogenesis: Putative Roles for Inflammation and Oxidative Stress. Future Virol. 2011, 6, 45–57. [Google Scholar] [CrossRef]
- Solares, A.M.; Santana, A.; Baladrón, I.; Valenzuela, C.; González, C.A.; Díaz, A.; Castillo, D.; Ramos, T.; Gómez, R.; Alonso, D.F.; et al. Safety and Preliminary Efficacy Data of a Novel Casein Kinase 2 (CK2) Peptide Inhibitor Administered Intralesionally at Four Dose Levels in Patients with Cervical Malignancies. BMC Cancer 2009, 9, 146. [Google Scholar] [CrossRef]
- Nouri, K.; Moll, J.M.; Milroy, L.-G.; Hain, A.; Dvorsky, R.; Amin, E.; Lenders, M.; Nagel-Steger, L.; Howe, S.; Smits, S.H.J.; et al. Biophysical Characterization of Nucleophosmin Interactions with Human Immunodeficiency Virus Rev and Herpes Simplex Virus US11. PLoS ONE 2015, 10, e0143634. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Agostino, M.; Di Cecco, M.; Marani, C.; Vigili, M.G.; Sileno, S.; Volpi, C.C.; Gloghini, A.; Avitabile, D.; Magenta, A.; Rahimi, S. Positive Linear Relationship between Nucleophosmin Protein Expression and the Viral Load in HPV-Associated Oropharyngeal Squamous Cell Carcinoma: A Possible Tool for Stratification of Patients. Int. J. Mol. Sci. 2023, 24, 3482. https://doi.org/10.3390/ijms24043482
D’Agostino M, Di Cecco M, Marani C, Vigili MG, Sileno S, Volpi CC, Gloghini A, Avitabile D, Magenta A, Rahimi S. Positive Linear Relationship between Nucleophosmin Protein Expression and the Viral Load in HPV-Associated Oropharyngeal Squamous Cell Carcinoma: A Possible Tool for Stratification of Patients. International Journal of Molecular Sciences. 2023; 24(4):3482. https://doi.org/10.3390/ijms24043482
Chicago/Turabian StyleD’Agostino, Marco, Marco Di Cecco, Carla Marani, Maurizio Giovanni Vigili, Sara Sileno, Chiara Costanza Volpi, Annunziata Gloghini, Daniele Avitabile, Alessandra Magenta, and Siavash Rahimi. 2023. "Positive Linear Relationship between Nucleophosmin Protein Expression and the Viral Load in HPV-Associated Oropharyngeal Squamous Cell Carcinoma: A Possible Tool for Stratification of Patients" International Journal of Molecular Sciences 24, no. 4: 3482. https://doi.org/10.3390/ijms24043482
APA StyleD’Agostino, M., Di Cecco, M., Marani, C., Vigili, M. G., Sileno, S., Volpi, C. C., Gloghini, A., Avitabile, D., Magenta, A., & Rahimi, S. (2023). Positive Linear Relationship between Nucleophosmin Protein Expression and the Viral Load in HPV-Associated Oropharyngeal Squamous Cell Carcinoma: A Possible Tool for Stratification of Patients. International Journal of Molecular Sciences, 24(4), 3482. https://doi.org/10.3390/ijms24043482







