Olive Polyphenol Oxidase Gene Family
Abstract
1. Introduction
2. Results and Discussion
2.1. Polyphenol Oxidase Gene Family in Olive
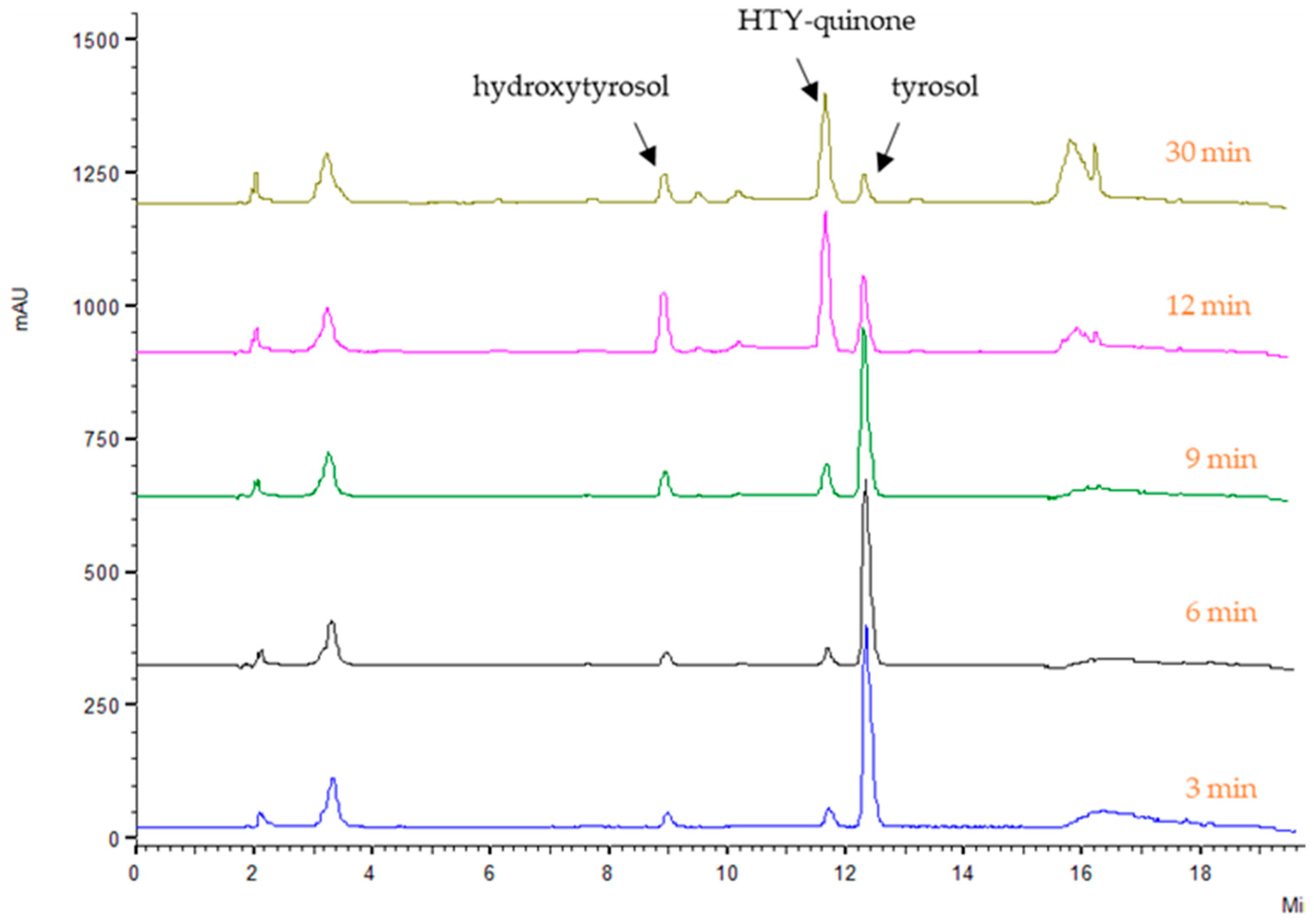
2.2. Enzymatic Activities of Encoded Olive PPO Proteins
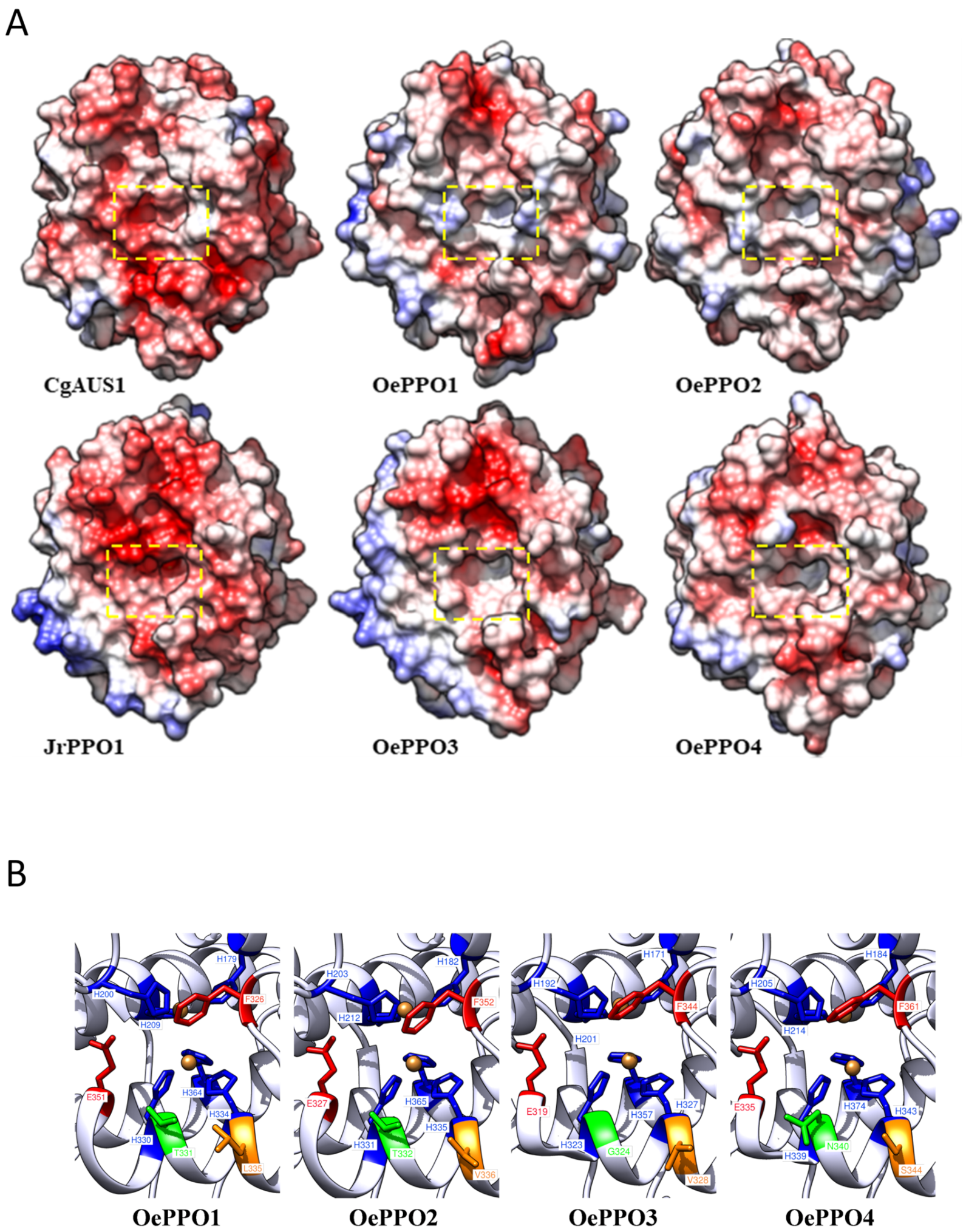
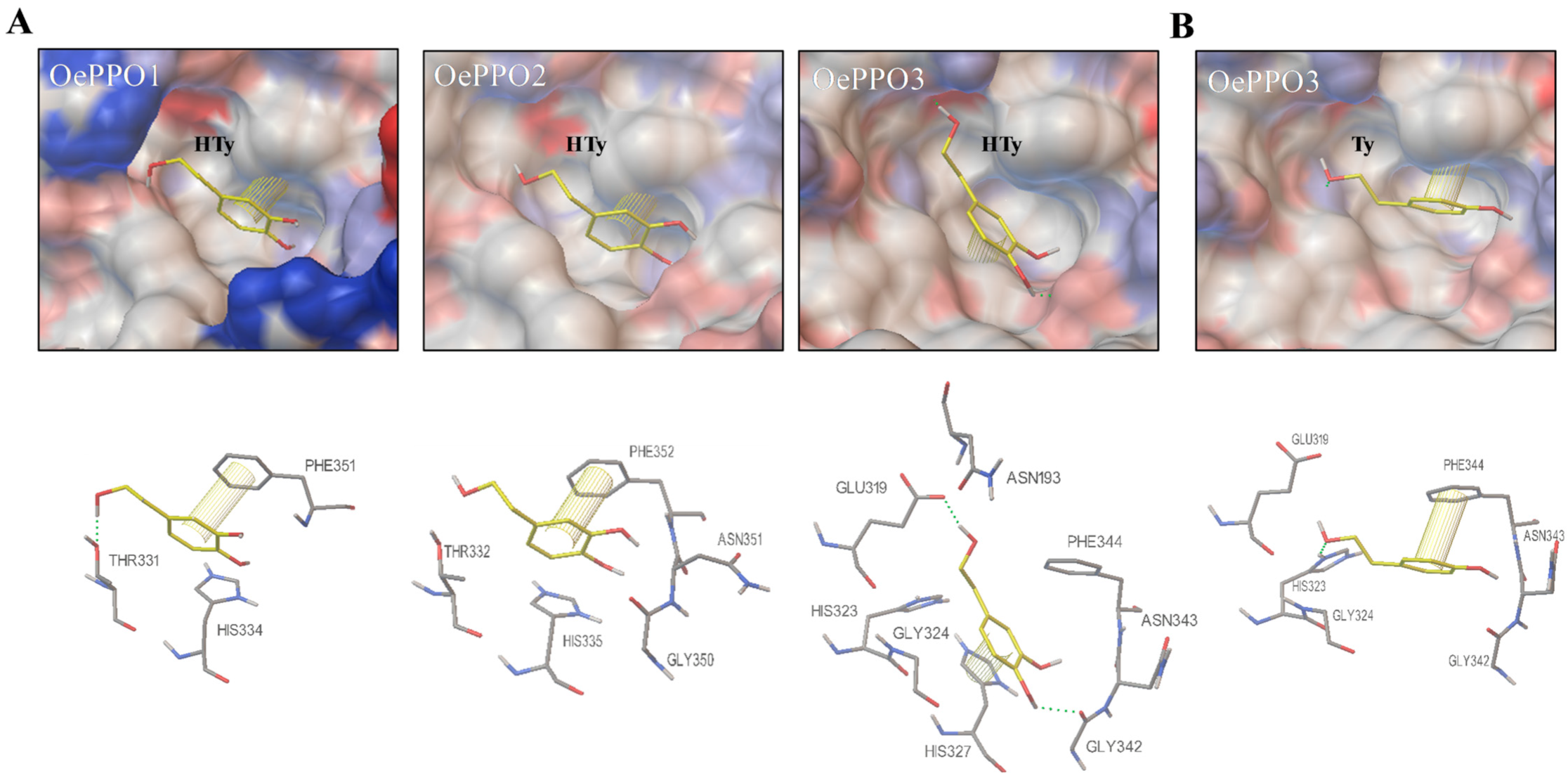
2.3. In Silico OePPOs Modelling and Substrates Docking
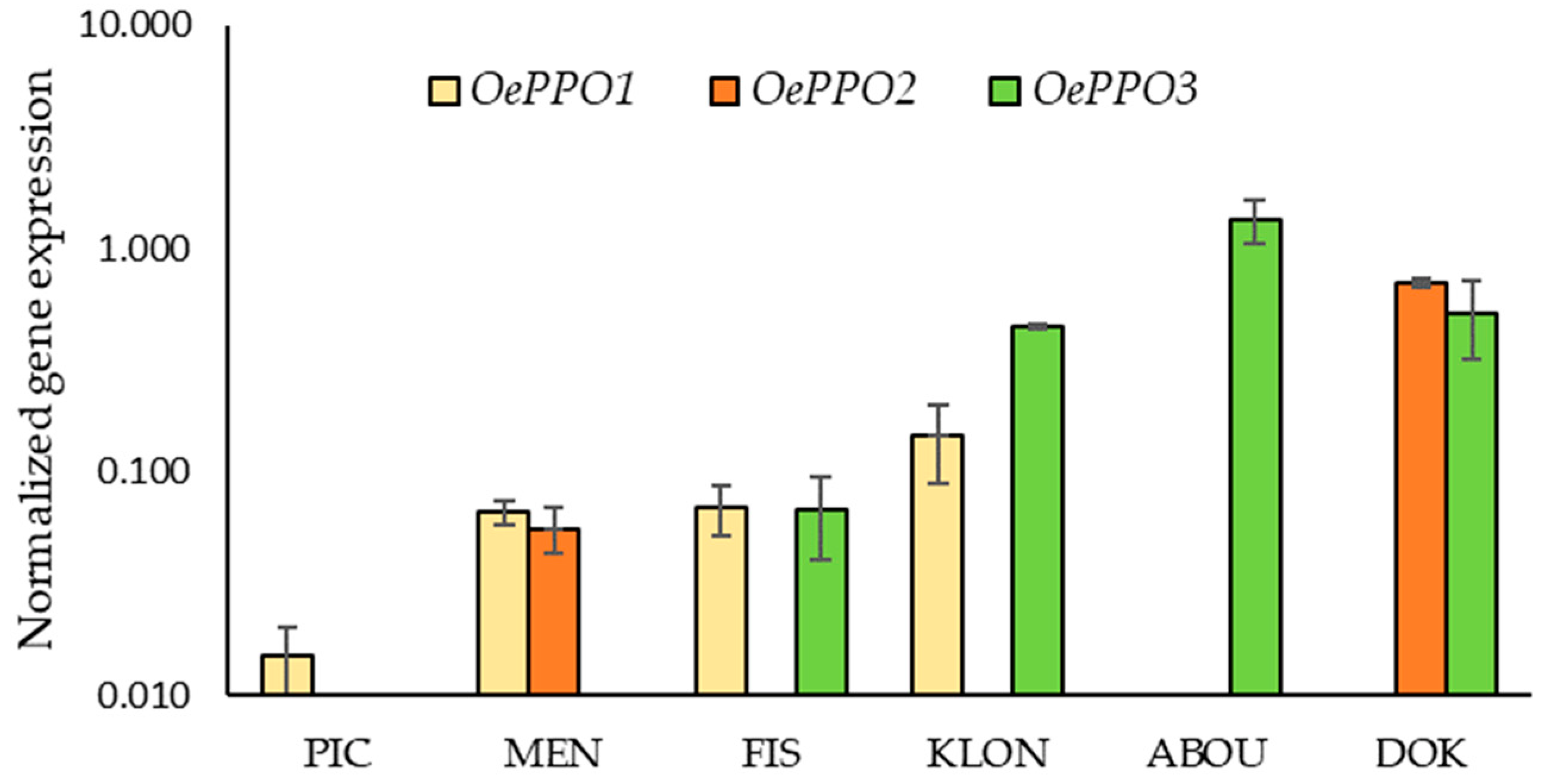
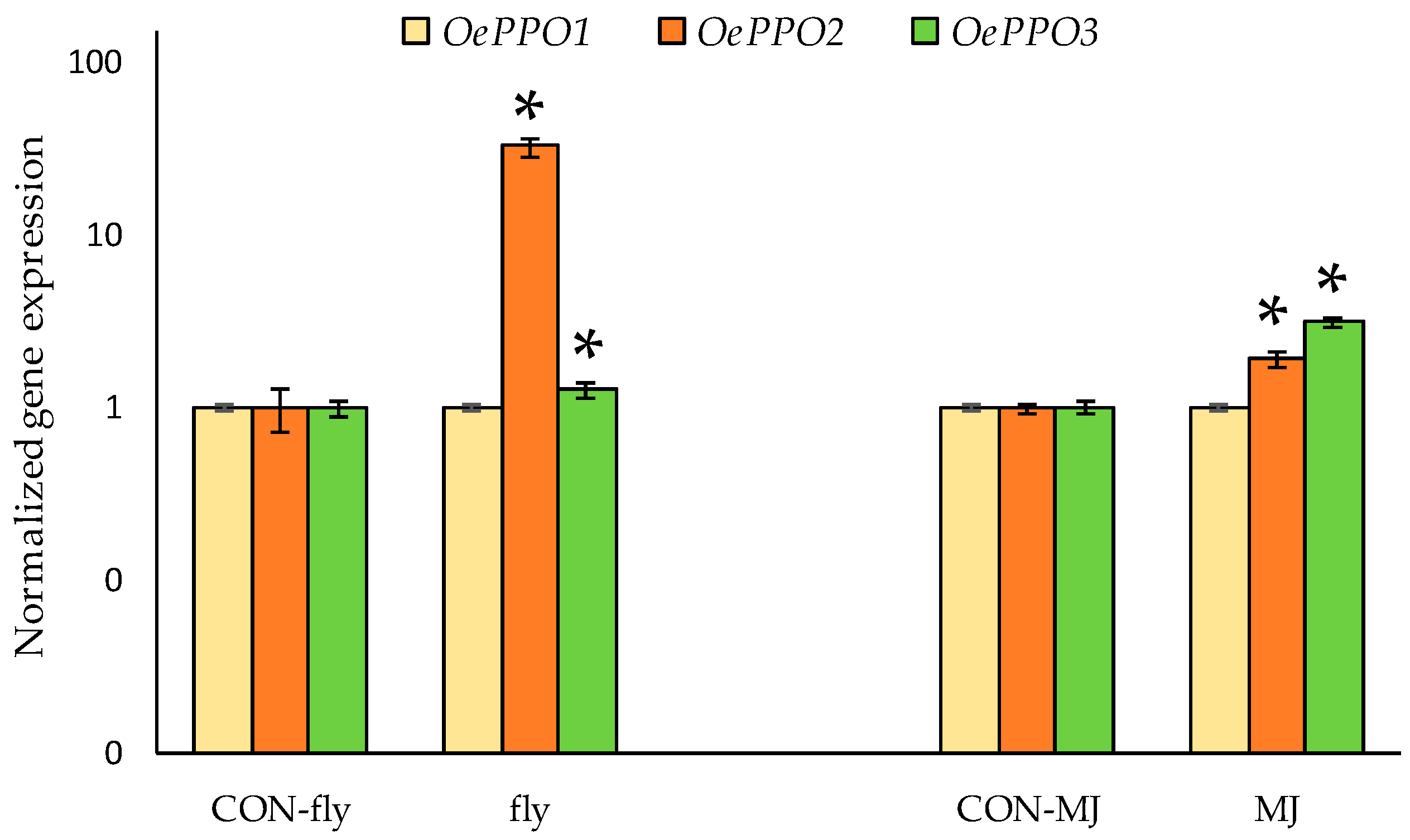
2.4. Gene Expression Analysis of Olive PPOs
2.4.1. OePPOs Expression Is Cultivar Dependent
2.4.2. Olive PPO Genes Are Involved in Plant Defense Mechanisms
3. Materials and Methods
3.1. Plant Material
3.2. Identification of Putative Olive Polyphenol Oxidase Genes
3.3. Sequence Alignment and Phylogenetic Analysis
3.4. OePPO Genes Cloning, Heterologous Protein Expression and Purification
3.5. Polyphenol Oxidase Activity Assays
3.5.1. HPLC Method
3.5.2. Spectrophotometric Method
3.6. Olive Oil Extraction
3.7. Extraction and Analysis of Phenolic Compounds from Olive Fruit and Oil
3.8. Modelling 3D Structures and Molecular Docking
3.9. Total RNA Extraction and Gene Expression Analysis by Real-Time Quantitative PCR (RT-qPCR)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gouvinhas, I.; Machado, N.; Sobreira, C.; Domínguez-Perles, R.; Gomes, S.; Rosa, E.; Barros, A. Critical review on the significance of olive phytochemicals in plant physiology and human health. Molecules 2017, 22, 1986. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.C.; Tomé-Carneiro, J.; Dávalos, A.; Visioli, F. Pharma-nutritional properties of olive oil phenols. Transfer of new findings to human nutrition. Foods 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Parra-Pérez, A.M.; Pérez-Jiménez, A.; Gris-Cárdenas, I.; Bonel-Pérez, G.C.; Carrasco-Díaz, L.M.; Mokhtari, K.; García-Salguero, L.; Lupiáñez, J.A.; Rufino-Palomares, E.E. Involvement of the PI3K/AKT intracellular signaling pathway in the anticancer activity of hydroxytyrosol, a polyphenol from Olea europaea, in hematological cells and implication of HSP60 levels in its anti-inflamatory activity. Int. J. Mol. Sci. 2022, 23, 7053. [Google Scholar] [CrossRef]
- Laghezza Masci, V.; Bernini, R.; Villanova, N.; Clemente, M.; Cicaloni, V.; Tinti, L.; Salvini, L.; Taddei, A.R.; Tiezzi, A.; Ovidi, E. In vitro anti-poliferative and apoptotic effects of hydroxytyrosyl oleate on SH-SY5Y human neuroblastoma cells. Int. J. Mol. Sci. 2022, 23, 12348. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission Regulation (EU) No 1018/2013 amending Regulation (EU) No 432/2012 establishing a list of permitted health claims made on foods other than those referring to the reduction of disease risk and to children’s development and heal. Off. J. Eur. Union 2012, 282, 43–45. [Google Scholar]
- Navarro, A.; Ruiz-Méndez, M.V.; Sanz, C.; Martínez, M.; Rego, D.; Pérez, A.G. Application of pulsed electric fields to pilot and industrial scale virgin olive oil extraction: Impact on organoleptic and functional quality. Foods 2022, 11, 2022. [Google Scholar] [CrossRef] [PubMed]
- Romero-Segura, C.; García-Rodríguez, R.; Sanz, C.; Pérez, A.G. Virgin olive phenolic profile as a result of the anabolic and catabolic enzymes status in the olive fruit. Acta Hortic. 2011, 924, 379–384. [Google Scholar] [CrossRef]
- Pérez, A.G.; León, L.; Sanz, C.; De la Rosa, R. Fruit phenolic profiling: A new selection criterion in olive breeding programs. Front. Plant Sci. 2018, 8, 241. [Google Scholar] [CrossRef]
- García-Vico, L.; Sánchez, R.; Rodríguez, R.; Sanz, C.; Pérez, A. Study of the olive β-glucosidase gene family putatively involved in the synthesis of phenolic compounds of virgin olive oil. J. Sci. Food Agric. 2021, 101, 5409–5418. [Google Scholar] [CrossRef]
- García-Rodríguez, R.; Romero-Segura, C.; Sanz, C.; Sánchez-Ortiz, A.; Pérez, A.G. Role of polyphenol oxidase and peroxidase in shaping the phenolic profile of virgin olive oil. Food Res. Int. 2011, 44, 629–635. [Google Scholar] [CrossRef]
- Allouche, N.; Fki, I.; Sayadi, S. Toward a high yield recovery of antioxidants and purified hydroxytyrosol from olive mill wastewaters. J. Agric. Food Chem. 2004, 52, 267–273. [Google Scholar] [CrossRef] [PubMed]
- García-García, M.I.; Hernández-García, S.; Sánchez-Ferrer, A.; García-Carmona, F. Kinetic study of hydroxytyrosol oxidation and its related compounds by red globe grape polyphenol oxidase. J. Agric. Food Chem. 2013, 61, 6050–6055. [Google Scholar] [CrossRef] [PubMed]
- Kampatsikas, I.; Bijelic, A.; Pretzler, M.; Rompel, A. Three recombinantly expressed apple tyrosinases suggest the amino acids responsible for mono- versus diphenolase activity in plant polyphenol oxidases. Sci. Rep. 2017, 7, 8860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, X. Recent advances in polyphenol oxidase-mediated plant stress responses. Phytochemistry 2021, 181, 112588. [Google Scholar] [CrossRef]
- Ortega-García, F.; Blanco, S.; Peinado, M.A.; Peragón, J. Polyphenol oxidase and its relationship with oleuropein concentration in fruits and leaves of olive (Olea europaea) cv. ’Picual’ trees during fruit ripening. Tree Physiol. 2008, 28, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Dirks-Hofmeister, M.E.; Kolkenbrock, S.; Moerschbacher, B.M. Parameters That enhance the bacterial expression of active plant polyphenol oxidases. PLoS ONE 2013, 8, e77291. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, R.; García-Vico, L.; Sanz, C.; Pérez, A. An aromatic aldehyde synthase controls the synthesis of hydroxytyrosol derivatives present in virgin olive oil. Antioxidants 2019, 8, 352. [Google Scholar] [CrossRef]
- Notario, A.; Sánchez, R.; Luaces, P.; Sanz, C.; Pérez, A.G. The infestation of olive fruits by Bactrocera oleae (Rossi) modifies the expression of key genes in the biosynthesis of volatile and phenolic compounds and alters the composition of virgin olive oil. Molecules 2022, 27, 1650. [Google Scholar] [CrossRef]
- Kampatsikas, I.; Bijelic, A.; Rompel, A. Biochemical and structural characterization of tomato polyphenol oxidases provide novel insights into their substrate specificity. Sci. Rep. 2019, 9, 4022. [Google Scholar] [CrossRef]
- Panis, F.; Kampatsikas, I.; Bijelic, A.; Rompel, A. Conversion of walnut tyrosinase into catechol oxidase by site directed mutagenesis. Sci. Rep. 2020, 7, 8860. [Google Scholar] [CrossRef]
- Prexler, S.M.; Frassek, M.; Moerschbacher, B.; Dirks-Hofmeister, M.E. Catechol oxidase versus tyrosinase classification revisited by site-directed mutagenesis studies. Angew. Chem. Int. Ed. 2019, 58, 8757–8761. [Google Scholar] [CrossRef] [PubMed]
- Kaintz, C.; Molitor, C.; Thill, J.; Kampatsikas, I.; Michael, C.; Halbwirth, H.; Rompel, A. Cloning and functional expression in E. coli of a polyphenol oxidase transcript from Coreopsis grandiflora involved in aurone formation. FEBS Lett. 2014, 588, 3417–3426. [Google Scholar] [CrossRef] [PubMed]
- Derardja, A.E.; Pretzler, M.; Kampatsikas, L.; Barkat, M.; Rompel, A. Purification and characterization of latent polyphenol oxidase from apricot (Prunus armeniaca L.). J. Agric. Food Chem. 2017, 65, 8203–8212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Garde-Cerdán, T.; Portu, J.; López, R.; Santamaría, P. Effect of methyl jasmonate application to grapevine leaves on grape amino acid content. Food Chem. 2016, 203, 536–539. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; Mcwilliam, H.P.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, L.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Garcia Molina, F.; Muñoz, J.L.; Varon, R.; Rodriguez-López, J.N.; Garcia-Cánovas, F.; Tudela, J. A review on spectrophotometric methods for measuring the monophenolase and diphenolase activities of tyrosinase. J. Agric. Food Chem. 2007, 55, 9739–9749. [Google Scholar] [CrossRef]
- Fernández, G.; García-Vico, L.; Sanz, C.; Peérez, A.G. Optimization of a simplified method for fruit phenolic extraction and analysis to be used in olive breeding. Acta Hortic. 2020, 1292, 357–363. [Google Scholar] [CrossRef]
- Mateos, R.; Espartero, J.L.; Trujillo, M.; Ríos, J.J.; León-Camacho, M.; Alcudia, F.; Cert, A. Determination of phenols, flavones, and lignans in virgin olive oils by SPE and high-performance liquid chromatography with diode array ultra-violet detection. J. Agric. Food Chem. 2001, 49, 2185–2192. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]




| Substrate | Specific Act. (U/mg) | Optimum pH | Optimum Temp. (˚C) | KM (mM) | Vmax (U/mg) | Vmax/KM | |
|---|---|---|---|---|---|---|---|
| OePPO1 | TBC | 11.1 | 7.0 | 25 | |||
| tyramine | nd | ||||||
| OePPO2 | TBC | 44.2 | 7.0 | 25 | 12.7 | 41.6 | 3.3 |
| tyramine | nd | ||||||
| OePPO3 | TBC | 96.8 | 6.8 | 25 | 0.8 | 129.9 | 162.4 |
| tyramine | 1.7 | 0.7 | 2.7 | 3.9 | |||
| OePPO4 | TBC | nd | |||||
| tyramine | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, R.; Arroyo, L.; Luaces, P.; Sanz, C.; Pérez, A.G. Olive Polyphenol Oxidase Gene Family. Int. J. Mol. Sci. 2023, 24, 3233. https://doi.org/10.3390/ijms24043233
Sánchez R, Arroyo L, Luaces P, Sanz C, Pérez AG. Olive Polyphenol Oxidase Gene Family. International Journal of Molecular Sciences. 2023; 24(4):3233. https://doi.org/10.3390/ijms24043233
Chicago/Turabian StyleSánchez, Rosario, Laura Arroyo, Pilar Luaces, Carlos Sanz, and Ana G. Pérez. 2023. "Olive Polyphenol Oxidase Gene Family" International Journal of Molecular Sciences 24, no. 4: 3233. https://doi.org/10.3390/ijms24043233
APA StyleSánchez, R., Arroyo, L., Luaces, P., Sanz, C., & Pérez, A. G. (2023). Olive Polyphenol Oxidase Gene Family. International Journal of Molecular Sciences, 24(4), 3233. https://doi.org/10.3390/ijms24043233







