Analysis of Electric Field Stimulation in Blue Light Stressed 661W Cells
Abstract
1. Introduction
2. Results
2.1. Effects of EF Stimulation in Li Exposed Cells at Cellular Level
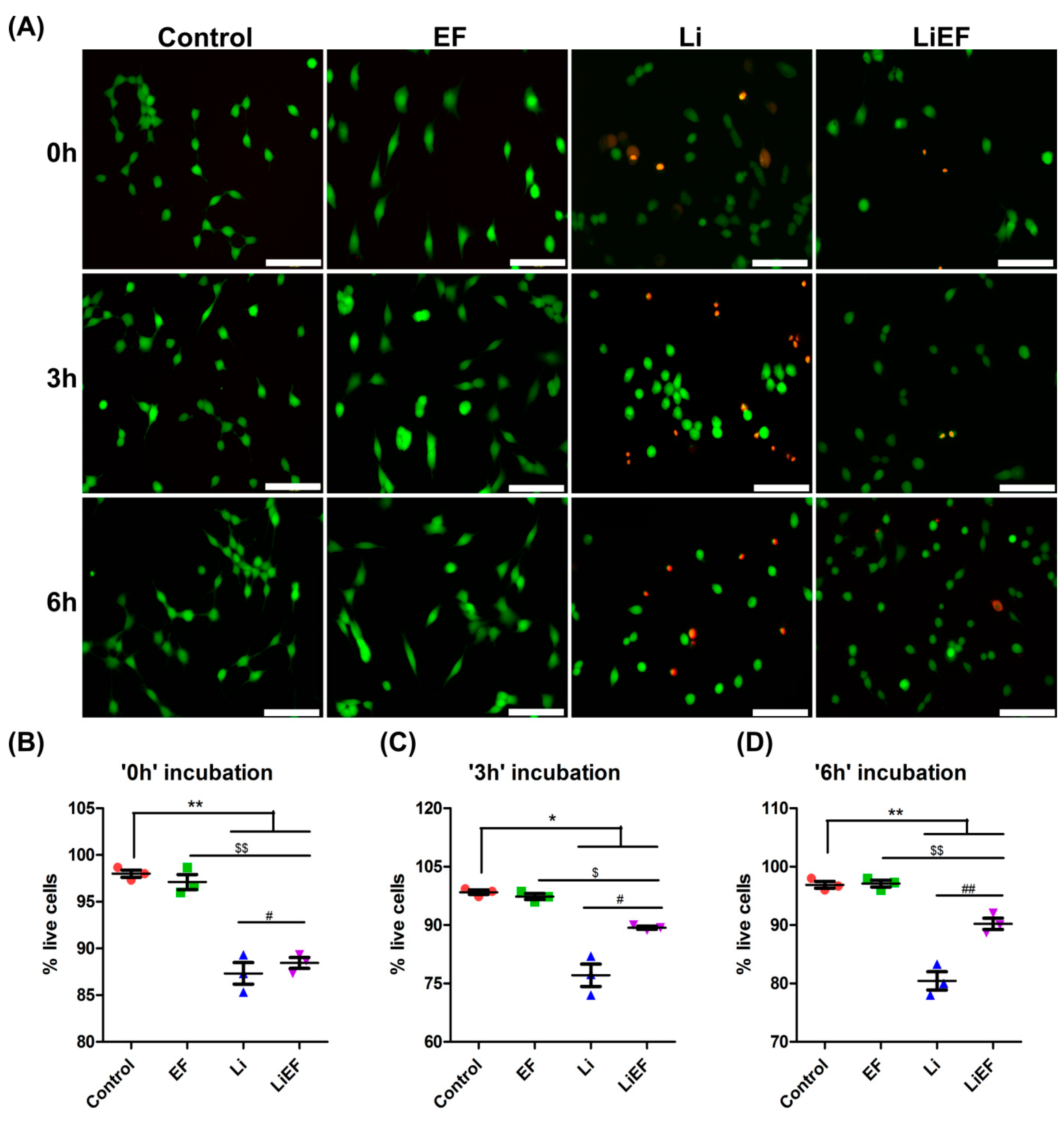
2.1.1. EF Induces Sustained Overall Cell Viability
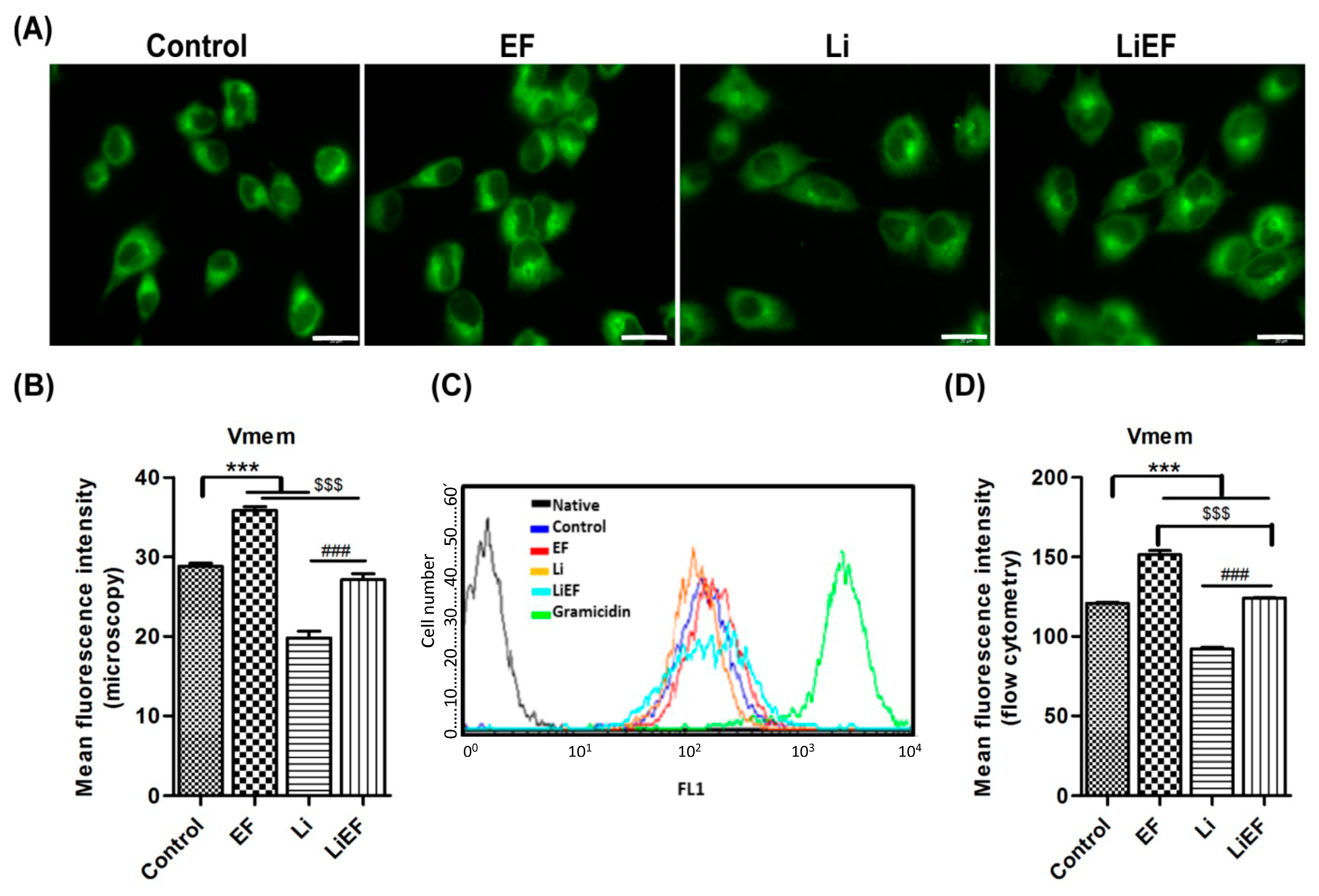
2.1.2. EF Depolarizes the Plasma Membrane Potential
2.1.3. EF Decreases Oxidative Stress
2.1.4. EF Enhances Cell Migration
2.2. Effects of EF on Mitochondria and Its Activity in Li Exposed Cells
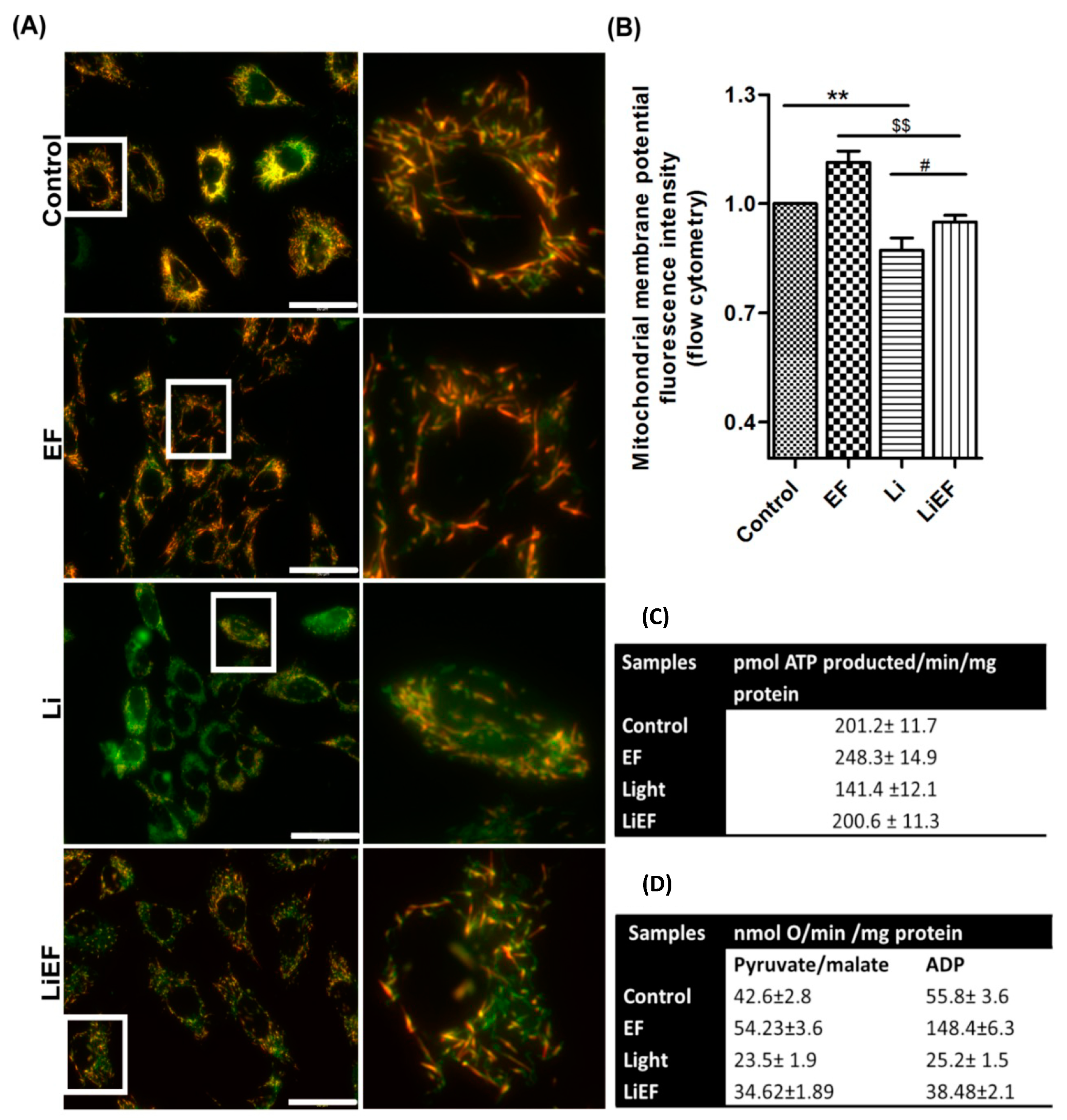
2.2.1. EF Promotes Gain in Mitochondrial Membrane Potential
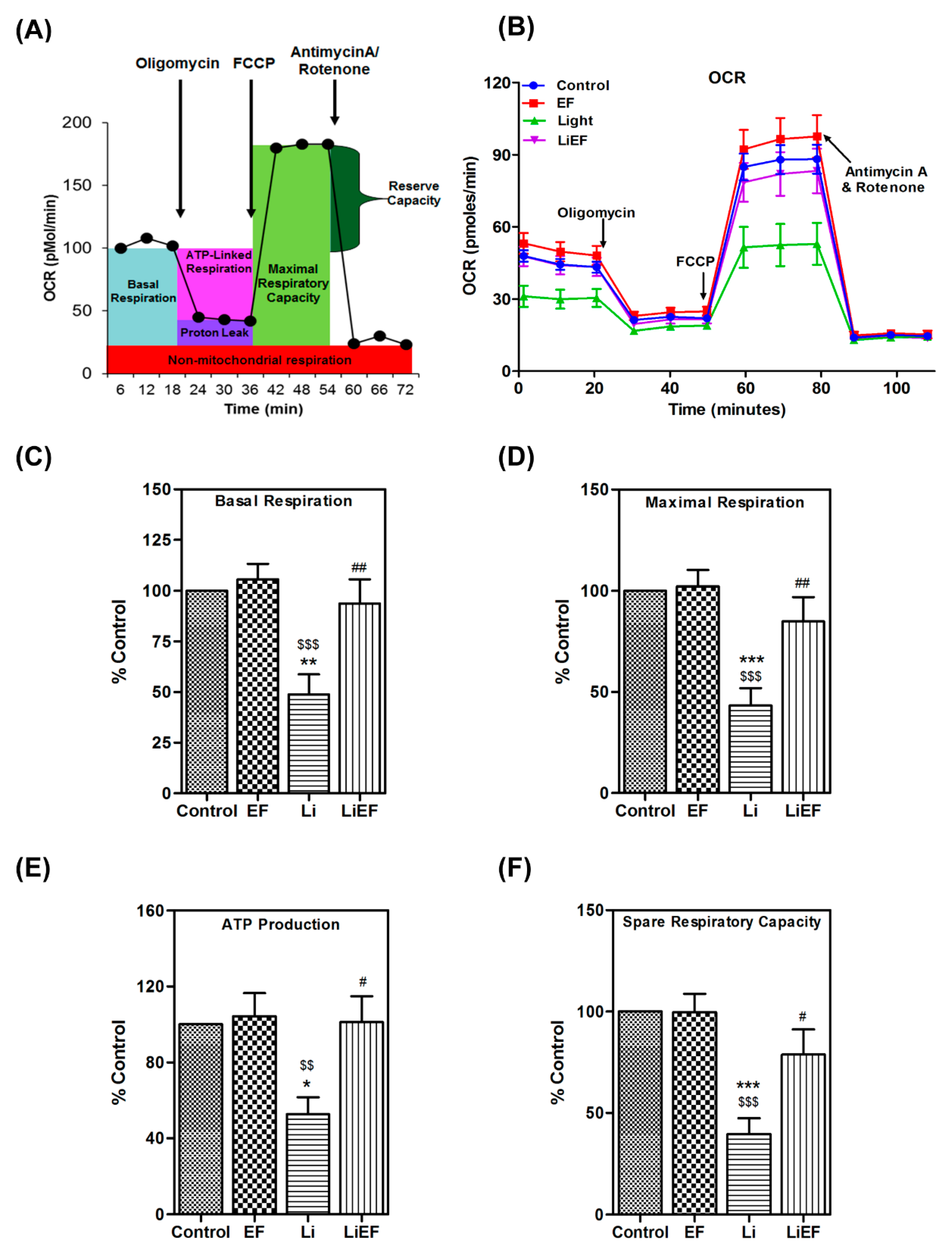
2.2.2. EF Increases Mitochondrial Respiratory Capacity
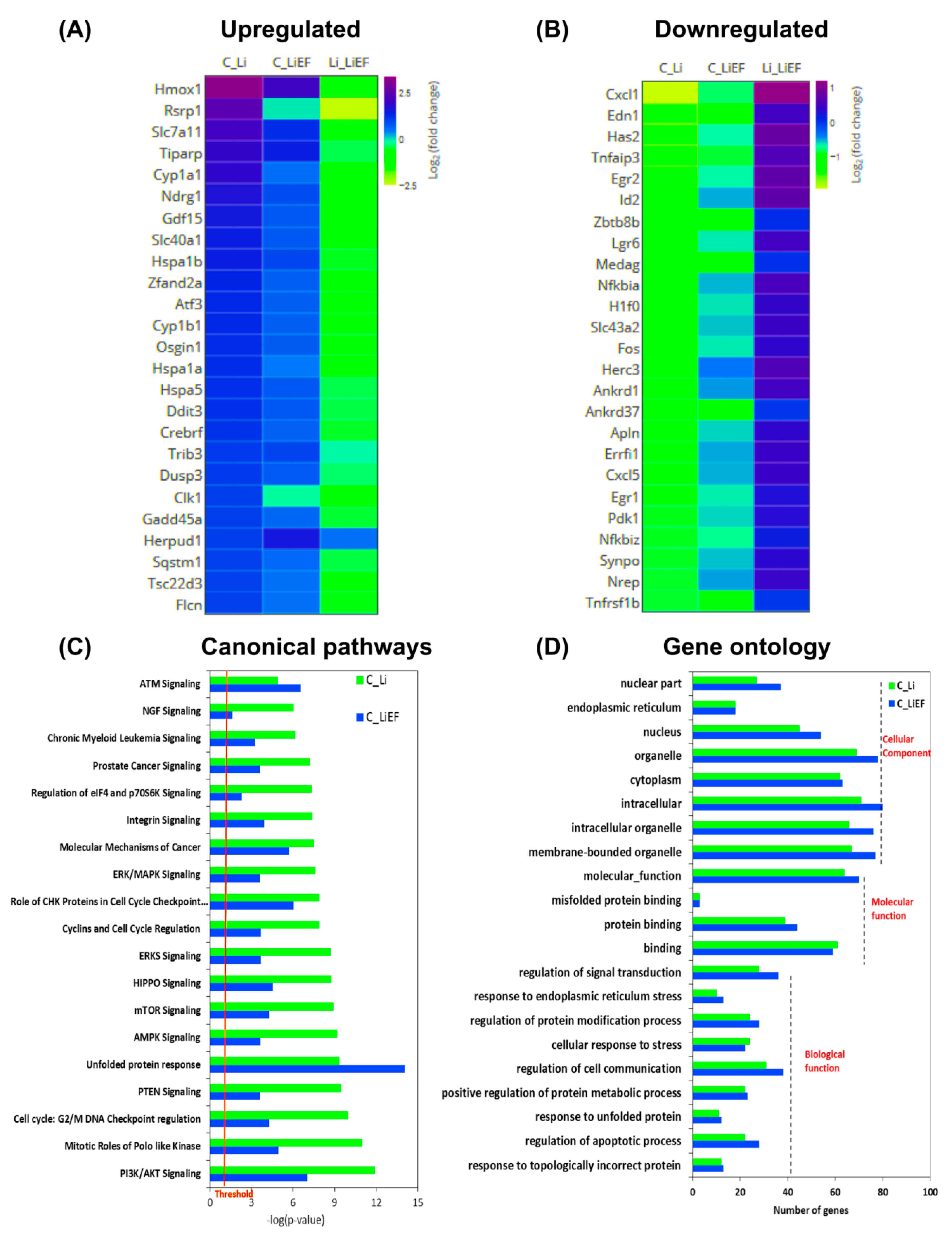
2.3. Effects of EF on Li Exposed Cells at Signaling Level
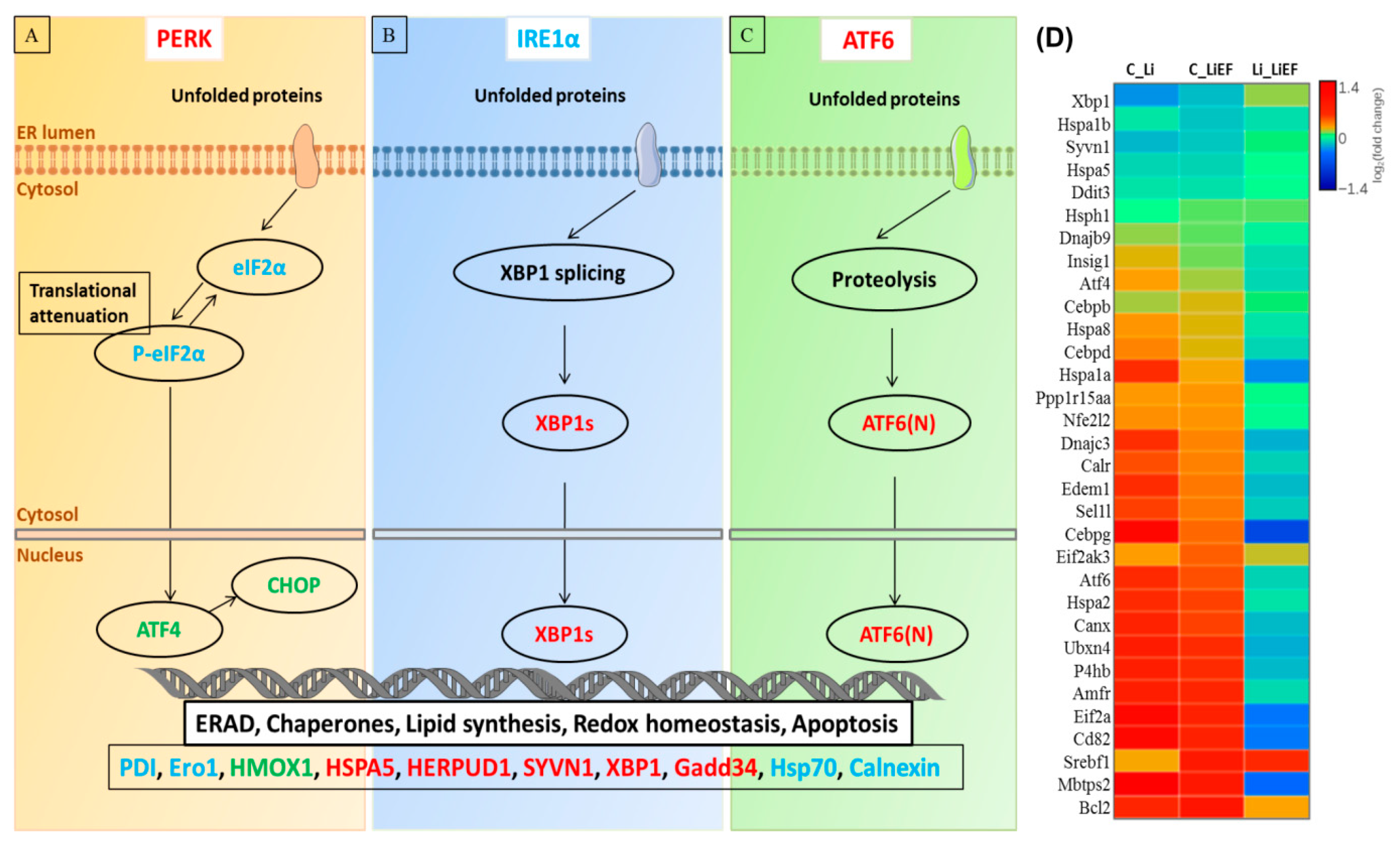
EF Upregulates Unfolded Protein Response (UPR) Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Experimental Setup
4.3. Quantitative Analysis of Electrotaxis
4.4. Antibodies
4.5. Live/Dead Assay
4.6. Membrane Potential (Vm) Measurements
4.7. Mitochondrial Membrane Potential (Δψm) Measurements
4.8. Superoxide Measurement
4.9. Mitochondrial ROS Measurement
4.10. Oxygen Consumption Measurements
4.11. ATP Synthesis Assay
4.12. Mitochondrial Bioenergetics
4.13. Immunofluorescence
4.14. Cell Cycle Analysis
4.15. RNA Isolation and Sequencing
4.16. Bioinformatics Analysis
4.17. Differential Genes Expression Analyses
4.18. qRT-PCR
4.19. Western Blot
4.20. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levin, M. Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. J. Physiol. 2014, 592, 2295–2305. [Google Scholar]
- McLaughlin, K.A.; Levin, M. Bioelectric signaling in regeneration: Mechanisms of ionic controls of growth and form. Dev. Biol. 2018, 433, 177–189. [Google Scholar] [CrossRef]
- Funk, R.H.W. Endogenous electric fields as guiding cue for cell migration. Front. Physiol. 2015, 6, 143. [Google Scholar] [CrossRef]
- McCaig, C.D.; Rajnicek, A.M. Electrical fields, nerve growth and nerve regeneration. Exp. Physiol. 1991, 76, 473–494. [Google Scholar] [PubMed]
- Levin, M. Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 2021, 184, 1971–1989. [Google Scholar] [PubMed]
- Levin, M.; Selberg, J.; Rolandi, M. Endogenous Bioelectrics in Development, Cancer, and Regeneration: Drugs and Bioelectronic Devices as Electroceuticals for Regenerative Medicine. iScience 2019, 22, 519–533. [Google Scholar] [CrossRef]
- Castaldi, E.; Cicchini, G.M.; Cinelli, L.; Biagi, L.; Rizzo, S.; Morrone, M.C. Visual BOLD Response in Late Blind Subjects with Argus II Retinal Prosthesis. PLoS Biol. 2016, 14, e1002569. [Google Scholar] [CrossRef]
- Yue, L.; Weiland, J.D.; Roska, B.; Humayun, M.S. Retinal stimulation strategies to restore vision: Fundamentals and systems. Prog. Retin. Eye Res. 2016, 53, 21–47. [Google Scholar] [CrossRef] [PubMed]
- Willmann, G.; Schaferhoff, K.; Fischer, M.D.; Arango-Gonzalez, B.; Bolz, S.; Naycheva, L.; Rock, T.; Bonin, M.; Bartz-Schmidt, K.U.; Zrenner, E.; et al. Gene expression profiling of the retina after transcorneal electrical stimulation in wild-type Brown Norway rats. Investig. Ophthalmol. Vis. Sci. 2011, 52, 7529–7537. [Google Scholar] [CrossRef]
- Sehic, A.; Guo, S.; Cho, K.-S.; Corraya, R.M.; Chen, D.F.; Utheim, T.P. Electrical Stimulation as a Means for Improving Vision. Am. J. Pathol. 2016, 186, 2783–2797. [Google Scholar] [CrossRef]
- Shen, J.; Wang, Y.; Yao, K. Protection of retinal ganglion cells in glaucoma: Current status and future. Exp. Eye Res. 2021, 205, 108506. [Google Scholar] [PubMed]
- Hu, Y.; Grodzki, L.M.; Bartsch, S.; Bartsch, U. Cell-Based Neuroprotection of Retinal Ganglion Cells in Animal Models of Optic Neuropathies. Biology 2021, 10, 1181. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Kwok, S.S.; Chan, Y.K.; Ming Lai, J.S.; Pan, W.; Nie, L.; Shih, K.C. Therapeutic Strategies for Attenuation of Retinal Ganglion Cell Injury in Optic Neuropathies: Concepts in Translational Research and Therapeutic Implications. Biomed. Res. Int. 2019, 2019, 8397521. [Google Scholar]
- Lee, S.; Park, J.; Kwon, J.; Kim, D.H.; Im, C.H. Multi-channel transorbital electrical stimulation for effective stimulation of posterior retina. Sci. Rep. 2021, 11, 9745. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.C.; Zhou, Y.; Tan, G.; Li, J. Research progress about the effect and prevention of blue light on eyes. Int. J. Ophthalmol. 2018, 11, 1999–2003. [Google Scholar]
- Ouyang, X.; Yang, J.; Hong, Z.; Wu, Y.; Xie, Y.; Wang, G. Mechanisms of blue light-induced eye hazard and protective measures: A review. Biomed. Pharmacother. 2020, 130, 110577. [Google Scholar]
- Krigel, A.; Berdugo, M.; Picard, E.; Levy-Boukris, R.; Jaadane, I.; Jonet, L.; Dernigoghossian, M.; Andrieu-Soler, C.; Torriglia, A.; Behar-Cohen, F. Light-induced retinal damage using different light sources, protocols and rat strains reveals LED phototoxicity. Neuroscience 2016, 339, 296–307. [Google Scholar] [CrossRef]
- Nakanishi-Ueda, T.; Majima, H.J.; Watanabe, K.; Ueda, T.; Indo, H.P.; Suenaga, S.; Hisamitsu, T.; Ozawa, T.; Yasuhara, H.; Koide, R. Blue LED light exposure develops intracellular reactive oxygen species, lipid peroxidation, and subsequent cellular injuries in cultured bovine retinal pigment epithelial cells. Free Radic. Res. 2013, 47, 774–780. [Google Scholar] [CrossRef]
- Roehlecke, C.; Schumann, U.; Ader, M.; Brunssen, C.; Bramke, S.; Morawietz, H.; Funk, R.H. Stress reaction in outer segments of photoreceptors after blue light irradiation. PLoS ONE 2013, 8, e71570. [Google Scholar]
- Heinig, N.; Schumann, U.; Calzia, D.; Panfoli, I.; Ader, M.; Schmidt, M.H.H.; Funk, R.H.W.; Roehlecke, C. Photobiomodulation Mediates Neuroprotection against Blue Light Induced Retinal Photoreceptor Degeneration. Int. J. Mol. Sci. 2020, 21, 2370. [Google Scholar] [CrossRef]
- Gorgidze, L.A.; Oshemkova, S.A.; Vorobjev, I.A. Blue light inhibits mitosis in tissue culture cells. Biosci. Rep. 1998, 18, 215–224. [Google Scholar] [CrossRef]
- Pflaum, M.; Kielbassa, C.; Garmyn, M.; Epe, B. Oxidative DNA damage induced by visible light in mammalian cells: Extent, inhibition by antioxidants and genotoxic effects. Mutat. Res. 1998, 408, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Wenzel, A.; Obin, M.S.; Chen, C.K.; Brill, E.; Krasnoperova, N.V.; Eversole-Cire, P.; Kleyner, Y.; Taylor, A.; Simon, M.I.; et al. Evidence for two apoptotic pathways in light-induced retinal degeneration. Nat. Genet. 2002, 32, 254–260. [Google Scholar] [PubMed]
- Roehlecke, C.; Schumann, U.; Ader, M.; Knels, L.; Funk, R.H. Influence of blue light on photoreceptors in a live retinal explant system. Mol. Vis. 2011, 17, 876–884. [Google Scholar] [PubMed]
- Sundelacruz, S.; Levin, M.; Kaplan, D.L. Role of membrane potential in the regulation of cell proliferation and differentiation. Stem Cell Rev. Rep. 2009, 5, 231–246. [Google Scholar] [PubMed]
- Kutty, R.K.; Kutty, G.; Wiggert, B.; Chader, G.J.; Darrow, R.M.; Organisciak, D.T. Induction of heme oxygenase 1 in the retina by intense visible light: Suppression by the antioxidant dimethylthiourea. Proc. Natl. Acad. Sci. USA 1995, 92, 1177–1181. [Google Scholar] [CrossRef]
- Demontis, G.C.; Longoni, B.; Marchiafava, P.L. Molecular steps involved in light-induced oxidative damage to retinal rods. Investig. Ophthalmol. Vis. Sci. 2002, 43, 2421–2427. [Google Scholar]
- Murphy, M.P. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab. 2013, 18, 145–146. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Gokoffski, K.K.; Jia, X.; Shvarts, D.; Xia, G.; Zhao, M. Physiologic Electrical Fields Direct Retinal Ganglion Cell Axon Growth In Vitro. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3659–3668. [Google Scholar]
- Morimoto, T.; Miyoshi, T.; Matsuda, S.; Tano, Y.; Fujikado, T.; Fukuda, Y. Transcorneal electrical stimulation rescues axotomized retinal ganglion cells by activating endogenous retinal IGF-1 system. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2147–2155. [Google Scholar] [CrossRef]
- Pardue, M.T.; Phillips, M.J.; Yin, H.; Sippy, B.D.; Webb-Wood, S.; Chow, A.Y.; Ball, S.L. Neuroprotective effect of subretinal implants in the RCS rat. Investig. Ophthalmol. Vis. Sci. 2005, 46, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Politis, M.J.; Zanakis, M.F.; Albala, B.J. Mammalian optic nerve regeneration following the application of electric fields. J. Trauma 1988, 28, 1548–1552. [Google Scholar] [PubMed]
- Ni, Y.Q.; Gan, D.K.; Xu, H.D.; Xu, G.Z.; Da, C.D. Neuroprotective effect of transcorneal electrical stimulation on light-induced photoreceptor degeneration. Exp. Neurol. 2009, 219, 439–452. [Google Scholar] [CrossRef]
- Schatz, A.; Arango-Gonzalez, B.; Fischer, D.; Enderle, H.; Bolz, S.; Röck, T.; Naycheva, L.; Grimm, C.; Messias, A.; Zrenner, E.; et al. Transcorneal Electrical Stimulation Shows Neuroprotective Effects in Retinas of Light-Exposed Rats. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5552–5561. [Google Scholar] [CrossRef]
- Zhou, W.T.; Ni, Y.Q.; Jin, Z.B.; Zhang, M.; Wu, J.H.; Zhu, Y.; Xu, G.Z.; Gan, D.K. Electrical stimulation ameliorates light-induced photoreceptor degeneration in vitro via suppressing the proinflammatory effect of microglia and enhancing the neurotrophic potential of Muller cells. Exp. Neurol. 2012, 238, 192–208. [Google Scholar] [CrossRef]
- Ciavatta, V.T.; Mocko, J.A.; Kim, M.K.; Pardue, M.T. Subretinal electrical stimulation preserves inner retinal function in RCS rat retina. Mol. Vis. 2013, 19, 995–1005. [Google Scholar]
- Sato, T.; Lee, T.S.; Takamatsu, F.; Fujikado, T. Induction of fibroblast growth factor-2 by electrical stimulation in cultured retinal Mueller cells. Neuroreport 2008, 19, 1617–1621. [Google Scholar]
- Al-Majed, A.A.; Tam, S.L.; Gordon, T. Electrical stimulation accelerates and enhances expression of regeneration-associated genes in regenerating rat femoral motoneurons. Cell Mol. Neurobiol. 2004, 24, 379–402. [Google Scholar] [CrossRef]
- Gao, R.C.; Zhang, X.D.; Sun, Y.H.; Kamimura, Y.; Mogilner, A.; Devreotes, P.N.; Zhao, M. Different roles of membrane potentials in electrotaxis and chemotaxis of dictyostelium cells. Eukaryot. Cell 2011, 10, 1251–1256. [Google Scholar] [CrossRef]
- Kobelt, L.J.; Wilkinson, A.E.; McCormick, A.M.; Willits, R.K.; Leipzig, N.D. Short duration electrical stimulation to enhance neurite outgrowth and maturation of adult neural stem progenitor cells. Ann. Biomed. Eng. 2014, 42, 2164–2176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, G.; Rong, W.; Wang, A.; Wu, C.; Huo, X. Early applied electric field stimulation attenuates secondary apoptotic responses and exerts neuroprotective effects in acute spinal cord injury of rats. Neuroscience 2015, 291, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Groeger, G.; Mackey, A.M.; Pettigrew, C.A.; Bhatt, L.; Cotter, T.G. Stress-induced activation of Nox contributes to cell survival signalling via production of hydrogen peroxide. J. Neurochem. 2009, 109, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, T.; Hu, S.; Lin, J.; Hu, R.; Feng, H. Superoxide Mediates Direct Current Electric Field-Induced Directional Migration of Glioma Cells through the Activation of AKT and ERK. PLoS ONE 2013, 8, e61195. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 323–354. [Google Scholar] [CrossRef]
- Oplander, C.; Hidding, S.; Werners, F.B.; Born, M.; Pallua, N.; Suschek, C.V. Effects of blue light irradiation on human dermal fibroblasts. J. Photochem. Photobiol. B 2011, 103, 118–125. [Google Scholar] [CrossRef]
- Lascaratos, G.; Ji, D.; Wood, J.P.; Osborne, N.N. Visible light affects mitochondrial function and induces neuronal death in retinal cell cultures. Vision Res. 2007, 47, 1191–1201. [Google Scholar] [CrossRef]
- Osborne, N.N.; Nunez-Alvarez, C.; Del Olmo-Aguado, S. The effect of visual blue light on mitochondrial function associated with retinal ganglions cells. Exp. Eye Res. 2014, 128, 8–14. [Google Scholar] [CrossRef]
- De Giorgi, F.; Lartigue, L.; Ichas, F. Electrical coupling and plasticity of the mitochondrial network. Cell Calcium 2000, 28, 365–370. [Google Scholar] [CrossRef]
- Skulachev, V.P. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 2001, 26, 23–29. [Google Scholar] [PubMed]
- Peng, T.I.; Jou, M.J. Mitochondrial swelling and generation of reactive oxygen species induced by photoirradiation are heterogeneously distributed. Ann. N. Y. Acad. Sci. 2004, 1011, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Jou, M.J.; Peng, T.I.; Reiter, R.J.; Jou, S.B.; Wu, H.Y.; Wen, S.T. Visualization of the antioxidative effects of melatonin at the mitochondrial level during oxidative stress-induced apoptosis of rat brain astrocytes. J. Pineal Res. 2004, 37, 55–70. [Google Scholar] [PubMed]
- Funk, R.H.; Monsees, T.; Ozkucur, N. Electromagnetic effects—From cell biology to medicine. Prog. Histochem. Cytochem. 2009, 43, 177–264. [Google Scholar] [CrossRef]
- Ozkucur, N.; Song, B.; Bola, S.; Zhang, L.; Reid, B.; Fu, G.; Funk, R.H.W.; Zhao, M. NHE3 phosphorylation via PKCeta marks the polarity and orientation of directionally migrating cells. Cell Mol. Life Sci. 2014, 71, 4653–4663. [Google Scholar] [CrossRef]
- Zhao, M.; Song, B.; Pu, J.; Wada, T.; Reid, B.; Tai, G.; Wang, F.; Guo, A.; Walczysko, P.; Gu, Y.; et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-γ and PTEN. Nature 2006, 442, 457–460. [Google Scholar]
- Nakanishi, T.; Shimazawa, M.; Sugitani, S.; Kudo, T.; Imai, S.; Inokuchi, Y.; Tsuruma, K.; Hara, H. Role of endoplasmic reticulum stress in light-induced photoreceptor degeneration in mice. J. Neurochem. 2013, 125, 111–124. [Google Scholar] [CrossRef]
- Kuse, Y.; Ogawa, K.; Tsuruma, K.; Shimazawa, M.; Hara, H. Damage of photoreceptor-derived cells in culture induced by light emitting diode-derived blue light. Sci. Rep. 2014, 4, 5223. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Morimoto, T.; Miyoshi, T.; Fujikado, T.; Tano, Y.; Fukuda, Y. Electrical stimulation enhances the survival of axotomized retinal ganglion cells in vivo. Neuroreport 2002, 13, 227–230. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk Is Essential for Translational Regulation and Cell Survival during the Unfolded Protein Response. Molecular. Cell 2000, 5, 897–904. [Google Scholar] [PubMed]
- Paredes, F.; Parra, V.; Torrealba, N.; Navarro-Marquez, M.; Gatica, D.; Bravo-Sagua, R.; Troncoso, R.; Pennanen, C.; Quiroga, C.; Chiong, M.; et al. HERPUD1 protects against oxidative stress-induced apoptosis through downregulation of the inositol 1,4,5-trisphosphate receptor. Free Radic. Biol. Med. 2016, 90, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Ishiguro, M.; Niinuma, Y.; Uesugi, M.; Nomura, Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation1. FEBS Lett. 2002, 532, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Gu, Y.; Pu, J.; Reid, B.; Zhao, Z.; Zhao, M. Application of direct current electric fields to cells and tissues in vitro and modulation of wound electric field in vivo. Nat. Protoc. 2007, 2, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef]
- Sgarbi, G.; Baracca, A.; Lenaz, G.; Valentino Lucia, M.; Carelli, V.; Solaini, G. Inefficient coupling between proton transport and ATP synthesis may be the pathogenic mechanism for NARP and Leigh syndrome resulting from the T8993G mutation in mtDNA. Biochem. J. 2006, 395, 493–500. [Google Scholar] [CrossRef]
- Rose, S.; Frye, R.E.; Slattery, J.; Wynne, R.; Tippett, M.; Pavliv, O.; Melnyk, S.; James, S.J. Oxidative stress induces mitochondrial dysfunction in a subset of autism lymphoblastoid cell lines in a well-matched case control cohort. PLoS ONE 2014, 9, e85436. [Google Scholar] [CrossRef]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Royal Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bola, S.; Subramanian, P.; Calzia, D.; Dahl, A.; Panfoli, I.; Funk, R.H.W.; Roehlecke, C. Analysis of Electric Field Stimulation in Blue Light Stressed 661W Cells. Int. J. Mol. Sci. 2023, 24, 3433. https://doi.org/10.3390/ijms24043433
Bola S, Subramanian P, Calzia D, Dahl A, Panfoli I, Funk RHW, Roehlecke C. Analysis of Electric Field Stimulation in Blue Light Stressed 661W Cells. International Journal of Molecular Sciences. 2023; 24(4):3433. https://doi.org/10.3390/ijms24043433
Chicago/Turabian StyleBola, Sharanya, Pallavi Subramanian, Daniela Calzia, Andreas Dahl, Isabella Panfoli, Richard H. W. Funk, and Cora Roehlecke. 2023. "Analysis of Electric Field Stimulation in Blue Light Stressed 661W Cells" International Journal of Molecular Sciences 24, no. 4: 3433. https://doi.org/10.3390/ijms24043433
APA StyleBola, S., Subramanian, P., Calzia, D., Dahl, A., Panfoli, I., Funk, R. H. W., & Roehlecke, C. (2023). Analysis of Electric Field Stimulation in Blue Light Stressed 661W Cells. International Journal of Molecular Sciences, 24(4), 3433. https://doi.org/10.3390/ijms24043433








