Neddylation of EphB1 Regulates Its Activity and Associates with Liver Fibrosis
Abstract
1. Introduction
2. Results
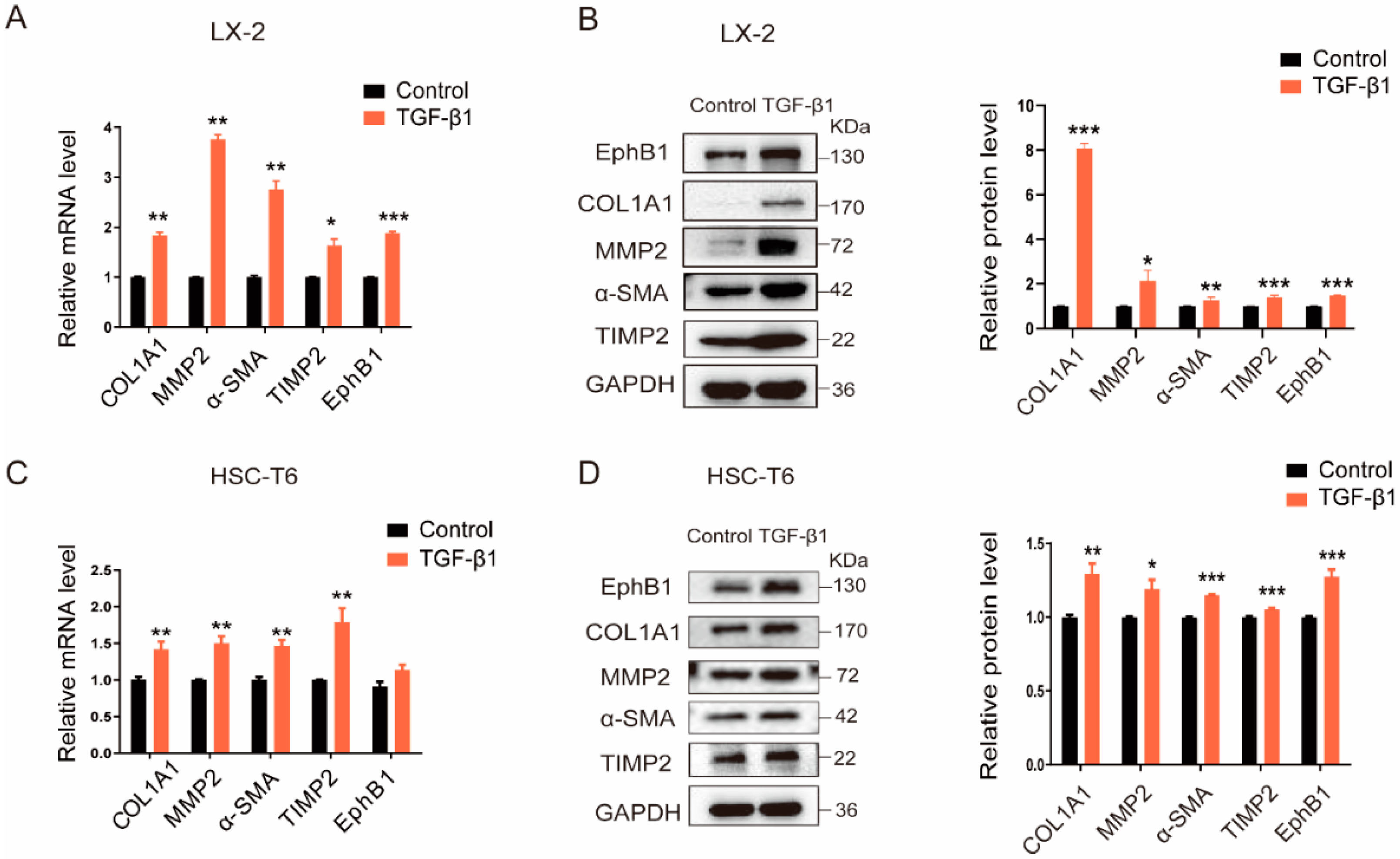
2.1. EphB1 Expression Is Increased in Activated HSCs
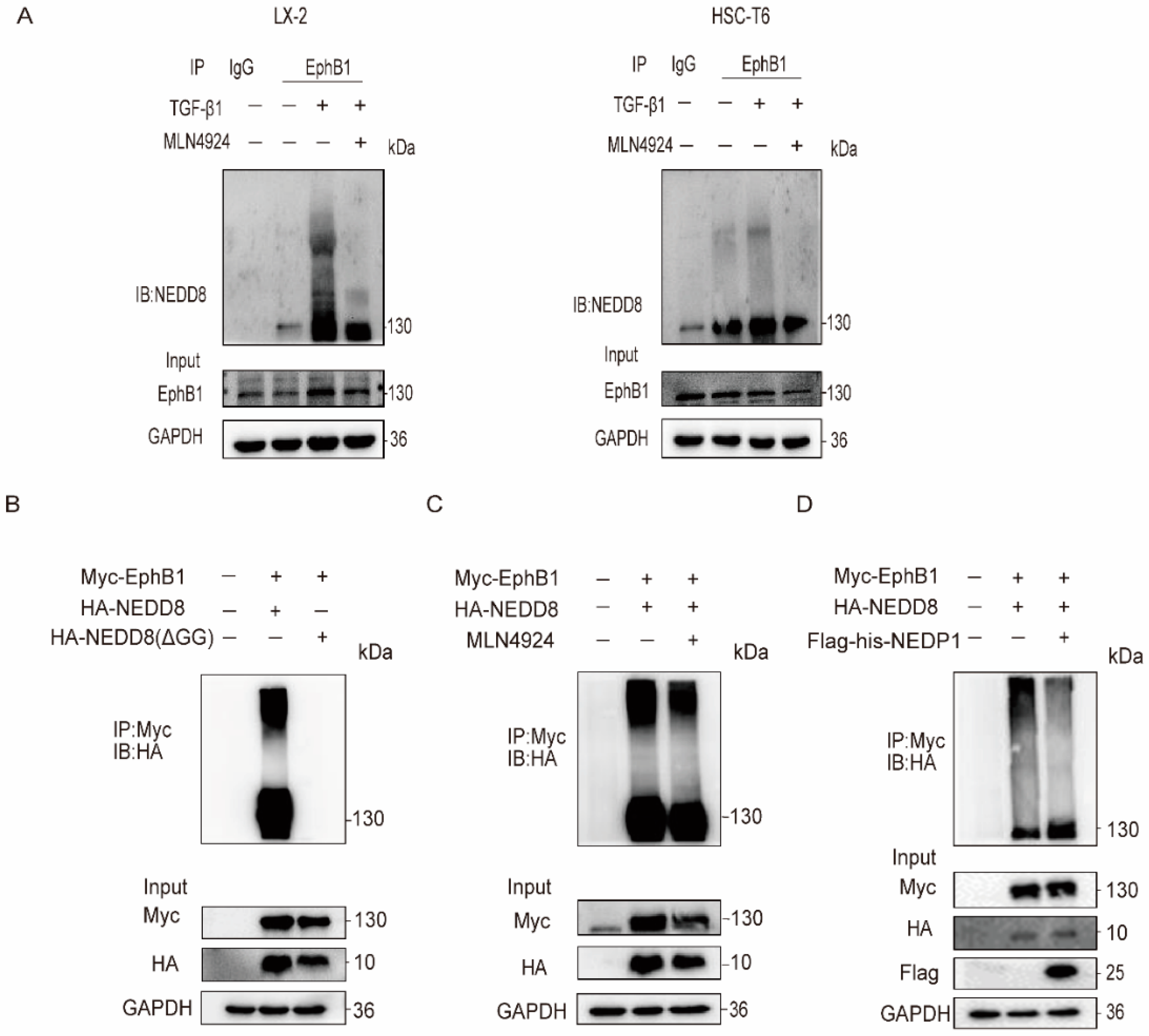
2.2. EphB1 Undergoes Augmented Neddylation in Activated HSCs
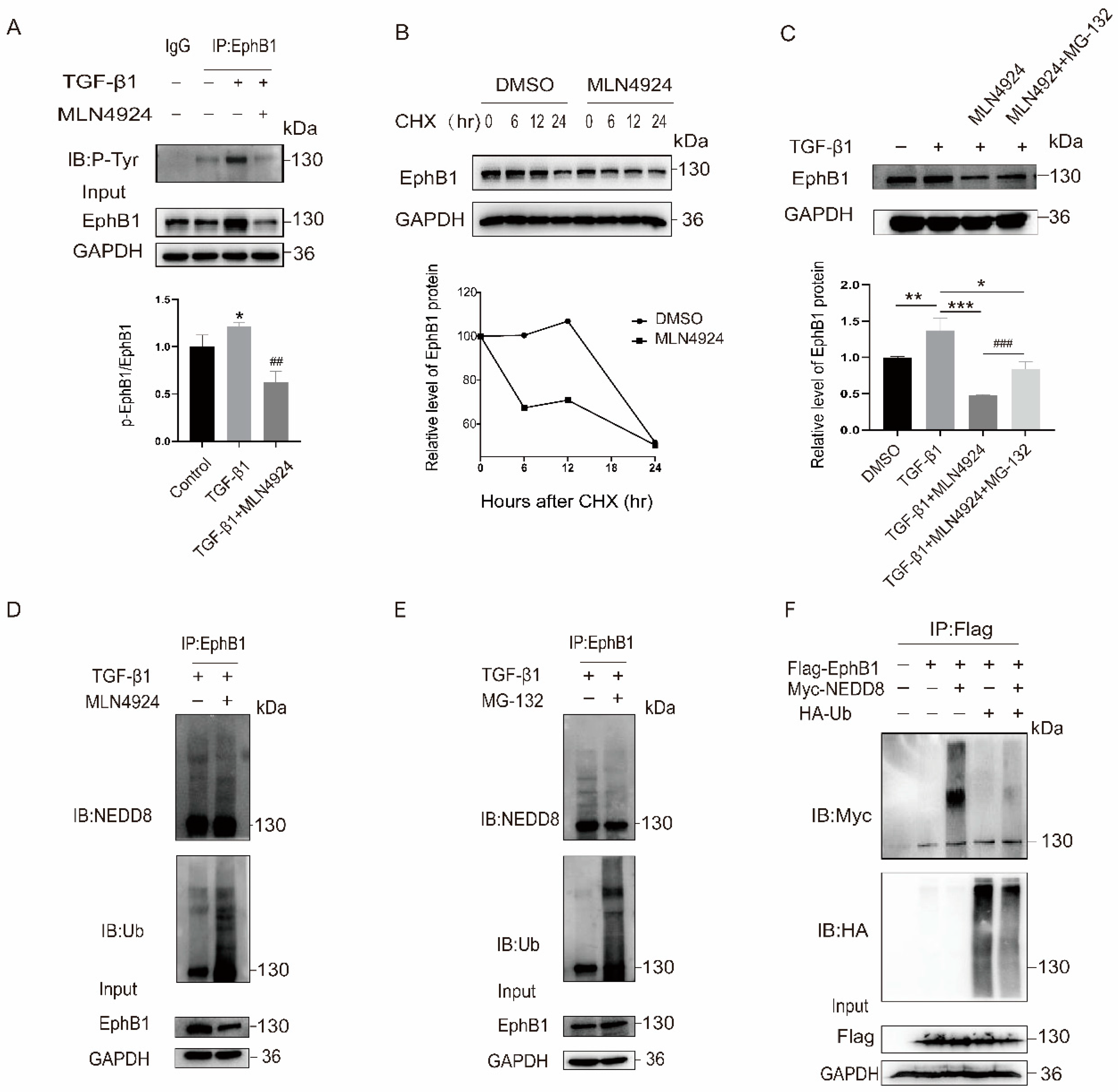
2.3. EphB1 Neddylation Enhances Its Kinase Activity and Inhibits Its Degradation
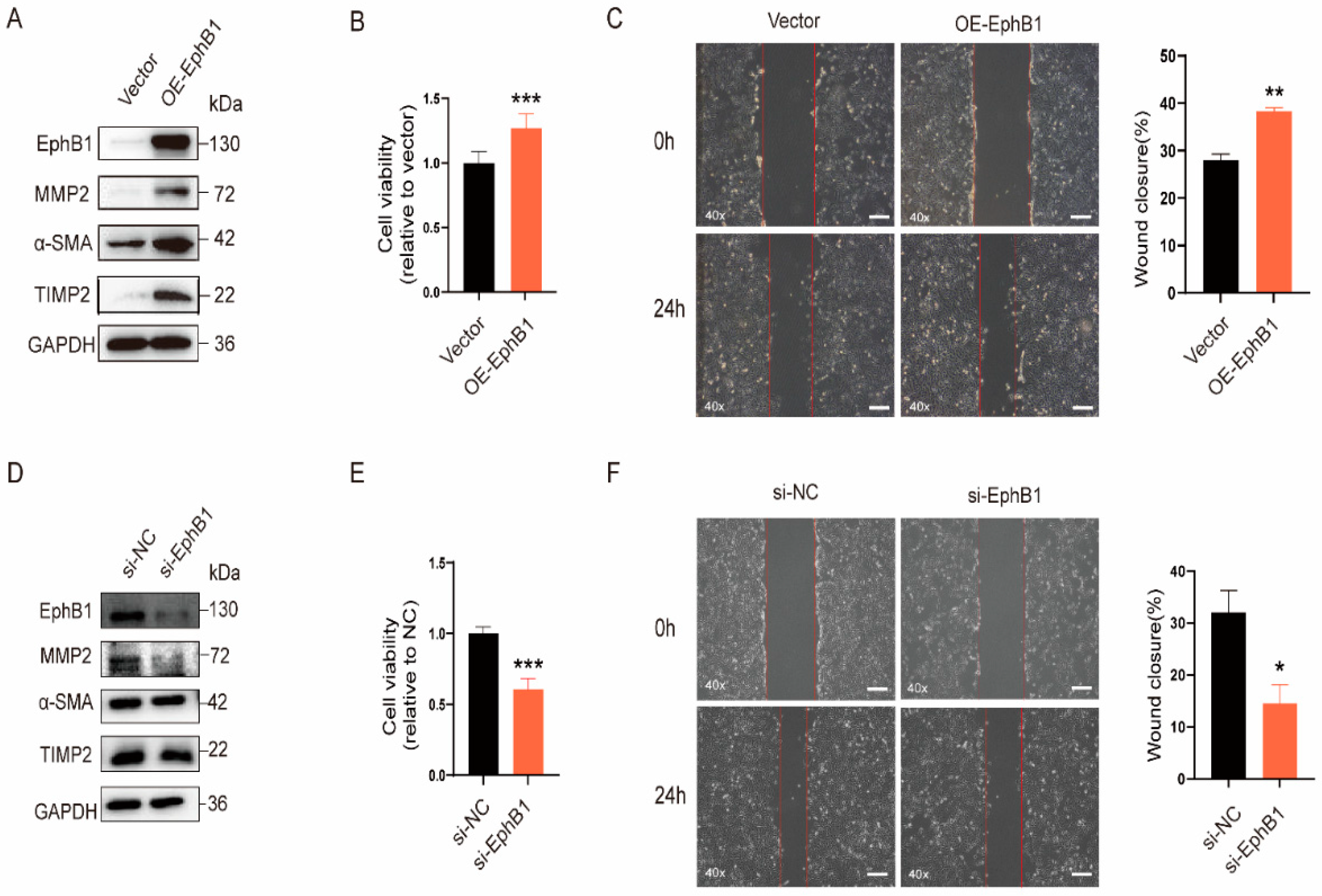
2.4. EphB1 Promotes the Proliferation, Migration, and Activation of HSCs
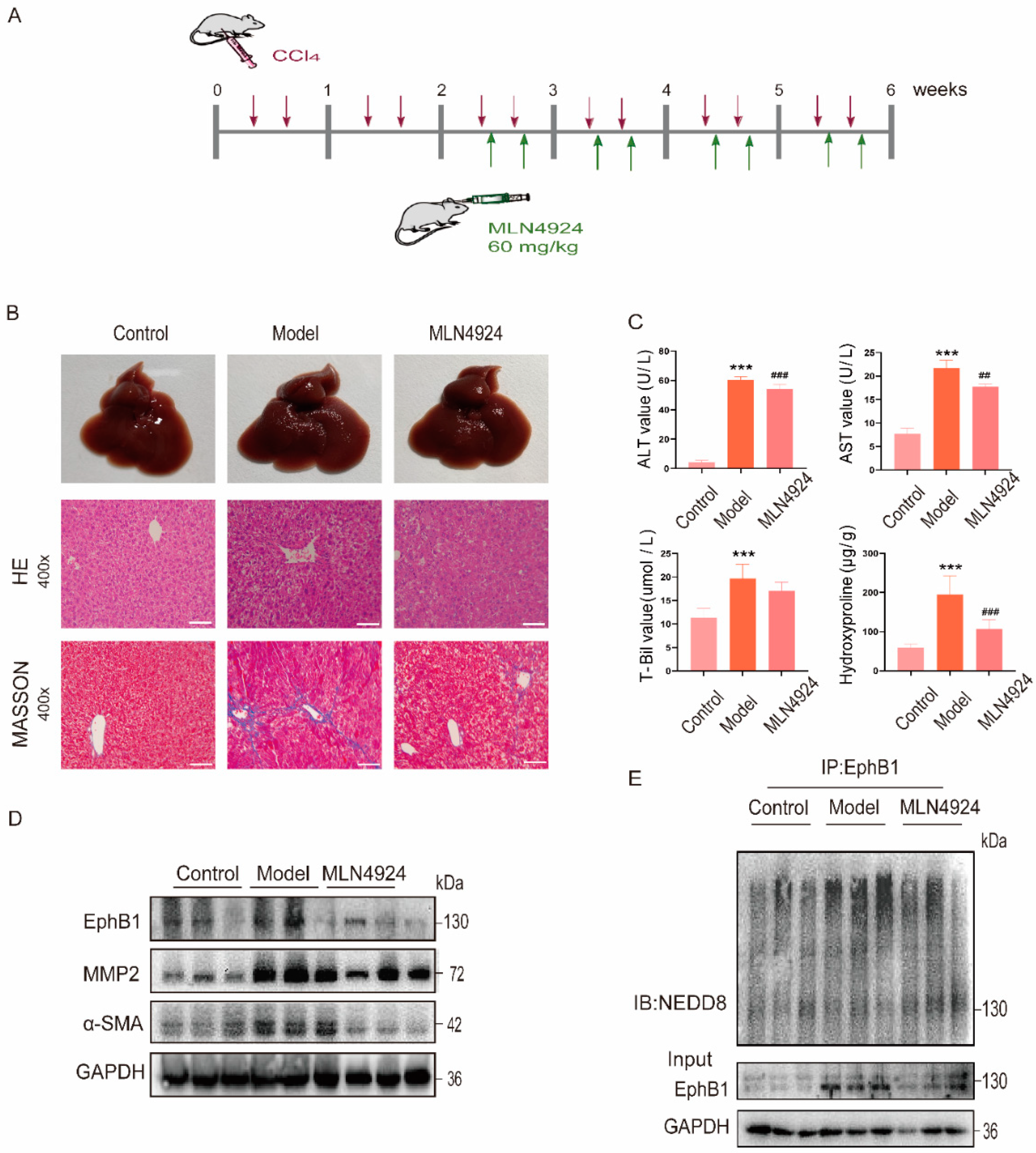
2.5. EphB1 Expression and Its Neddylation Elevated in CCl4-Induced Liver Fibrosis Mice
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Lines and Culture
4.3. Animal Models
4.4. Plasmids, siRNAs and Transfection
4.5. Western Blotting
4.6. Immunoprecipitation
4.7. Quantitative RT-PCR (qRT-PCR)
4.8. Biochemical Assays
4.9. H&E and Masson’s Trichrome Staining
4.10. Cell Proliferation Assay
4.11. Wound Healing Assay
4.12. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parola, M.; Pinzani, M. Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Asp. Med. 2019, 65, 37–55. [Google Scholar] [CrossRef]
- Pellicoro, A.; Ramachandran, P.; Iredale, J.P.; Fallowfield, J.A. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 2014, 14, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Friedman, S.L.; Lee, Y.A. Antifibrotic therapies: Where are we now? Semin. Liver. Dis. 2016, 36, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Pinzani, M. Hepatic fibrosis 2022: Unmet needs and a blueprint for the future. Hepatology 2022, 75, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Distler, J.H.; Distler, O. Intracellular tyrosine kinases as novel targets for anti-fibrotic therapy in systemic sclerosis. Rheumatology 2008, 47 (Suppl. 5), v10–v11. [Google Scholar] [CrossRef]
- Qu, K.; Huang, Z.; Lin, T.; Liu, S.; Chang, H.; Yan, Z.; Zhang, H.; Liu, C. New Insight into the anti-liver fibrosis effect of multitargeted tyrosine kinase inhibitors: From molecular target to clinical trials. Front. Pharmacol. 2016, 6, 300. [Google Scholar] [CrossRef] [PubMed]
- Kania, A.; Klein, R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell. Biol. 2016, 17, 240–256. [Google Scholar] [CrossRef]
- Wu, B.; Rockel, J.S.; Lagares, D.; Kapoor, M. Ephrins and Eph receptor signaling in tissue repair and fibrosis. Curr. Rheumatol. Rep. 2019, 21, 23. [Google Scholar] [CrossRef]
- Mimche, P.N.; Brady, L.M.; Bray, C.F.; Lee, C.M.; Thapa, M.; King, T.P.; Quicke, K.; McDermott, C.D.; Mimche, S.M.; Grakoui, A.; et al. The receptor tyrosine kinase EphB2 promotes hepatic fibrosis in mice. Hepatology 2015, 62, 900–914. [Google Scholar] [CrossRef]
- Mimche, P.N.; Lee, C.M.; Mimche, S.M.; Thapa, M.; Grakoui, A.; Henkemeyer, M.; Lamb, T.J. EphB2 receptor tyrosine kinase promotes hepatic fibrogenesis in mice via activation of hepatic stellate cells. Sci. Rep. 2018, 8, 2532. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Wang, Y.; Chen, K.; Zhao, L.; Xu, Y.; Jiang, H.; Wang, S. Synergistic antifibrotic effects of miR-451 with miR-185 partly by co-targeting EphB2 on hepatic stellate cells. Cell Death Dis. 2020, 11, 402. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, H.; Ji, S. Paradoxes of the EphB1 receptor in malignant brain tumors. Cancer Cell Int. 2017, 17, 21. [Google Scholar] [CrossRef]
- Teng, L.; Nakada, M.; Furuyama, N.; Sabit, H.; Furuta, T.; Hayashi, Y.; Takino, T.; Dong, Y.; Sato, H.; Sai, Y.; et al. Ligand-dependent EphB1 signaling suppresses glioma invasion and correlates with patient survival. Neuro Oncol. 2013, 15, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.C.; Liang, Y.; Li, L.L.; Tang, J.Z.; Yang, Y.P.; Geng, Y.C.; He, J.; Luo, L.Y.; Li, W.X.; Sun, Z.W.; et al. Bioinformatics analysis identifies protein tyrosine kinase 7 (PTK7) as a potential prognostic and therapeutic biomarker in stages I to IV hepatocellular carcinoma. Med. Sci. Monit. 2019, 25, 8618–8627. [Google Scholar] [CrossRef] [PubMed]
- Rabut, G.; Peter, M. Function and regulation of protein neddylation. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Rep. 2008, 9, 969–976. [Google Scholar] [CrossRef]
- Mendoza, H.M.; Shen, L.N.; Botting, C.; Lewis, A.; Chen, J.; Ink, B.; Hay, R.T. NEDP1, a highly conserved cysteine protease that deNEDDylates cullins. J. Biol. Chem. 2003, 278, 25637–25643. [Google Scholar] [CrossRef]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef]
- Zubiete-Franco, I.; Fernández-Tussy, P.; Barbier-Torres, L.; Simon, J.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; Gutiérrez-de Juan, V.; de Davalillo, S.L.; Duce, A.M.; Iruzubieta, P.; et al. Deregulated neddylation in liver fibrosis. Hepatology 2017, 65, 694–709. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Guan, J.; Yu, S.; Zheng, X. NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase η in response to oxidative DNA damage. Protein Cell 2018, 9, 365–379. [Google Scholar] [CrossRef]
- Osna, N.A.; Carter, W.G.; Ganesan, M.; Kirpich, I.A.; McClain, C.J.; Petersen, D.R.; Shearn, C.T.; Tomasi, M.L.; Kharbanda, K.K. Aberrant post-translational protein modifications in the pathogenesis of alcohol-induced liver injury. World J. Gastroenterol. 2016, 22, 6192–6200. [Google Scholar] [CrossRef]
- Lachiondo-Ortega, S.; Mercado-Gómez, M.; Serrano-Maciá, M.; Lopitz-Otsoa, F.; Salas-Villalobos, T.B.; Varela-Rey, M.; Delgado, T.C.; Martínez-Chantar, M.L. Ubiquitin-like post-translational modifications (Ubl-PTMs): Small peptides with huge impact in liver fibrosis. Cells 2019, 8, 1575. [Google Scholar] [CrossRef]
- Banner, B.F.; Savas, L.; Zivny, J.; Tortorelli, K.; Bonkovsky, H.L. Ubiquitin as a marker of cell injury in nonalcoholic steatohepatitis. Am. J. Clin. Pathol. 2000, 114, 860–866. [Google Scholar] [CrossRef]
- Wilson, C.L.; Murphy, L.B.; Leslie, J.; Kendrick, S.; French, J.; Fox, C.R.; Sheerin, N.S.; Fisher, A.; Robinson, J.H.; Tiniakos, D.G.; et al. Ubiquitin C-terminal hydrolase 1: A novel functional marker for liver myofibroblasts and a therapeutic target in chronic liver disease. J. Hepatol. 2015, 63, 1421–1428. [Google Scholar] [CrossRef]
- Zhou, J.; Cui, S.; He, Q.; Guo, Y.; Pan, X.; Zhang, P.; Huang, N.; Ge, C.; Wang, G.; Gonzalez, F.J.; et al. SUMOylation inhibitors synergize with FXR agonists in combating liver fibrosis. Nat. Commun. 2020, 11, 240. [Google Scholar] [CrossRef]
- Yao, J.; Liang, X.; Liu, Y.; Zheng, M. Neddylation: A versatile pathway takes on chronic liver diseases. Front. Med. 2020, 7, 586881. [Google Scholar] [CrossRef]
- Csizmok, V.; Forman-Kay, J.D. Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications. Curr. Opin. Struct. Biol. 2018, 48, 58–67. [Google Scholar] [CrossRef]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein language: Post-translational modifications talking to each other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, J.; Pei, Y.; Wang, Q.; Wu, Y.; Qiu, G.; Zhang, D.; Lv, M.; Li, W.; Zhang, J. Neddylation controls basal MKK7 kinase activity in breast cancer cells. Oncogene 2016, 35, 2624–2633. [Google Scholar] [CrossRef]
- Barbier-Torres, L.; Delgado, T.C.; García-Rodríguez, J.L.; Zubiete-Franco, I.; Fernández-Ramos, D.; Buqué, X.; Cano, A.; Gutiérrez-de Juan, V.; Fernández-Domínguez, I.; Lopitz-Otsoa, F.; et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 2015, 6, 2509–2523. [Google Scholar] [CrossRef]
- Henkemeyer, M.; Itkis, O.S.; Ngo, M.; Hickmott, P.W.; Ethell, I.M. Multiple EphB receptor tyrosine kinases shape dendritic spines in the hippocampus. J. Cell Biol. 2003, 163, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.Y.; Patel, O.; Janes, P.W.; Murphy, J.M.; Lucet, I.S. Eph receptor signalling: From catalytic to non-catalytic functions. Oncogene 2019, 38, 6567–6584. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Morgan, M.A.; Sun, Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 2014, 21, 2383–2400. [Google Scholar] [CrossRef] [PubMed]
- Oved, S.; Mosesson, Y.; Zwang, Y.; Santonico, E.; Shtiegman, K.; Marmor, M.D.; Kochupurakkal, B.S.; Katz, M.; Lavi, S.; Cesareni, G.; et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 2006, 281, 21640–21651. [Google Scholar] [CrossRef]
- Zuo, W.; Huang, F.; Chiang, Y.J.; Li, M.; Du, J.; Ding, Y.; Zhang, T.; Lee, H.W.; Jeong, L.S.; Chen, Y.; et al. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol. Cell 2013, 49, 499–510. [Google Scholar] [CrossRef]
- Lee, G.W.; Park, J.B.; Park, S.Y.; Seo, J.; Shin, S.H.; Park, J.W.; Kim, S.J.; Watanabe, M.; Chun, Y.S. The E3 ligase C-CBL inhibits cancer cell migration by neddylating the proto-oncogene c-Src. Oncogene 2018, 37, 5552–5568. [Google Scholar] [CrossRef]
- Fasen, K.; Cerretti, D.P.; Huynh-Do, U. Ligand binding induces Cbl-dependent EphB1 receptor degradation through the lysosomal pathway. Traffic 2008, 9, 251–266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Zhang, D.; Han, Y.; Chen, K.; Guo, W.; Chen, Y.; Wang, S. Neddylation of EphB1 Regulates Its Activity and Associates with Liver Fibrosis. Int. J. Mol. Sci. 2023, 24, 3415. https://doi.org/10.3390/ijms24043415
Li R, Zhang D, Han Y, Chen K, Guo W, Chen Y, Wang S. Neddylation of EphB1 Regulates Its Activity and Associates with Liver Fibrosis. International Journal of Molecular Sciences. 2023; 24(4):3415. https://doi.org/10.3390/ijms24043415
Chicago/Turabian StyleLi, Rongxin, Dan Zhang, Yueqing Han, Ke Chen, Weiran Guo, Yijun Chen, and Shuzhen Wang. 2023. "Neddylation of EphB1 Regulates Its Activity and Associates with Liver Fibrosis" International Journal of Molecular Sciences 24, no. 4: 3415. https://doi.org/10.3390/ijms24043415
APA StyleLi, R., Zhang, D., Han, Y., Chen, K., Guo, W., Chen, Y., & Wang, S. (2023). Neddylation of EphB1 Regulates Its Activity and Associates with Liver Fibrosis. International Journal of Molecular Sciences, 24(4), 3415. https://doi.org/10.3390/ijms24043415






