Antioxidant, Tyrosinase, α-Glucosidase, and Elastase Enzyme Inhibition Activities of Optimized Unripe Ajwa Date Pulp (Phoenix dactylifera) Extracts by Response Surface Methodology
Abstract
1. Introduction
2. Results and Discussion
2.1. Fitting of the RSM Models
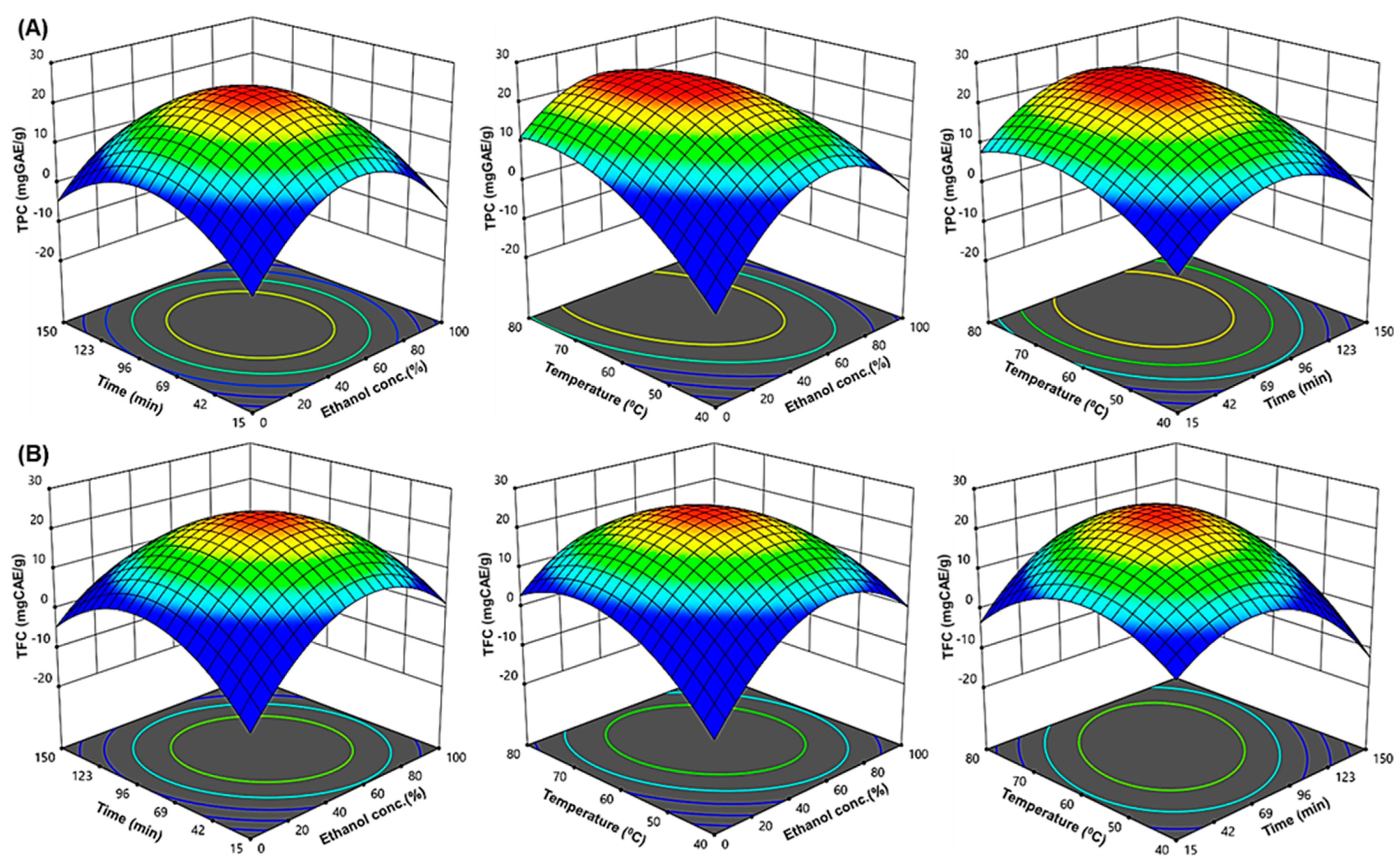
2.2. Effect of Extraction Parameters on TPC and TFC
2.3. Model Validation
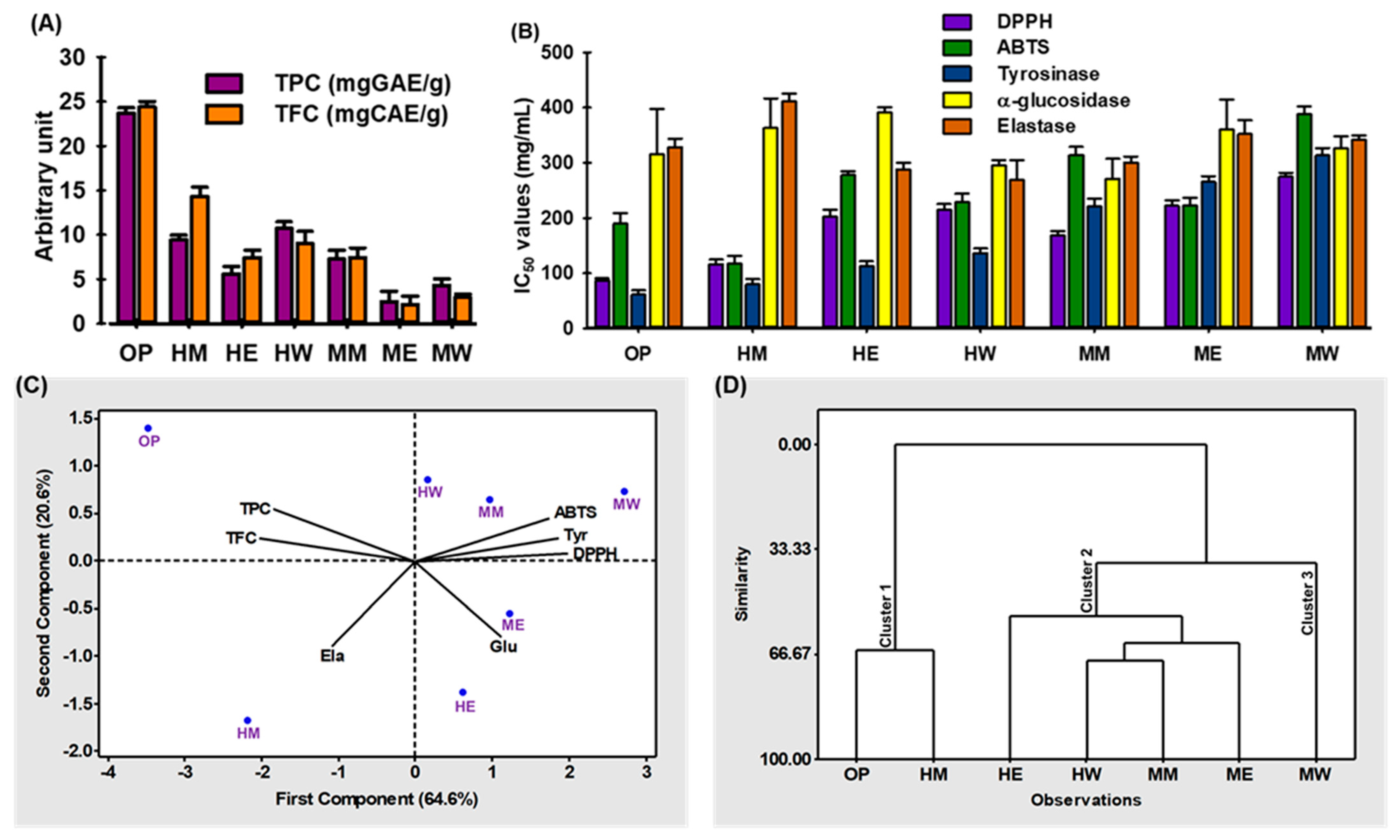
2.4. Comparison of Optimized Extraction Condition with Other Extraction Methods Using Different Solvents
2.5. Chemometric Analysis
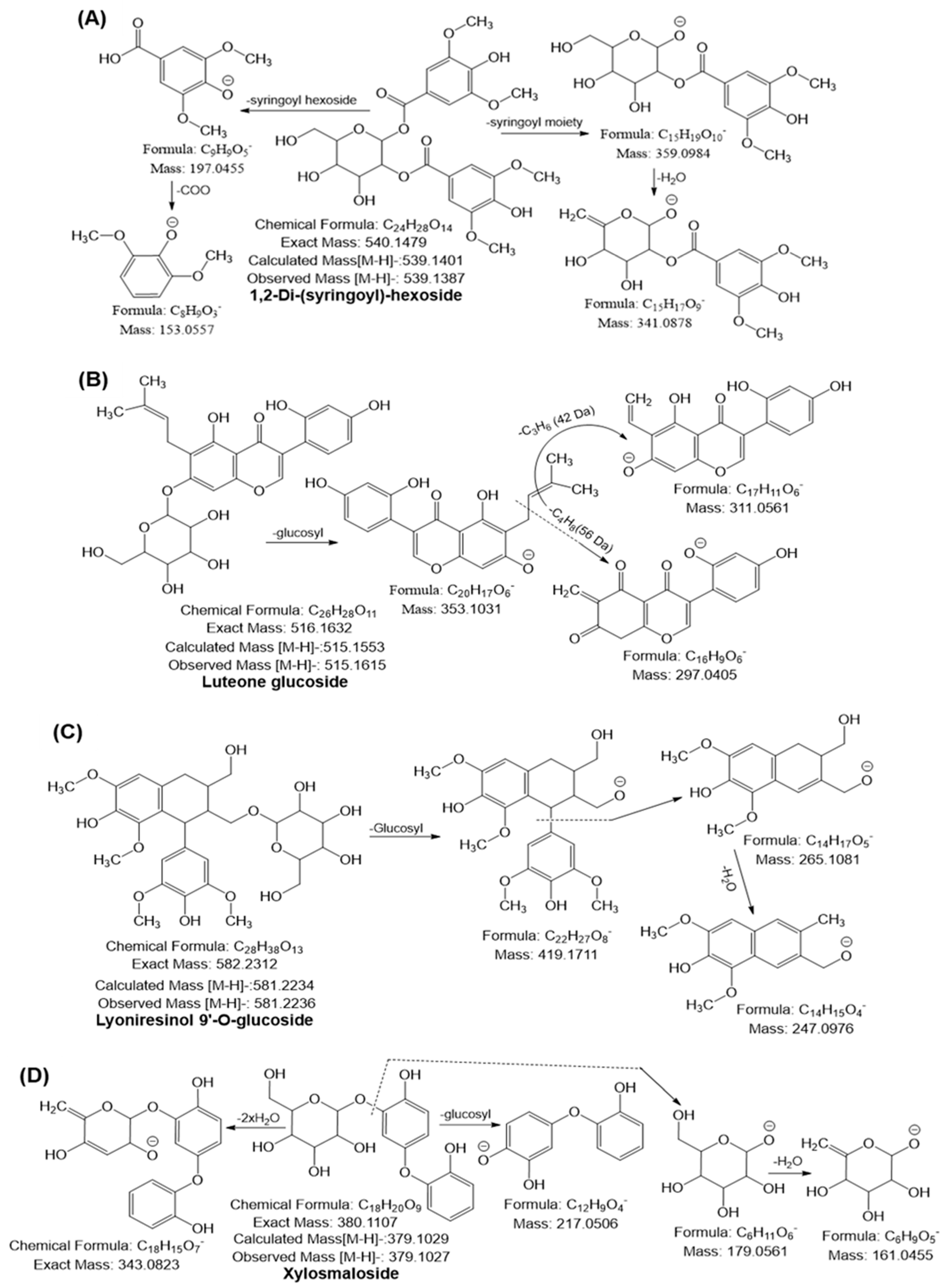
2.6. Secondary Metabolites Profiling in URADP by High-Resolution Mass Spectrometry
2.6.1. Phenolic Acids
2.6.2. Flavonoids
2.6.3. Sugar Molecules
2.6.4. Carboxylic Acids and Fatty Acids
3. Materials and Methods
3.1. Sample Collection and Preparation
3.2. Extraction Methods
3.3. Total Phenolic Content (TPC) and Total Flavonoid Content (TFC)
3.4. Antioxidant Assay and Enzyme Inhibitory Effects
3.5. Experimental Design of RSM for the Extraction Process
3.6. Optimal Extraction Condition and Validation of the Model
3.7. Analysis of Chemical Compounds by ESI-MS/MS
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simić, V.M.; Rajković, K.M.; Stojičević, S.S.; Veličković, D.T.; Nikolić, N.Č.; Lazić, M.L.; Karabegović, I.T. Optimization of microwave-assisted extraction of total polyphenolic compounds from chokeberries by response surface methodology and artificial neural network. Sep. Purif. Technol. 2016, 160, 89–97. [Google Scholar] [CrossRef]
- Bochi, V.C.; Barcia, M.T.; Rodrigues, D.; Speroni, C.S.; Giusti, M.M.; Godoy, H.T. Polyphenol extraction optimisation from Ceylon gooseberry (Dovyalis hebecarpa) pulp. Food Chem. 2014, 164, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Samaram, S.; Mirhosseini, H.; Tan, C.P.; Ghazali, H.M.; Bordbar, S.; Serjouie, A. Optimisation of ultrasound-assisted extraction of oil from papaya seed by response surface methodology: Oil recovery, radical scavenging antioxidant activity, and oxidation stability. Food Chem. 2015, 172, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sedraoui, S.; Badr, A.; Barba, M.G.M.; Doyen, A.; Tabka, Z.; Desjardins, Y. Optimization of the Ultrahigh-Pressure–Assisted Extraction of Phenolic Compounds and Antioxidant Activity from Palm Dates (Phoenix dactylifera L.). Food Anal. Methods 2020, 13, 1556–1569. [Google Scholar] [CrossRef]
- Choi, H.-J.; Naznin, M.; Alam, M.B.; Javed, A.; Alshammari, F.H.; Kim, S.; Lee, S.-H. Optimization of the extraction conditions of Nypa fruticans Wurmb. using response surface methodology and artificial neural network. Food Chem. 2022, 381, 132086. [Google Scholar] [CrossRef]
- Javed, A.; Naznin, M.; Alam, M.B.; Fanar, A.; Song, B.-R.; Kim, S.; Lee, S.-H. Metabolite Profiling of Microwave-Assisted Sargassum fusiforme Extracts with Improved Antioxidant Activity Using Hybrid Response Surface Methodology and Artificial Neural Networking-Genetic Algorithm. Antioxidants 2022, 11, 2246. [Google Scholar] [CrossRef]
- Hou, M.; Hu, W.; Wang, A.; Xiu, Z.; Shi, Y.; Hao, K.; Sun, X.; Cao, D.; Lu, R.; Sun, J. Ultrasound-Assisted Extraction of Total Flavonoids from Pteris cretica L.: Process Optimization, HPLC Analysis, and Evaluation of Antioxidant Activity. Antioxidants 2019, 8, 425. [Google Scholar] [CrossRef]
- Alam, M.B.; Ahmed, A.; Motin, M.A.; Kim, S.; Lee, S.H. Attenuation of melanogenesis by Nymphaea nouchali (Burm. f) flower extract through the regulation of cAMP/CREB/MAPKs/MITF and proteasomal degradation of tyrosinase. Sci. Rep. 2018, 8, 13928. [Google Scholar] [CrossRef]
- Kim, J.H.; Jang, D.H.; Lee, K.W.; Kim, K.D.; Shah, A.B.; Zhumanova, K.; Park, K.H. Tyrosinase Inhibition and Kinetic Details of Puerol A Having But-2-Enolide Structure from Amorpha fruticosa. Molecules 2020, 25. [Google Scholar] [CrossRef]
- Kim, D.S.; Cha, S.B.; Park, M.C.; Park, S.A.; Kim, H.S.; Woo, W.H.; Mun, Y.J. Scopoletin Stimulates Melanogenesis via cAMP/PKA Pathway and Partially p38 Activation. Biol. Pharm. Bull. 2017, 40, 2068–2074. [Google Scholar] [CrossRef]
- Shah, A.B.; Yoon, S.; Kim, J.H.; Zhumanova, K.; Ban, Y.J.; Lee, K.W.; Park, K.H. Effectiveness of cyclohexyl functionality in ugonins from Helminthostachys zeylanica to PTP1B and α-glucosidase inhibitions. Int. J. Biol. Macromol. 2020, 165, 1822–1831. [Google Scholar] [CrossRef]
- Jakimiuk, K.; Gesek, J.; Atanasov, A.G.; Tomczyk, M. Flavonoids as inhibitors of human neutrophil elastase. J. Enzym. Inhib. Med. Chem. 2021, 36, 1016–1028. [Google Scholar] [CrossRef]
- Alam, S.R.; Newby, D.E.; Henriksen, P.A. Role of the endogenous elastase inhibitor, elafin, in cardiovascular injury: From epithelium to endothelium. Biochem. Pharm. 2012, 83, 695–704. [Google Scholar] [CrossRef]
- Ban, Y.J.; Baiseitova, A.; Nafiah, M.A.; Kim, J.Y.; Park, K.H. Human neutrophil elastase inhibitory dihydrobenzoxanthones and alkylated flavones from the Artocarpus elasticus root barks. Appl. Biol. Chem. 2020, 63, 63. [Google Scholar] [CrossRef]
- von Nussbaum, F.; Li, V.M. Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases: Into clinical testing with pre-adaptive pharmacophores. Bioorg. Med. Chem. Lett. 2015, 25, 4370–4381. [Google Scholar] [CrossRef]
- Delgado-Rizo, V.; Martínez-Guzmán, M.A.; Iñiguez-Gutierrez, L.; García-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef]
- Yang, H.; Biermann, M.H.; Brauner, J.M.; Liu, Y.; Zhao, Y.; Herrmann, M. New Insights into Neutrophil Extracellular Traps: Mechanisms of Formation and Role in Inflammation. Front. Immunol. 2016, 7, 302. [Google Scholar] [CrossRef]
- Tabaraki, R.; Nateghi, A. Optimization of ultrasonic-assisted extraction of natural antioxidants from rice bran using response surface methodology. Ultrason. Sonochem. 2011, 18, 1279–1286. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef]
- Yasin, B.R.; El-Fawal, H.A.; Mousa, S.A. Date (Phoenix dactylifera) Polyphenolics and Other Bioactive Compounds: A Traditional Islamic Remedy’s Potential in Prevention of Cell Damage, Cancer Therapeutics and Beyond. Int. J. Mol. Sci. 2015, 16, 30075–30090. [Google Scholar] [CrossRef]
- Al-Yahya, M.; Raish, M.; AlSaid, M.S.; Ahmad, A.; Mothana, R.A.; Al-Sohaibani, M.; Al-Dosari, M.S.; Parvez, M.K.; Rafatullah, S. ‘Ajwa’ dates (Phoenix dactylifera L.) extract ameliorates isoproterenol-induced cardiomyopathy through downregulation of oxidative, inflammatory and apoptotic molecules in rodent model. Phytomed. Int. J. Phytother. Phytopharm. 2016, 23, 1240–1248. [Google Scholar] [CrossRef] [PubMed]
- Almatroodi, S.A.; Khan, A.A.; Aloliqi, A.A.; Ali Syed, M.; Rahmani, A.H. Therapeutic Potential of Ajwa Dates (Phoenix dactylifera) Extract in Prevention of Benzo(a)pyrene-Induced Lung Injury through the Modulation of Oxidative Stress, Inflammation, and Cell Signalling Molecules. Appl. Sci. 2022, 12, 6784. [Google Scholar] [CrossRef]
- Abdelghffar, E.A.; Obaid, W.A.; Mohammedsaleh, Z.M.; Ouchari, W.; Eldahshan, O.A.; Sobeh, M. Ajwa dates (Phoenix dactylifera L.) attenuate cisplatin-induced nephrotoxicity in rats via augmenting Nrf2, modulating NADPH oxidase-4 and mitigating inflammatory/apoptotic mediators. Biomed. Pharmacother. Biomed. Pharmacother. 2022, 156, 113836. [Google Scholar] [CrossRef] [PubMed]
- Al-Shahib, W.; Marshall, R.J. The fruit of the date palm: Its possible use as the best food for the future? Int. J. Food Sci. Nutr. 2003, 54, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, A.H.; Aly, S.M.; Ali, H.; Babiker, A.Y.; Srikar, S.; Khan, A.A. Therapeutic effects of date fruits (Phoenix dactylifera) in the prevention of diseases via modulation of anti-inflammatory, anti-oxidant and anti-tumour activity. Int. J. Clin. Exp. Med. 2014, 7, 483–491. [Google Scholar]
- Boulenouar, N.; Marouf, A.; Cheriti, A. Antifungal activity and phytochemical screening of extracts from Phoenix dactylifera L. cultivars. Nat. Prod. Res. 2011, 25, 1999–2002. [Google Scholar] [CrossRef]
- Almusallam, I.A.; Ahmed, I.A.M.; Babiker, E.E.; Al Juhaimi, F.Y.; Fadimu, G.J.; Osman, M.A.; Al Maiman, S.A.; Ghafoor, K.; Alqah, H.A. Optimization of ultrasound-assisted extraction of bioactive properties from date palm (Phoenix dactylifera L.) spikelets using response surface methodology. LWT 2021, 140, 110816. [Google Scholar] [CrossRef]
- Ghafoor, K.; Sarker, M.Z.I.; Al-Juhaimi, F.Y.; Babiker, E.E.; Alkaltham, M.S.; Almubarak, A.K. Extraction and Evaluation of Bioactive Compounds from Date (Phoenix dactylifera) Seed Using Supercritical and Subcritical CO2 Techniques. Foods 2022, 11, 1806. [Google Scholar] [CrossRef]
- Pourshoaib, S.J.; Ghatrami, E.R.; Shamekhi, M.A. Comparing ultrasonic-and microwave-assisted methods for extraction of phenolic compounds from Kabkab date seed (Phoenix dactylifera L.) and stepwise regression analysis of extracts antioxidant activity. Sustain. Chem. Pharm. 2022, 30, 100871. [Google Scholar] [CrossRef]
- Al-Turki, S.; Shahba, M.A.; Stushnoff, C. Diversity of antioxidant properties and phenolic content of date palm (Phoenix dactylifera L.) fruits as affected by cultivar and location. J. Food Agric. Environ. 2010, 8, 253–260. [Google Scholar]
- Mohamed, S.A.; Awad, M.A.; El-Dengawy, E.-R.F.A.; Abdel-Mageed, H.M.; El-Badry, M.O.; Salah, H.A.; Abdel-Aty, A.M.; Fahmy, A.S. Total phenolic and flavonoid contents and antioxidant activities of sixteen commercial date cultivars grown in Saudi Arabia. RSC Adv. 2016, 6, 44814–44819. [Google Scholar] [CrossRef]
- Hamad, I.; AbdElgawad, H.; Al Jaouni, S.; Zinta, G.; Asard, H.; Hassan, S.; Hegab, M.; Hagagy, N.; Selim, S. Metabolic Analysis of Various Date Palm Fruit (Phoenix dactylifera L.) Cultivars from Saudi Arabia to Assess Their Nutritional Quality. Molecules 2015, 20, 13620–13641. [Google Scholar] [CrossRef]
- Eid, N.M.S.; Al-Awadi, B.; Vauzour, D.; Oruna-Concha, M.J.; Spencer, J.P.E. Effect of Cultivar Type and Ripening on the Polyphenol Content of Date Palm Fruit. J. Agric. Food Chem. 2013, 61, 2453–2460. [Google Scholar] [CrossRef]
- Hassan, S.M.A.; Aboonq, M.S.; Albadawi, E.A.; Aljehani, Y.; Abdel-Latif, H.M.; Mariah, R.A.; Shafik, N.M.; Soliman, T.M.; Abdel-Gawad, A.R.; Omran, F.M.; et al. The Preventive and Therapeutic Effects of Ajwa Date Fruit Extract Against Acute Diclofenac Toxicity-Induced Colopathy: An Experimental Study. Drug Des. Devel. 2022, 16, 2601–2616. [Google Scholar] [CrossRef]
- Xi, J.; Wang, B. Optimization of Ultrahigh-Pressure Extraction of Polyphenolic Antioxidants from Green Tea by Response Surface Methodology. Food Bioprocess Technol. 2013, 6, 2538–2546. [Google Scholar] [CrossRef]
- Gurtovenko, A.A.; Anwar, J. Interaction of Ethanol with Biological Membranes: The Formation of Non-bilayer Structures within the Membrane Interior and their Significance. J. Phys. Chem. B 2009, 113, 1983–1992. [Google Scholar] [CrossRef]
- Derrien, M.; Badr, A.; Gosselin, A.; Desjardins, Y.; Angers, P. Optimization of a green process for the extraction of lutein and chlorophyll from spinach by-products using response surface methodology (RSM). LWT-Food Sci. Technol. 2017, 79, 170–177. [Google Scholar] [CrossRef]
- Xu, S.; Li, X.; Liu, S.; Tian, P.; Li, D. Juniperus sabina L. as a Source of Podophyllotoxins: Extraction Optimization and Anticholinesterase Activities. Int. J. Mol. Sci. 2022, 23, 10205. [Google Scholar] [CrossRef]
- Alam, M.B.; Ju, M.K.; Lee, S.H. DNA Protecting Activities of Nymphaea nouchali (Burm. f) Flower Extract Attenuate t-BHP-Induced Oxidative Stress Cell Death through Nrf2-Mediated Induction of Heme Oxygenase-1 Expression by Activating MAP-Kinases. Int. J. Mol. Sci. 2017, 18, 2069. [Google Scholar] [CrossRef]
- Rebollo-Hernanz, M.; Cañas, S.; Taladrid, D.; Segovia, Á.; Bartolomé, B.; Aguilera, Y.; Martín-Cabrejas, M.A. Extraction of phenolic compounds from cocoa shell: Modeling using response surface methodology and artificial neural networks. Sep. Purif. Technol. 2021, 270, 118779. [Google Scholar] [CrossRef]
- Sunarwidhi, A.L.; Hernawan, A.; Frediansyah, A.; Widyastuti, S.; Martyasari, N.W.R.; Abidin, A.S.; Padmi, H.; Handayani, E.; Utami, N.W.P.; Maulana, F.A.; et al. Multivariate Analysis Revealed Ultrasonic-Assisted Extraction Improves Anti-Melanoma Activity of Non-Flavonoid Compounds in Indonesian Brown Algae Ethanol Extract. Molecules 2022, 27, 7509. [Google Scholar] [CrossRef] [PubMed]
- Naznin, M.; Badrul Alam, M.; Alam, R.; Islam, S.; Rakhmat, S.; Lee, S.-H.; Kim, S. Metabolite profiling of Nymphaea rubra (Burm. f.) flower extracts using cyclic ion mobility–mass spectrometry and their associated biological activities. Food Chem. 2023, 404, 134544. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.B.; Naznin, M.; Islam, S.; Alshammari, F.H.; Choi, H.J.; Song, B.R.; Kim, S.; Lee, S.H. High Resolution Mass Spectroscopy-Based Secondary Metabolite Profiling of Nymphaea nouchali (Burm. f) Stem Attenuates Oxidative Stress via Regulation of MAPK/Nrf2/HO-1/ROS Pathway. Antioxidants 2021, 10, 719. [Google Scholar] [CrossRef] [PubMed]
- Najm, O.A.; Addnan, F.H.; Mohd-Manzor, N.F.; Elkadi, M.A.; Abdullah, W.O.; Ismail, A.; Mansur, F.A.F. Identification of Phytochemicals of Phoenix dactylifera L. Cv Ajwa with UHPLC-ESI-QTOF-MS/MS. Int. J. Fruit Sci. 2021, 21, 848–867. [Google Scholar] [CrossRef]
- Nematallah, K.A.; Ayoub, N.A.; Abdelsattar, E.; Meselhy, M.R.; Elmazar, M.M.; El-Khatib, A.H.; Linscheid, M.W.; Hathout, R.M.; Godugu, K.; Adel, A.; et al. Polyphenols LC-MS2 profile of Ajwa date fruit (Phoenix dactylifera L.) and their microemulsion: Potential impact on hepatic fibrosis. J. Funct. Foods 2018, 49, 401–411. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural Characterization of Flavonoid Glycoconjugates and Their Derivatives with Mass Spectrometric Techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef]
- Vukics, V.; Guttman, A. Structural characterization of flavonoid glycosides by multi-stage mass spectrometry. Mass Spectrom. Rev. 2010, 29, 1–16. [Google Scholar] [CrossRef]
- Wang, F.; Liigand, J.; Tian, S.; Arndt, D.; Greiner, R.; Wishart, D.S. CFM-ID 4.0: More Accurate ESI-MS/MS Spectral Prediction and Compound Identification. Anal. Chem. 2021, 93, 11692–11700. [Google Scholar] [CrossRef]
- Naveja, J.J.; Rico-Hidalgo, M.P.; Medina-Franco, J.L. Analysis of a large food chemical database: Chemical space, diversity, and complexity. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Guijas, C.; Billings, E.; Montenegro-Burke, J.R.; Uritboonthai, W.; Aisporna, A.E.; Chen, E.; Benton, H.P.; Siuzdak, G. The METLIN small molecule dataset for machine learning-based retention time prediction. Nat. Commun. 2019, 10, 5811. [Google Scholar] [CrossRef]
- Mollica, A.; Zengin, G.; Sinan, K.I.; Marletta, M.; Pieretti, S.; Stefanucci, A.; Etienne, O.K.; Jekő, J.; Cziáky, Z.; Bahadori, M.B.; et al. A Study on Chemical Characterization and Biological Abilities of Alstonia boonei Extracts Obtained by Different Techniques. Antioxidants 2022, 11, 2171. [Google Scholar] [CrossRef]
- Alam, M.B.; Bajpai, V.K.; Ra, J.S.; Lim, J.Y.; An, H.; Shukla, S.; Quan, K.T.; Khan, I.; Huh, Y.S.; Han, Y.K.; et al. Anthraquinone-type inhibitor of α-glucosidase enhances glucose uptake by activating an insulin-like signaling pathway in C2C12 myotubes. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 129, 337–343. [Google Scholar] [CrossRef]
- Zhao, P.; Alam, M.B.; Lee, S.-H. Protection of UVB-Induced Photoaging by Fuzhuan-Brick Tea Aqueous Extract via MAPKs/Nrf2-Mediated Down-Regulation of MMP-1. Nutrients 2019, 11, 60. [Google Scholar] [CrossRef]

| Run | Independent Variables | Responses | |||||
|---|---|---|---|---|---|---|---|
| (X1) | (X2) | (X3) | TPC (Y1) | TFC (Y2) | |||
| Exp. | Pred. | Exp. | Pred. | ||||
| 1 | 100 | 82.5 | 60 | 5.41 ± 0.28 | 5.70 | 11.02 ± 0.33 | 9.90 |
| 2 | 50 | 82.5 | 60 | 23.69 ± 0.43 | 23.34 | 21.05 ± 0.62 | 23.10 |
| 3 | 75 | 120 | 70 | 13.75 ± 0.54 | 14.17 | 12.9 ± 0.15 | 13.44 |
| 4 | 50 | 15 | 60 | 10.24 ± 0.76 | 9.75 | 11.52 ± 0.25 | 10.24 |
| 5 | 75 | 120 | 50 | 10.21 ± 0.61 | 10.60 | 7.83 ± 0.39 | 9.31 |
| 6 | 50 | 82.5 | 60 | 23.12 ± 0.12 | 23.34 | 24.20 ± 0.20 | 23.10 |
| 7 | 0 | 82.5 | 60 | 6.53 ± 0.12 | 6.73 | 6.85 ± 0.16 | 6.04 |
| 8 | 25 | 45 | 70 | 17.50 ± 0.69 | 17.62 | 11.52 ± 0.46 | 11.97 |
| 9 | 25 | 120 | 50 | 8.88 ± 0.45 | 8.99 | 6.81 ± 0.35 | 7.39 |
| 10 | 50 | 82.5 | 80 | 23.00 ± 0.43 | 22.89 | 16.05 ± 0.75 | 15.28 |
| 11 | 50 | 82.5 | 60 | 23.10 ± 0.72 | 23.34 | 23.59 ± 0.36 | 23.10 |
| 12 | 50 | 82.5 | 60 | 23.51 ± 0.16 | 23.34 | 24.01 ± 0.43 | 23.10 |
| 13 | 50 | 82.5 | 60 | 23.92 ± 0.54 | 23.34 | 24.01 ± 0.63 | 23.10 |
| 14 | 25 | 45 | 50 | 7.85 ± 0.72 | 7.94 | 7.85 ± 0.55 | 9.25 |
| 15 | 50 | 150 | 60 | 10.11 ± 0.46 | 9.97 | 9.25 ± 0.25 | 8.14 |
| 16 | 75 | 45 | 50 | 11.01 ± 0.04 | 11.74 | 15.25 ± 0.80 | 16.05 |
| 17 | 25 | 120 | 70 | 19.22 ± 0.58 | 19.00 | 15.25 ± 0.92 | 16.38 |
| 18 | 50 | 82.5 | 40 | 10.05 ± 0.18 | 9.64 | 9.59 ± 0.22 | 8.43 |
| 19 | 50 | 82.5 | 60 | 23.10 ± 0.53 | 23.34 | 23.25 ± 0.59 | 23.10 |
| 20 | 75 | 45 | 70 | 14.58 ± 0.54 | 14.98 | 12.56 ± 0.27 | 13.92 |
| ANOVA for Quadratic Model for TPC | |||||||
|---|---|---|---|---|---|---|---|
| Source | RC | SS | DF | MS | F Value | p Value | |
| Model | 843.91 | 9 | 93.77 | 347.63 | <0.0001 | Significant | |
| Intercept | 23.39 | ||||||
| Linear terms | |||||||
| X1 | −0.3838 | 2.36 | 1 | 2.36 | 8.74 | 0.0144 | Significant |
| X2 | 0.0612 | 0.0542 | 1 | 0.0542 | 0.2010 | 0.6635 | Nonsignificant |
| X3 | 3.06 | 150.31 | 1 | 150.31 | 557.25 | <0.0001 | Significant |
| Interaction terms | |||||||
| X1X2 | −0.5475 | 2.40 | 1 | 2.40 | 8.89 | 0.0138 | Significant |
| X1X3 | −1.61 | 20.74 | 1 | 20.74 | 76.88 | <0.0001 | Significant |
| X2X3 | 0.0825 | 0.0544 | 1 | 0.0544 | 0.2019 | 0.6628 | Nonsignificant |
| Quadratic terms | |||||||
| X12 | −4.37 | 484.62 | 1 | 484.62 | 1796.67 | <0.0001 | Significant |
| X22 | −4.10 | 292.30 | 1 | 292.30 | 1083.65 | <0.0001 | Significant |
| X32 | −1.98 | 99.41 | 1 | 99.41 | 368.54 | <0.0001 | Significant |
| Lack of Fit | 2.07 | 5 | 0.4145 | 3.32 | 0.1071 | Nonsignificant | |
| Pure error | 0.6247 | 5 | 0.1249 | ||||
| R2 | 0.9968 | ||||||
| Adjusted R2 | 0.9939 | ||||||
| Adeq Precision | 49.6969 | ||||||
| C.V.% | 3.39 | ||||||
| ANOVA for quadratic model for TFC | |||||||
| Model | 751.10 | 9 | 83.46 | 36.64 | <0.0001 | Significant | |
| Intercept | 23.10 | ||||||
| Linear terms | |||||||
| X1 | 0.9656 | 14.92 | 1 | 14.92 | 6.55 | 0.0284 | Significant |
| X2 | −0.5854 | 4.96 | 1 | 4.96 | 2.18 | 0.1708 | Nonsignificant |
| X3 | 1.71 | 46.96 | 1 | 46.96 | 20.61 | 0.0011 | Significant |
| Interaction terms | |||||||
| X1X2 | −1.22 | 11.93 | 1 | 11.93 | 5.24 | 0.0451 | Significant |
| X1X3 | −1.22 | 11.83 | 1 | 11.83 | 5.20 | 0.0458 | Significant |
| X2X3 | 1.57 | 19.63 | 1 | 19.63 | 8.62 | 0.0149 | Significant |
| Quadratic terms | |||||||
| X12 | −3.78 | 363.27 | 1 | 363.27 | 159.47 | <0.0001 | Significant |
| X22 | −4.29 | 320.24 | 1 | 320.24 | 140.58 | <0.0001 | Significant |
| X32 | −2.81 | 200.73 | 1 | 200.73 | 88.12 | <0.0001 | Significant |
| Lack of Fit | 15.83 | 5 | 3.17 | 2.28 | 0.1938 | Nonsignificant | |
| Pure error | 6.95 | 5 | 1.39 | ||||
| R2 | 0.9706 | ||||||
| Adjusted R2 | 0.9441 | ||||||
| Adeq Precision | 15.9930 | ||||||
| C.V.% | 10.25 | ||||||
| Response | Exp. | Pred. | Std | RSD (%) |
|---|---|---|---|---|
| TPC (mgGAE/g) | 24.25 ± 1.02 | 23.97 | 0.20 | 0.82 |
| TFC (mgCAE/g) | 23.98 ± 0.65 | 23.39 | 0.42 | 1.76 |
| Group | No. | Compound Name | EF | OM (m/z)– | CM (m/z)– | MS/MS (Negative Mode) | CE | CL |
|---|---|---|---|---|---|---|---|---|
| Phenolic acids and derivatives | 1 | 4-Hydroxybenzoyl glucose | C13H16O8 | 299.0773 | 299.0766 | 137.02, 163.02 | 20 | 2 |
| 2 | Coumaroylshikimic acid | C16H16O7 | 319.0824 | 319.0817 | 173.04, 163.03, 145.02 | 20 | 2 | |
| 3 | Vanillic acid glucoside | C14H18O9 | 329.0873 | 329.0872 | 167.03, 152.02, 123.04 | 20 | 2 | |
| 4 | Caffeoylshikimic acid | C16H16O8 | 335.0776 | 335.0772 | 179.01, 161.03, 155.03, 137.05 | 20 | 2 | |
| 5 | Quinic acid hexoside | C13H22O11 | 353.1085 | 353.1084 | 191.05, 173.04, 179.05 | 20 | 2 | |
| 6 | 5-Feruloylquinic acid | C17H20O9 | 367.1046 | 367.1029 | 191.08, 173.04, 127.01 | 30 | 2 | |
| 5 | Caffeic acid derivatives | C18H18O9 | 377.0885 | 377.0878 | 341.10, 215.03, 179.06, 161.04, 135.05 | 10 | 2 | |
| 6 | Sinapic acid hexoside | C17H22O10 | 385.1141 | 385.1135 | 223.06, 205.05 | 10 | 2 | |
| 7 | Caffeoyl shikimic acid hexoside | C22H26O13 | 497.1297 | 497.1295 | 335.01, 178.02, 161.03, 155.03, 135.02 | 20 | 2 | |
| 8 | Quinic acid derivatives | C19H34O17 | 533.1718 | 533.1718 | 341.10, 191.05 | 30 | 2 | |
| 9 | 1,2-di-(syringoyl)-hexoside # | C24H28O14 | 539.1377 | 539.1401 | 359.09, 341.08, 197.04, 153.05 | 30 | 3 | |
| Flavonoids and derivatives | 10 | Luteolin | C15H10O6 | 285.0405 | 285.0399 | 267.05, 241.03, 151.00, 133.02 | 20 | 2 |
| 11 | Catechin/Epicatechin | C15H14O6 | 289.0718 | 289.0712 | 245.04, 205.05, 179, 151.04, 137.02 | 20 | 2 | |
| 12 | Chrysoeriol | C13H16O8 | 299.0561 | 299.0555 | 285.03, 153.01, 135.03, 125.03 | 20 | 2 | |
| 13 | Quercetin | C15H10O7 | 301.0354 | 301.0348 | 273.02, 229.05, 179.01, 151.01 | 20 | 2 | |
| 14 | Epigallocatechin | C15H14O7 | 305.0644 | 305.0661 | 287.05, 137.02, 125.02 | 20 | 2 | |
| 15 | Methoxysinensetin # | C21H22O8 | 401.1299 | 401.1236 | 371.11, 339.08, 191.71 | 20 | 2 | |
| 16 | Epicatechin hydroxybenzoate # | C22H18O8 | 409.0924 | 409.0923 | 289.07, 271.06, 137.02, 119.01 | 30 | 2 | |
| 17 | Naringenin rhamnoside | C21H22O9 | 417.1245 | 417.1186 | 271.06, 187.03, 151.00, 119.05 | 20 | 2 | |
| 18 | Epicatechin-3-gallate | C22H18O10 | 441.081 | 441.0821 | 371.04, 273.02, 135.10, 169.02 | 30 | 2 | |
| 19 | Biochanin A 7-glucoside # | C22H22O10 | 445.1195 | 445.1135 | 283.06, 239.03, 211.04, 132.02 | 30 | 2 | |
| 20 | Epicatechin 3-(-methylgallate) # | C23H20O10 | 455.1015 | 455.0978 | 289.02, 183.05, 124.01 | 30 | 2 | |
| 21 | Afrormosin 7-glucoside | C23H24O10 | 459.1354 | 459.1291 | 297.07, 281.04, 267.06 | 20 | 2 | |
| 22 | Chrysoeriol hexoside | C22H22O11 | 461.1085 | 461.1083 | 299.07, 283.02, 269.06 | 20 | 2 | |
| 23 | Isoquercitrin | C21H20O12 | 463.0878 | 463.0876 | 301.05, 268.01, 179.02, 151.01 | 20 | 2 | |
| 24 | Epicatechin 4’-glucuronide# | C21H22O12 | 465.1036 | 465.1033 | 289.15, 151.10, 137.08, 123.10 | 20 | 2 | |
| 25 | Isorhamnetin hexoside | C22H22O12 | 477.1035 | 477.1033 | 315.05, 300.01, 179.05, 151.02 | 20 | 2 | |
| 26 | Luteone glucoside | C26H28O11 | 515.1611 | 515.1553 | 353.10, 311.05, 297.04 | 20 | 3 | |
| 27 | Luteolin hexosyl sulfate | C21H20O14S | 527.0491 | 527.0495 | 447.05, 285.01, 241.06 | 20 | 2 | |
| 28 | Chrysoeriol hexosyl sulfate | C22H22O14S | 541.0645 | 541.0652 | 299.05, 284.05, 241.02 | 20 | 2 | |
| 29 | Isoquercitrin sulfate | C21H20O15S | 543.0441 | 543.0444 | 463.05, 301.01, 179.02, 151.01 | 20 | 2 | |
| 30 | Procyanidin B2 # | C30H26O12 | 577.1347 | 577.1346 | 451.10, 407.07, 289.07, 287.05, 125.02 | 20 | 2 | |
| 31 | Lyoniresinol 9-glucoside # | C28H37O13 | 581.2236 | 581.2234 | 419.17, 265.10, 247.09 | 20 | 2 | |
| 32 | Luteolin rhamnosyl hexoside | C27H30O15 | 593.1507 | 593.1506 | 447.09, 285.03, 153.01, 135.04 | 20 | 2 | |
| 33 | Chrysoeriol rhamnosyl hexoside | C28H32O15 | 607.1669 | 607.1663 | 461.10, 299.05, 153.01, 149.05 | 20 | 2 | |
| 34 | Isorhamnetin rhamnosyl hexoside | C28H32O16 | 623.1617 | 623.1612 | 477.10, 315.05, 299.05, 165.05 | 20 | 2 | |
| 35 | Isorhamnetin diglucoside | C28H32O17 | 639.1563 | 639.1561 | 447.01, 315.01 | 20 | 2 | |
| 36 | Quercetin xylosyl rutinoside # | C32H38O20 | 741.1846 | 741.1878 | 609.14, 301.03 | 10 | 2 | |
| 37 | Luteolin rhamnosyl dihexoside | C33H40O20 | 755.2046 | 755.2034 | 709.16, 593.10, 575.05, 285.01 | 20 | 2 | |
| 38 | Quercetin glucosyl-rutinoside | C33H40O21 | 771.1981 | 771.1983 | 609.14, 591.05, 301.03, 153.02, 125.00 | 20 | 2 | |
| 39 | Isorhamnetin rhamnosyl dihexoside | C34H42O21 | 785.211 | 785.214 | 623.16, 477.10, 315.05 | 20 | 2 | |
| 40 | Epicatechin-(2α→7,4α→8)-epicatechin glucoside # | C36H34O17 | 737.1721 | 737.1718 | 721.02, 577.05, 425.05, 195.02 | 30 | 2 | |
| Sugar molecules | 41 | Ribonic acid | C5H10O6 | 165.0421 | 165.0418 | 149.04, 105.01, 87.00, 75.00 | 10 | 2 |
| 42 | L-Galactose | C6H12O6 | 179.0572 | 179.0561 | 161.04, 143.03, 113.02, 101.02, | 10 | 2 | |
| 43 | Gluconic acid | C6H12O7 | 195.0522 | 195.0504 | 177.05, 159.02, 129.05, 98.90 | 10 | 2 | |
| 48 | Sedoheptulose | C7H14O7 | 209.0679 | 209.068 | 191.05, 179.05, 149.04, | 20 | 2 | |
| 49 | Xylosmaloside # | C18H20O9 | 379.1027 | 379.1029 | 343.08, 217.05, 179.05, 161.04 | 20 | 3 | |
| Carboxylic acids | 50 | Fumaric acid | C4H4O4 | 115.005 | 115.0037 | 71.01 | 10 | 2 |
| 51 | Glutaconic acid | C5H6O4 | 129.0203 | 129.0203 | 111.00, 85.02 | 10 | 2 | |
| 52 | Glutaric acid | C5H8O4 | 131.0355 | 131.035 | 113.00, 87.02 | 10 | 2 | |
| 53 | 3-Methylglutaconic acid | C6H8O4 | 143.0367 | 143.0361 | 99.03 | 20 | 2 | |
| 54 | Methyl glutaric acid | C6H10O4 | 145.0521 | 145.0506 | 127.02, 101.02 | 10 | 2 | |
| 55 | 2-Hydroxyglutaric acid | C5H8O5 | 147.0301 | 147.0299 | 129.01, 99.03 | 10 | 2 | |
| 56 | Hydroxymethyl glutaric acid | C6H10O5 | 161.0459 | 161.0455 | 143.03, 117.05, 99.04 | 10 | 2 | |
| 58 | Citric acid | C6H8O7 | 191.0197 | 191.0197 | 173.00, 129.01, 111.00 | 20 | 2 | |
| Fatty acids | 59 | Palmitic acid | C16H32O2 | 255.233 | 255.233 | 237.23, 211.24, 197.22 | 20 | 2 |
| 60 | Linolenic acid | C18H30O2 | 277.2165 | 277.2169 | 259.20, 233.22, 205.21, 179.25, | 10 | 2 | |
| 61 | α-Linoleic acid | C18H32O2 | 279.2331 | 279.2330 | 261.22 | 10 | 2 | |
| 62 | Oleic acid | C18H34O2 | 281.2487 | 281.2486 | 263.25, 181.21, 127.25 | 10 | 2 | |
| 63 | Hydroxy octadecatrienoic acid # | C18H30O3 | 293.212 | 293.0216 | 275.22 | 20 | 3 | |
| 64 | Hydroxy octadecadienoic acid | C18H32O3 | 295.2276 | 295.2273 | 277.23 | 20 | 2 | |
| 65 | Hydroxy octadecenoic acid | C18H34O3 | 297.2433 | 297.2429 | 279.23 | 20 | 2 | |
| 66 | Dihydroxy octadecadienoic acid | C18H32O4 | 311.2246 | 311.2239 | 293.22, 275.23 | 20 | 2 | |
| 67 | Dihydroxy octadecenoic acid | C18H34O4 | 313.2381 | 313.2378 | 295.23, 277.25, 183.32 | 20 | 2 | |
| 68 | Dihydroxy octadecanoic acid | C18H36O4 | 315.2538 | 315.2535 | 297.23, 279.25, | 20 | 2 | |
| 69 | Trihydroxy octadecadienoic acid | C18H32O5 | 327.2176 | 327.2171 | 309.23, 291.25, 273.23 | 20 | 2 | |
| 70 | Trihydroxy octadecenoic acid | C18H34O5 | 329.2346 | 329.2333 | 311.25, 293.26, 275.23 | 20 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammari, F.; Alam, M.B.; Song, B.-R.; Lee, S.-H. Antioxidant, Tyrosinase, α-Glucosidase, and Elastase Enzyme Inhibition Activities of Optimized Unripe Ajwa Date Pulp (Phoenix dactylifera) Extracts by Response Surface Methodology. Int. J. Mol. Sci. 2023, 24, 3396. https://doi.org/10.3390/ijms24043396
Alshammari F, Alam MB, Song B-R, Lee S-H. Antioxidant, Tyrosinase, α-Glucosidase, and Elastase Enzyme Inhibition Activities of Optimized Unripe Ajwa Date Pulp (Phoenix dactylifera) Extracts by Response Surface Methodology. International Journal of Molecular Sciences. 2023; 24(4):3396. https://doi.org/10.3390/ijms24043396
Chicago/Turabian StyleAlshammari, Fanar, Md Badrul Alam, Bo-Rim Song, and Sang-Han Lee. 2023. "Antioxidant, Tyrosinase, α-Glucosidase, and Elastase Enzyme Inhibition Activities of Optimized Unripe Ajwa Date Pulp (Phoenix dactylifera) Extracts by Response Surface Methodology" International Journal of Molecular Sciences 24, no. 4: 3396. https://doi.org/10.3390/ijms24043396
APA StyleAlshammari, F., Alam, M. B., Song, B.-R., & Lee, S.-H. (2023). Antioxidant, Tyrosinase, α-Glucosidase, and Elastase Enzyme Inhibition Activities of Optimized Unripe Ajwa Date Pulp (Phoenix dactylifera) Extracts by Response Surface Methodology. International Journal of Molecular Sciences, 24(4), 3396. https://doi.org/10.3390/ijms24043396







