Emerging Roles of Salicylic Acid in Plant Saline Stress Tolerance
Abstract
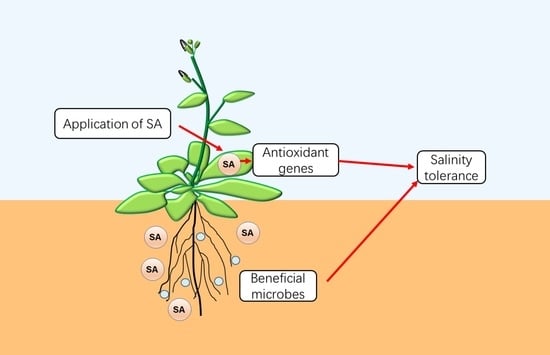
:1. Introduction
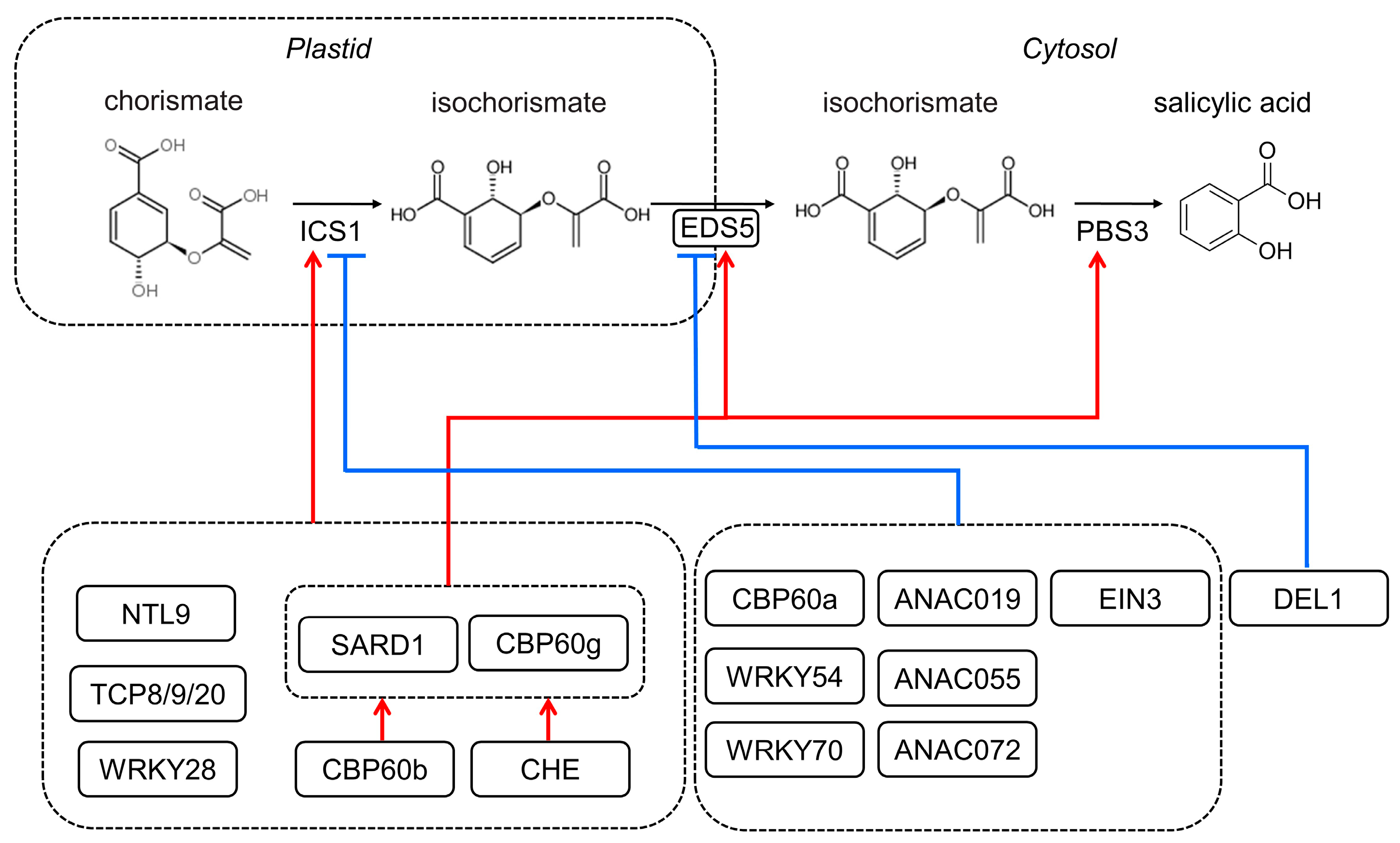
2. SA Biosynthesis in Plants
3. Signal Transduction Pathway of SA
4. Physiological Responses by SA during Saline Stress in Plants
5. Molecular Mechanisms Controlled by SA Regulation in Response to Saline Stress
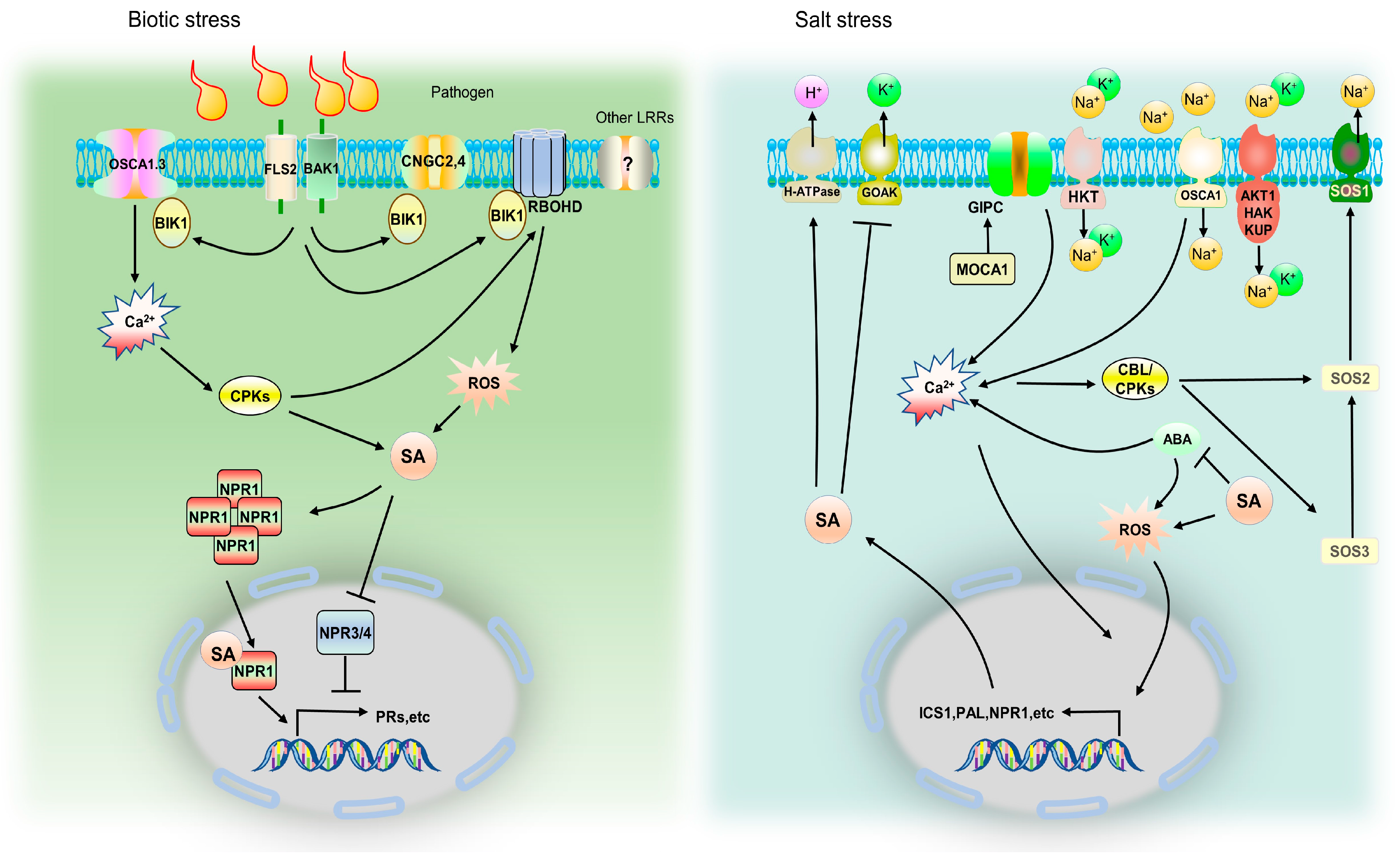
6. Mechanism Comparison of SA in Saline and Biotic Stress
7. Summary and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Fritschi, F.B.; Zandalinas, S.I.; Mittler, R. The impact of stress combination on reproductive processes in crops. Plant Sci. 2021, 311, 111007. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Panta, S.; Flowers, T.; Lane, P.; Doyle, R.; Haros, G.; Shabala, S. Halophyte agriculture: Success stories. Environ. Exp. Bot. 2014, 107, 71–83. [Google Scholar] [CrossRef]
- Verma, K.; Agrawal, S.B. Salicylic acid-mediated defence signalling in respect to its perception, alteration and transduction. In Salicylic Acid: A Multifaceted Hormone; Springer: Singapore, 2017; pp. 97–122. [Google Scholar]
- Pokotylo, I.; Hodges, M.; Kravets, V.; Ruelland, E. A ménage à trois: Salicylic acid, growth inhibition, and immunity. Trends Plant Sci. 2021, 27, 460–471. [Google Scholar] [CrossRef]
- Khan, M.I.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Klessig, D.F.; Tian, M.; Choi, H.W. Multiple targets of salicylic acid and its derivatives in plants and animals. Front. Immunol. 2016, 7, 206. [Google Scholar] [CrossRef]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Anwar, S.; Iqbal, M.; Raza, S.H.; Iqbal, N. Efficacy of seed preconditioning with salicylic and ascorbic acid in increasing vigor of rice (Oryza sativa L.) seedling. Pak. J. Bot. 2013, 45, 157–162. [Google Scholar]
- Naser Alavi, S.M.; Arvin, M.J.; Manoochehri Kalantari, K. Salicylic acid and nitric oxide alleviate osmotic stress in wheat (Triticum aestivum L.) seedlings. J. Plant Interact. 2014, 9, 683–688. [Google Scholar] [CrossRef]
- Khan, M.I.; Asgher, M.; Khan, N.A. Alleviation of salt-induced photosynthesis and growth inhibition by salicylic acid involves glycinebetaine and ethylene in mungbean (Vigna radiata L.). Plant Physiol. Biochem. 2014, 80, 67–74. [Google Scholar] [CrossRef]
- Nazar, R.; Umar, S.; Khan, N.A. Exogenous salicylic acid improves photosynthesis and growth through increase in ascorbate-glutathione metabolism and S assimilation in mustard under salt stress. Plant Signal. Behav. 2015, 10, e1003751. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A. Effects of exogenously applied salicylic acid and putrescine alone and in combination with rhizobacteria on the phytoremediation of heavy metals and chickpea growth in sandy soil. Int. J. Phytoremediat. 2018, 20, 405–414. [Google Scholar] [CrossRef]
- Snyman, M.; Cronje, M.J. Modulation of heat shock factors accompanies salicylic acid-mediated potentiation of Hsp70 in tomato seedlings. J. Exp. Bot. 2008, 59, 2125–2132. [Google Scholar] [CrossRef]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic acid in relation to other phytohormones in plant: A study towards physiology and signal transduction under challenging environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Chavoushi, M.; Najafi, F.; Salimi, A.; Angaji, S.A. Effect of salicylic acid and sodium nitroprusside on growth parameters, photosynthetic pigments and secondary metabolites of safflower under drought stress. Sci. Hortic. 2020, 259, 108823. [Google Scholar] [CrossRef]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef]
- Strawn, M.A.; Marr, S.K.; Inoue, K.; Inada, N.; Zubieta, C.; Wildermuth, M.C. Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 2007, 282, 5919–5933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayakannan, M.; Bose, J.; Babourina, O.; Rengel, Z.; Shabala, S. Salicylic acid in plant salinity stress signalling and tolerance. Plant Growth Regul. 2015, 76, 25–40. [Google Scholar] [CrossRef]
- Serrano, M.; Wang, B.; Aryal, B.; Garcion, C.; Abou-Mansour, E.; Heck, S.; Geisler, M.; Mauch, F.; Nawrath, C.; Metraux, J.P. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 2013, 162, 1815–1821. [Google Scholar] [CrossRef] [PubMed]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; van der Drift, K.M.; Olsson, P.E.; Thomas-Oates, J.E.; van Loon, L.C.; Bakker, P.A. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 2001, 183, 1909–1920. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Yalpani, N.; León, J.; Lawton, M.A.; Raskin, I. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 1993, 103, 315–321. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, W.; Zhao, Q. Salicylic acid biosynthesis is not from phenylalanine in Arabidopsis. J. Integr. Plant Biol. 2022, 1–7. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, H.; Ruan, W.; Deng, M.; Wang, F.; Peng, J.; Luo, J.; Chen, Z.; Yi, K. ABNORMAL INFLORESCENCE MERISTEM1 functions in salicylic acid biosynthesis to maintain proper reactive oxygen species levels for root meristem activity in rice. Plant Cell 2017, 29, 560–574. [Google Scholar] [CrossRef]
- Leon, J.; Shulaev, V.; Yalpani, N.; Lawton, M.A.; Raskin, I. Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Natl. Acad. Sci. USA 1995, 92, 10413–10417. [Google Scholar] [CrossRef] [Green Version]
- Sawada, H.; Shim, I.S.; Usui, K. Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis modulation by salt stress in rice seedlings. Plant Sci. 2006, 171, 263–270. [Google Scholar] [CrossRef]
- Duan, L.; Liu, H.; Li, X.; Xiao, J.; Wang, S. Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol. Plant 2014, 152, 486–500. [Google Scholar] [CrossRef]
- Shine, M.B.; Yang, J.W.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.Q.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef]
- Huang, W.; Wang, Y.; Li, X.; Zhang, Y. Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 2020, 13, 31–41. [Google Scholar] [CrossRef]
- Wang, X.; Gao, J.; Zhu, Z.; Dong, X.; Wang, X.; Ren, G.; Zhou, X.; Kuai, B. TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 2015, 82, 151–162. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Li, L.S.; Ying, J.; Li, E.; Ma, T.; Li, M.; Gong, L.M.; Wei, G.; Zhang, Y.; Li, S. Arabidopsis CBP60b is a central transcriptional activator of immunity. Plant Physiol. 2021, 186, 1645–1659. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wu, Z.; Tian, H.; Li, X.; Zhang, Y. Arabidopsis CALMODULIN-BINDING PROTEIN 60b plays dual roles in plant immunity. Plant Commun. 2021, 2, 100213. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Truman, W.; Sreekanta, S.; Lu, Y.; Bethke, G.; Tsuda, K.; Katagiri, F.; Glazebrook, J. The CALMODULIN-BINDING PROTEIN60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 2013, 163, 1741–1751. [Google Scholar] [CrossRef]
- Chandran, D.; Rickert, J.; Huang, Y.; Steinwand, M.A.; Marr, S.K.; Wildermuth, M.C. Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe 2014, 15, 506–513. [Google Scholar] [CrossRef]
- Lai, Y.S.; Renna, L.; Yarema, J.; Ruberti, C.; He, S.Y.; Brandizzi, F. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2018, 115, E5203–E5212. [Google Scholar] [CrossRef]
- Mhamdi, A. NPR1 has everything under control. Plant Physiol. 2019, 181, 6–7. [Google Scholar] [CrossRef]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 2020, 182, 1093–1108. [Google Scholar] [CrossRef]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.L.; et al. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef]
- Jayakannan, M.; Bose, J.; Babourina, O.; Shabala, S.; Massart, A.; Poschenrieder, C.; Rengel, Z. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 1865–1875. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, T.; Sun, Y.; Zhang, Y.; Radojicic, A.; Ding, Y.; Tian, H.; Huang, X.; Lan, J.; Chen, S.; et al. Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 2020, 32, 4002–4016. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Withers, J.; Li, H.; Zwack, P.J.; Rusnac, D.V.; Shi, H.; Liu, L.; Yan, S.; Hinds, T.R.; Guttman, M.; et al. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature 2020, 586, 311–316. [Google Scholar] [CrossRef]
- Choudhary, A.; Senthil-Kumar, M. Drought attenuates plant defence against bacterial pathogens by suppressing the expression of CBP60g/SARD1 during combined stress. Plant Cell Environ. 2022, 45, 1127–1145. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Lapin, D.; Bhandari, D.D.; Parker, J.E. Origins and immunity networking functions of EDS1 family proteins. Annu. Rev. Phytopathol. 2020, 58, 253–276. [Google Scholar] [CrossRef]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Chang, M.; Zhao, J.; Chen, H.; Li, G.; Chen, J.; Li, M.; Palmer, A.; Song, J.; Alfano, J.; Liu, F.; et al. PBS3 protects EDS1 from proteasome-mediated degradation in plant immunity. Mol. Plant 2019, 12, 678–688. [Google Scholar] [CrossRef]
- Zewdu, S.; Suryabhagavan, K.V.; Balakrishnan, M. Land-use/land-cover dynamics in Sego Irrigation Farm, southern Ethiopia: A comparison of temporal soil salinization using geospatial tools. J. Saudi Soc. Agric. Sci. 2016, 15, 91–97. [Google Scholar] [CrossRef]
- Abbas, A.; Khan, S.; Hussain, N.; Hanjra, M.A.; Akbar, S. Characterizing soil salinity in irrigated agriculture using a remote sensing approach. Phys. Chem. Earth Parts A/B/C 2013, 55–57, 43–52. [Google Scholar] [CrossRef]
- Li, P.; Wu, J.; Qian, H. Regulation of secondary soil salinization in semi-arid regions: A simulation research in the Nanshantaizi area along the Silk Road, northwest China. Environ. Earth Sci. 2016, 75, 698. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.; Li, J.; Yahya, M.; Wang, M.; Ali, A.; Cheng, A.; Wang, X.; Ma, C. Grain legumes and fear of salt stress: Focus on mechanisms and management strategies. Int. J. Mol. Sci. 2019, 20, 799. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Chandra, A.; Anand, A.; Dubey, A. Effect of salicylic acid on morphological and biochemical attributes in cowpea. J. Environ. Biol. 2007, 28, 193–196. [Google Scholar]
- Jini, D.; Joseph, B. Physiological mechanism of salicylic acid for alleviation of salt stress in rice. Rice Sci. 2017, 24, 97–108. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Jayakannan, M.; Bose, J.; Babourina, O.; Rengel, Z.; Shabala, S. Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 2013, 64, 2255–2268. [Google Scholar] [CrossRef]
- Duarte, B.; Santos, D.; Marques, J.C.; Cacador, I. Ecophysiological adaptations of two halophytes to salt stress: Photosynthesis, PS II photochemistry and anti-oxidant feedback-implications for resilience in climate change. Plant Physiol. Biochem. 2013, 67, 178–188. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane lipid remodeling in response to salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef] [Green Version]
- Nadarajah, K.; Abdul Hamid, N.W.; Abdul Rahman, N.S.N. SA-mediated regulation and control of abiotic stress tolerance in rice. Int. J. Mol. Sci. 2021, 22, 5591. [Google Scholar] [CrossRef]
- Saeidnejad, A.H.; Mardani, H.; Naghibolghora, M. Protective effects of salicylic acid on physiological parameters and antioxidants response in maize seedlings under salinity stress. J. Appl. Environ. Biol. Sci. 2012, 2, 364–373. [Google Scholar]
- Li, G.; Peng, X.; Wei, L.; Kang, G. Salicylic acid increases the contents of glutathione and ascorbate and temporally regulates the related gene expression in salt-stressed wheat seedlings. Gene 2013, 529, 321–325. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Colmenero-Flores, J.M.; Franco-Navarro, J.D.; Cubero-Font, P.; Peinado-Torrubia, P.; Rosales, M.A. Chloride as a beneficial macronutrient in higher plants: New roles and regulation. Int. J. Mol. Sci. 2019, 20, 4686. [Google Scholar] [CrossRef]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and temporal profile of glycine betaine accumulation in plants under abiotic stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef]
- Syeed, S.; Sehar, Z.; Masood, A.; Anjum, N.A.; Khan, N.A. Control of elevated ion accumulation, oxidative stress, and lipid peroxidation with salicylic acid-induced accumulation of glycine betaine in salinity-exposed Vigna radiata L. Appl. Biochem. Biotech. 2021, 193, 3301–3320. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant 2021, 171, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, W.F.; Soares, L.H.; Fagan, E.B.; da Costa Mello, S.; Reichardt, K.; Dourado-Neto, D. Amino acids as stress reducers in soybean plant growth under different water-deficit conditions. J. Plant Growth Regul. 2020, 39, 905–919. [Google Scholar] [CrossRef]
- Misra, N.; Saxena, P. Effect of salicylic acid on proline metabolism in lentil grown under salinity stress. Plant Sci. 2009, 177, 181–189. [Google Scholar] [CrossRef]
- El-Tayeb, M.A. Response of barley grains to the interactive e.ect of salinity and salicylic acid. Plant Growth Regul. 2005, 45, 215–224. [Google Scholar] [CrossRef]
- Tufail, A.; Arfan, M.; Gurmani, A.R.; Khan, A.; Bano, A. Salicylic acid induced salinity tolerance in maize (Zea mays). Pak. J. Bot. 2013, 45, 75–82. [Google Scholar]
- Al-Whaibi, M.H.; Siddiqui, M.H.; Basalah, M.O. Salicylic acid and calcium-induced protection of wheat against salinity. Protoplasma 2012, 249, 769–778. [Google Scholar] [CrossRef]
- Jini, D.; Joseph, B. Salicylic acid mediated salt tolerance at different growth stages of Oryza sativa L. and its effect on salicylic acid biosynthetic pathway genes. Biotechnol. Ind. J. 2017, 13, 134. [Google Scholar]
- Nie, W.; Gong, B.; Chen, Y.; Wang, J.; Wei, M.; Shi, Q. Photosynthetic capacity, ion homeostasis and reactive oxygen metabolism were involved in exogenous salicylic acid increasing cucumber seedlings tolerance to alkaline stress. Sci. Hortic. 2018, 235, 413–423. [Google Scholar] [CrossRef]
- Gharbi, E.; Lutts, S.; Dailly, H.; Quinet, M. Comparison between the impacts of two different modes of salicylic acid application on tomato (Solanum lycopersicum) responses to salinity. Plant Signal. Behav. 2018, 13, e1469361. [Google Scholar] [CrossRef]
- Souana, K.; Taïbi, K.; Ait Abderrahim, L.; Amirat, M.; Achir, M.; Boussaid, M.; Mulet, J.M. Salt-tolerance in Vicia faba L. is mitigated by the capacity of salicylic acid to improve photosynthesis and antioxidant response. Sci. Hortic. 2020, 273, 109641. [Google Scholar] [CrossRef]
- Palma, F.; Lopez-Gomez, M.; Tejera, N.A.; Lluch, C. Salicylic acid improves the salinity tolerance of Medicago sativa in symbiosis with Sinorhizobium meliloti by preventing nitrogen fixation inhibition. Plant Sci. 2013, 208, 75–82. [Google Scholar] [CrossRef]
- Yastreb, T.O.; Kolupaev, Y.E.; Lugovaya, A.A.; Dmitriev, A.P. Hydrogen peroxide-induced salt tolerance in the Arabidopsis salicylate-deficient transformants NahG. Appl. Biochem. Microbiol. 2017, 53, 719–724. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H.M. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef]
- Dong, Q.; Wallrad, L.; Almutairi, B.O.; Kudla, J. Ca2+ signaling in plant responses to abiotic stresses. J. Integr. Plant Biol. 2022, 64, 287–300. [Google Scholar]
- Fang, S.; Hou, X.; Liang, X. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef]
- Bassil, E.; Zhang, S.; Gong, H.; Tajima, H.; Blumwald, E. Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiol. 2019, 179, 616–629. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.R.; Ansari, M.W.; Sahoo, R.K.; Wattal, R.K.; Tuteja, N.; Kumar, V.R. Salicylic acid modulates ACS, NHX1, sos1 and HKT1; 2 expression to regulate ethylene overproduction and Na+ ions toxicity that leads to improved physiological status and enhanced salinity stress tolerance in tomato plants cv. Pusa Ruby. Plant Signal. Behav. 2021, 16, 1950888. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, G.; Alli, A.; Yu, L. Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ragel, P.; Raddatz, N.; Leidi, E.O.; Quintero, F.J.; Pardo, J.M. Regulation of K+ nutrition in plants. Front. Plant Sci. 2019, 10, 281. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, X.; Wang, L.; Cao, Y.; Song, W.; Shi, J.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef]
- Csiszár, J.; Horvath, E.; Vary, Z.; Galle, A.; Bela, K.; Brunner, S.; Tari, I. Glutathione transferase supergene family in tomato: Salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol. Biochem. 2014, 78, 15–26. [Google Scholar] [CrossRef]
- Kim, Y.; Mun, B.G.; Khan, A.L.; Waqas, M.; Kim, H.H.; Shahzad, R.; Imran, M.; Yun, B.W.; Lee, I.J. Regulation of reactive oxygen and nitrogen species by salicylic acid in rice plants under salinity stress conditions. PLoS ONE 2018, 13, e0192650. [Google Scholar] [CrossRef]
- Rasheed, F.; Anjum, N.A.; Masood, A.; Sofo, A.; Khan, N.A. The key roles of salicylic acid and sulfur in plant salinity stress tolerance. J. Plant Growth Regul. 2022, 41, 1891–1904. [Google Scholar] [CrossRef]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Ni, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Thor, K.; Jiang, S.; Michard, E.; George, J.; Scherzer, S.; Huang, S.; Dindas, J.; Derbyshire, P.; Leitão, N.; DeFalco, T.A.; et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 2020, 585, 569–573. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef]
- Mao, J.; Mo, Z.; Yuan, G.; Xiang, H.; Visser, R.G.; Bai, Y.; Liu, H.; Wang, Q.; van der Linden, C.G. The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaptation. Plant Cell Environ. 2022, 1–11. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Ma, X.; Li, Q.H.; Yu, Y.N.; Qiao, Y.M.; Haq, S.U.; Gong, Z.H. The CBL-CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef] [Green Version]
- De la Torre, F.; Gutiérrez-Beltrán, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; del Pozo, O. The tomato calcium sensor CBL10 and its interacting protein kinase CIPK6 define a signaling pathway in plant immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar] [CrossRef]
- Liu, P.; Duan, Y.; Liu, C.; Xue, Q.; Guo, J.; Qi, T.; Kang, Z.; Guo, J. The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J. Exp. Bot. 2018, 69, 4443–4457. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, Q.; He, J.; Cao, J.; Liu, Z.; Wang, J.; Yang, Y. The kinase CIPK14 functions as a negative regulator of plant immune responses to Pseudomonas syringae in Arabidopsis. Plant Sci. 2021, 312, 111017. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Chang, M.; Chen, H.; Liu, F.; Fu, Z.Q. PTI and ETI: Convergent pathways with diverse elicitors. Trends Plant Sci. 2021, 27, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Tak, Y.; Asthir, B. Salicylic acid: A key signal molecule ameliorating plant stresses. Cereal Res. Commun. 2022, 50, 617–626. [Google Scholar] [CrossRef]
- Xie, Y.J.; Xu, S.; Han, B.; Wu, M.Z.; Yuan, X.X.; Han, Y.; Gu, Q.; Xu, D.K.; Yang, Q.; Shen, W.B. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J. 2011, 66, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.Y.; Wi, S.J.; Park, K.Y. Functional switching of NPR1 between chloroplast and nucleus for adaptive response to salt stress. Sci. Rep. 2020, 10, 4339. [Google Scholar] [CrossRef]
- Seo, S.Y.; Park, K.Y. Translocation of chloroplast NPR1 to the nucleus in retrograde signaling for adaptive response to salt stress in tobacco. BioRxiv 2021. [Google Scholar] [CrossRef]
- Li, B.; Liu, Y.; Cui, X.Y.; Fu, J.D.; Zhou, Y.B.; Zheng, W.J.; Lan, J.H.; Jin, L.G.; Chen, M.; Ma, Y.Z.; et al. Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 2019, 10, 549. [Google Scholar] [CrossRef] [Green Version]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef]
- Keswani, C.; Prakash, O.; Bharti, N.; Vilchez, J.I.; Sansinenea, E.; Lally, R.D.; Borriss, R.; Singh, S.P.; Gupta, V.K.; Fraceto, L.F.; et al. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 2019, 690, 841–852. [Google Scholar] [CrossRef]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Ran, L.; Mercado-Blanco, J. Rhizobacterial salicylate production provokes headaches! Plant Soil. 2014, 382, 1–16. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Zhou, Z.; Chu, Z. Emerging Roles of Salicylic Acid in Plant Saline Stress Tolerance. Int. J. Mol. Sci. 2023, 24, 3388. https://doi.org/10.3390/ijms24043388
Yang W, Zhou Z, Chu Z. Emerging Roles of Salicylic Acid in Plant Saline Stress Tolerance. International Journal of Molecular Sciences. 2023; 24(4):3388. https://doi.org/10.3390/ijms24043388
Chicago/Turabian StyleYang, Wei, Zhou Zhou, and Zhaohui Chu. 2023. "Emerging Roles of Salicylic Acid in Plant Saline Stress Tolerance" International Journal of Molecular Sciences 24, no. 4: 3388. https://doi.org/10.3390/ijms24043388