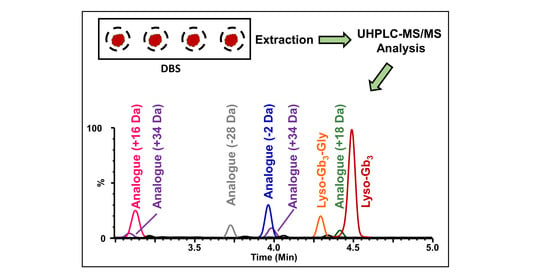
Mass Spectrometry Analysis of Globotriaosylsphingosine and Its Analogues in Dried Blood Spots
Abstract
1. Introduction
2. Results
2.1. Method Validation
2.1.1. Accuracy and Precision
2.1.2. Limits of Detection (LODs), Limits of Quantitation (LOQs), Recovery, and Linearity
2.1.3. Stability
2.1.4. Matrix Effect
2.2. Normal Reference Values
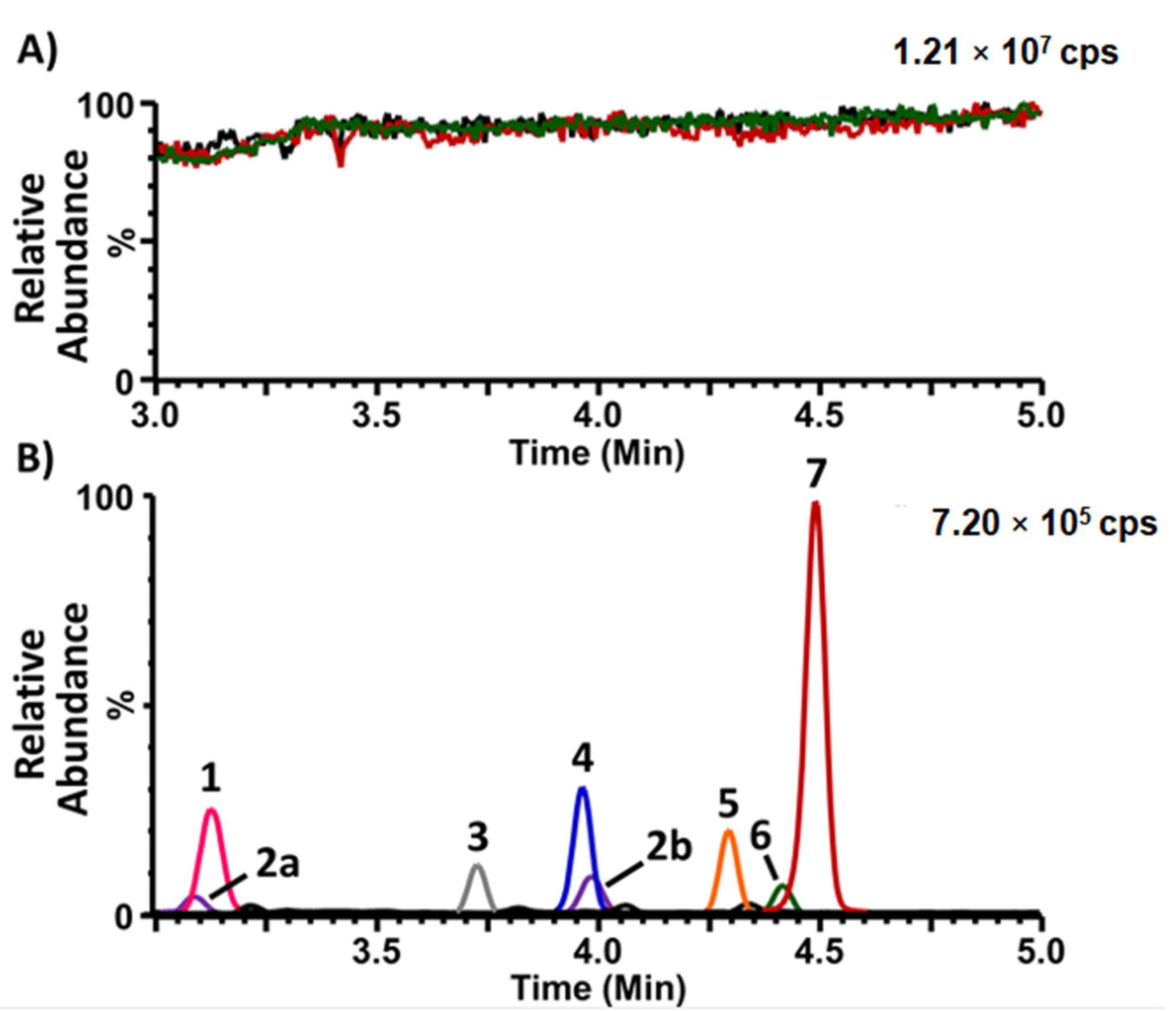
2.3. Analysis of Collected Specimens
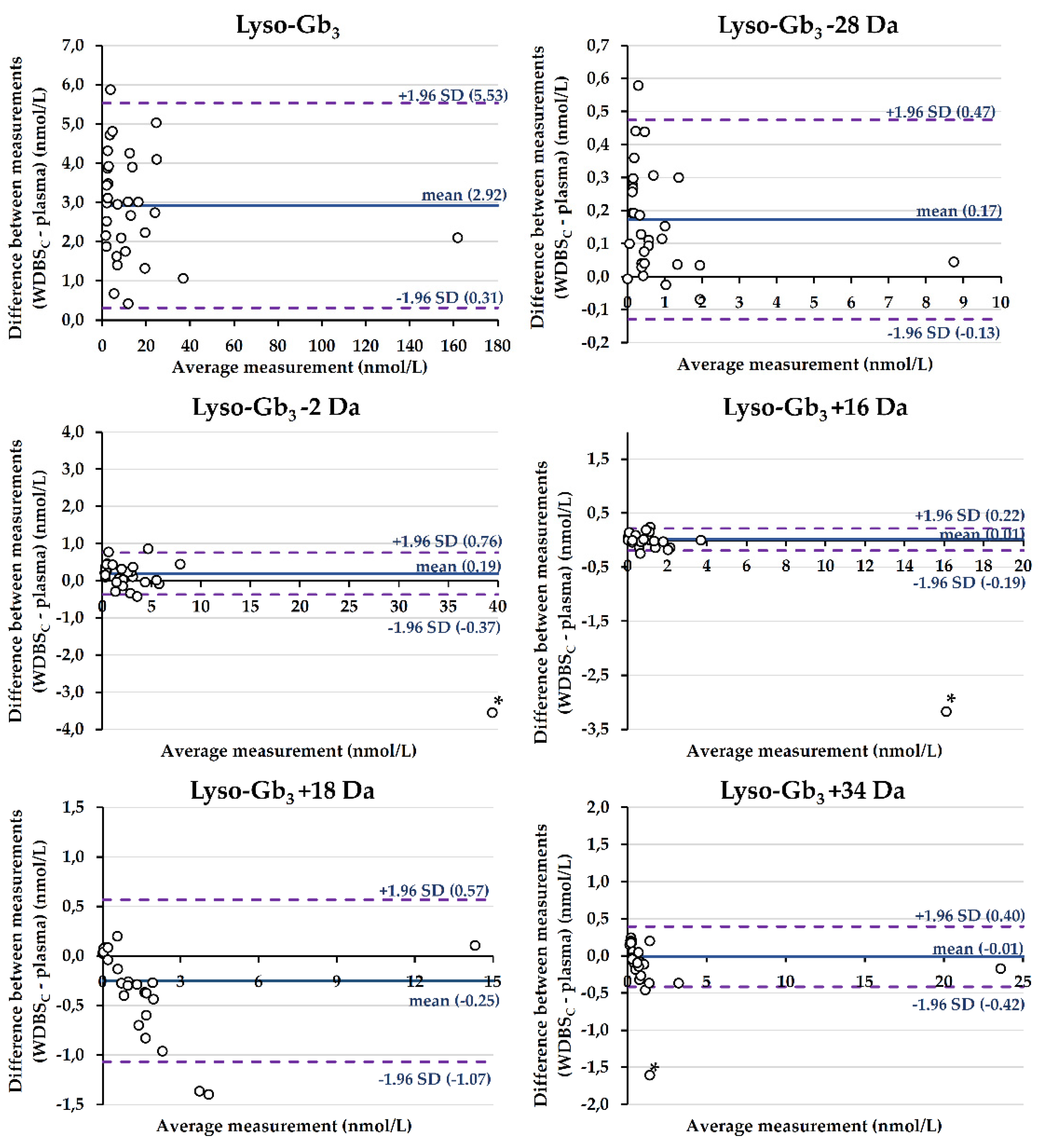
2.3.1. Comparison of Biomarker Levels between Plasma and W-DBSC Matrices
2.3.2. Comparison of Biomarker Levels between Venous and Capillary Blood Matrices
2.3.3. Evaluation of the Hematocrit Effect
3. Discussion
4. Materials and Methods
4.1. Subject Selection
4.2. Sample Collection and Storage
4.3. Reagents
4.4. Quality Controls and Calibration Curves
4.5. Sample Preparation
4.6. UHPLC-MS/MS Analysis
4.7. Method Validation
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clarke, J.T. Narrative review: Fabry disease. Ann. Intern. Med. 2007, 146, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Najafian, B.; Tøndel, C.; Svarstad, E.; Gubler, M.-C.; Oliveira, J.-P.; Mauer, M. Accumulation of globotriaosylceramide in podocytes in Fabry nephropathy is associated with progressive podocyte loss. J. Am. Soc. Nephrol. 2020, 31, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.-J.; Chang, F.-P.; Lu, Y.-H.; Hung, S.-C.; Wang, Y.-C.; Yang, A.-H.; Lee, H.-J.; Sung, S.-H.; Wang, Y.-F.; Yu, W.-C.; et al. Identification of lysosomal and extralysosomal globotriaosylceramide (Gb3) accumulations before the occurrence of typical pathological changes in the endomyocardial biopsies of Fabry disease patients. Genet. Med. 2019, 21, 22–232. [Google Scholar] [CrossRef]
- Toupin, A.; Lavoie, P.; Arthus, M.F.; Abaoui, M.; Boutin, M.; Fortier, C.; Ménard, C.; Bichet, D.G.; Auray-Blais, C. Analysis of globotriaosylceramide (Gb3) isoforms/analogs in unfractionated leukocytes, B lymphocytes and monocytes from Fabry patients using ultra-high performance liquid chromatography/tandem mass spectrometry. Anal. Chim. Acta 2018, 1015, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Manwaring, V.; Boutin, M.; Auray-Blais, C. A Metabolomic study to identify new globotriaosylceramide-related biomarkers in the plasma of Fabry disease patients. Anal. Chem. 2013, 85, 9039–9048. [Google Scholar] [CrossRef] [PubMed]
- Auray-Blais, C.; Boutin, M. Novel gb(3) isoforms detected in urine of fabry disease patients: A metabolomic study. Curr. Med. Chem. 2012, 19, 3241–3252. [Google Scholar] [CrossRef] [PubMed]
- Auray-Blais, C.; Boutin, M.; Gagnon, R.; Dupont, F.O.; Lavoie, P.; Clarke, J.T. Urinary globotriaosylsphingosine-related biomarkers for Fabry disease targeted by metabolomics. Anal. Chem. 2012, 84, 2745–2753. [Google Scholar] [CrossRef]
- Dupont, F.O.; Gagnon, R.; Boutin, M.; Auray-Blais, C. A metabolomic study reveals novel plasma lyso-Gb3 analogs as Fabry disease biomarkers. Curr. Med. Chem. 2013, 20, 280–288. [Google Scholar] [CrossRef]
- Boutin, M.; Auray-Blais, C. Metabolomic discovery of novel urinary galabiosylceramide analogs as Fabry disease biomarkers. J. Am. Soc. Mass Spectrom. 2015, 26, 499–510. [Google Scholar] [CrossRef]
- Touboul, D.; Roy, S.; Germain, D.P.; Baillet, A.; Brion, F.; Prognon, P.; Chaminade, P.; Laprévote, O. Fast fingerprinting by MALDI-TOF mass spectrometry of urinary sediment glycosphingolipids in Fabry disease. Anal. Bioanal. Chem. 2005, 382, 1209–1216. [Google Scholar] [CrossRef]
- Muntean, C.; Starcea, I.M.; Stoica, C.; Banescu, C. Clinical characteristics, renal involvement, and therapeutic options of pediatric patients with Fabry disease. Front. Pediatr. 2022, 10, 908657. [Google Scholar] [CrossRef] [PubMed]
- Paim-Marques, L.; de Oliveira, R.J.; Appenzeller, S. Multidisciplinary management of Fabry disease: Current perspectives. J. Multidiscip. Healthc. 2022, 15, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Lelis, A.; Mirocha, J.; Wilcox, W.R. Heterozygous Fabry women are not just carriers, but have a significant burden of disease and impaired quality of life. Genet. Med. 2007, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Lenders, M.; Brand, E. Fabry disease: The current treatment landscape. Drugs 2021, 81, 635–645. [Google Scholar] [CrossRef]
- Weidemann, F.; Jovanovic, A.; Herrmann, K.; Vardarli, I. Chaperone therapy in Fabry disease. Int. J. Mol. Sci. 2022, 23, 1887. [Google Scholar] [CrossRef] [PubMed]
- Domm, J.M.; Wootton, S.K.; Medin, J.A.; West, M.L. Gene therapy for Fabry disease: Progress, challenges, and outlooks on gene-editing. Mol. Genet. Metab. 2021, 134, 117–131. [Google Scholar] [CrossRef]
- van der Veen, S.J.; Hollak, C.E.M.; van Kuilenburg, A.B.P.; Langeveld, M. Developments in the treatment of Fabry disease. J. Inherit. Metab. Dis. 2020, 43, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, O.; Gago, M.F.; Miltenberger-Miltenyi, G.; Sousa, N.; Cunha, D. Fabry disease therapy: State-of-the-art and current challenges. Int. J. Mol. Sci. 2020, 22, 206. [Google Scholar] [CrossRef]
- Boutin, M.; Auray-Blais, C. Multiplex tandem mass spectrometry analysis of novel plasma lyso-Gb3-related analogues in Fabry disease. Anal. Chem. 2014, 86, 3476–3483. [Google Scholar] [CrossRef]
- Auray-Blais, C.; Lavoie, P.; Boutin, M.; Ntwari, A.; Hsu, T.R.; Huang, C.K.; Niu, D.M. Biomarkers associated with clinical manifestations in Fabry disease with a late-onset cardiac variant mutation. Clin. Chim. Acta 2017, 466, 185–193. [Google Scholar] [CrossRef]
- Nowak, A.; Beuschlein, F.; Sivasubramaniam, V.; Kasper, D.; Warnock, D.G. Lyso-Gb3 associates with adverse long-term outcome in patients with Fabry disease. J. Med. Genet. 2022, 59, 287–293. [Google Scholar] [CrossRef]
- Lenders, M.; Stappers, F.; Niemietz, C.; Schmitz, B.; Boutin, M.; Ballmaier, P.J.; Zibert, A.; Schmidt, H.; Brand, S.-M.; Auray-Blais, C.; et al. Mutation-specific Fabry disease patient-derived cell model to evaluate the amenability to chaperone therapy. J. Med. Genet. 2019, 56, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P.; Altarescu, G.; Barriales-Villa, R.; Mignani, R.; Pawlaczyk, K.; Pieruzzi, F.; Terryn, W.; Vujkovac, B.; Ortiz, A. An expert consensus on practical clinical recommendations and guidance for patients with classic Fabry disease. Mol. Genet. Metab. 2022, 137, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Malvagia, S.; Ferri, L.; Della Bona, M.; Borsini, W.; Cirami, C.L.; Dervishi, E.; Feriozzi, S.; Gasperini, S.; Motta, S.; Mignani, R.; et al. Multicenter evaluation of use of dried blood spot compared to conventional plasma in measurements of globotriaosylsphingosine (LysoGb3) concentration in 104 Fabry patients. Clin. Chem. Lab. Med. 2021, 59, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Delarosa-Rodríguez, R.; Santotoribio, J.D.; Paula, H.A.; González-Meneses, A.; García-Morillo, S.; Jiménez-Arriscado, P.; Guerrero, J.M.; Macher, H.C. Accuracy diagnosis improvement of Fabry disease from dried blood spots: Enzyme activity, lyso-Gb3 accumulation and GLA gene sequencing. Clin. Genet. 2021, 99, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Olivera, S.; Iñiguez, C.; García-Fernández, L.; Sierra, J.L.; Camón, A.M.; Menao, S.; Torralba, M.Á. Usefulness of lyso-globotriaosylsphingosine in dried blood spots in the differential diagnosis between multiple sclerosis and Anderson-Fabry’s disease. Mult. Scler. Relat. Disord. 2020, 38, 101466. [Google Scholar] [CrossRef]
- Nowak, A.; Mechtler, T.; Kasper, D.C.; Desnick, R.J. Correlation of Lyso-Gb3 levels in dried blood spots and sera from patients with classic and later-onset Fabry disease. Mol. Genet. Metab. 2017, 121, 320–324. [Google Scholar] [CrossRef]
- Gatterer, C.; Gaggl, M.; Mundigler, G.; Rommer, P.; Graf, S.; Sunder-Plassmann, G. Agreement of dried blood spot lyso-Gb3 concentrations obtained from different laboratories in patients with Fabry disease. Clin. Chem. Lab. Med. 2020, 58, e275–e278. [Google Scholar] [CrossRef]
- Zakaria, R.; Allen, K.J.; Koplin, J.J.; Roche, P.; Greaves, R.F. Advantages and challenges of dried blood spot analysis by mass spectrometry across the total testing process. EJIFCC 2016, 27, 288–317. [Google Scholar]
- Ostler, M.W.; Porter, J.H.; Buxton, O.M. Dried blood spot collection of health biomarkers to maximize participation in population studies. J. Vis. Exp. 2014, 83, e50973. [Google Scholar] [CrossRef]
- Malsagova, K.; Kopylov, A.; Stepanov, A.; Butkova, T.; Izotov, A.; Kaysheva, A. Dried blood spot in the laboratory: Directions and prospects. Diagnostics 2020, 10, 248. [Google Scholar] [CrossRef]
- Su, X.; Carlson, B.F.; Wang, X.; Li, X.; Zhang, Y.; Montgomery, J.P.; Ding, Y.; Wagner, A.L.; Gillespie, B.; Boulton, M.L. Dried blood spots: An evaluation of utility in the field. J. Infect. Public Health 2018, 11, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Velghe, S.; Delahaye, L.; Stove, C.P. Is the hematocrit still an issue in quantitative dried blood spot analysis? J. Pharm. Biomed. Anal. 2019, 163, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Neto, R.; Gooley, A.; Breadmore, M.C.; Hilder, E.F.; Lapierre, F. Precise, accurate and user-independent blood collection system for dried blood spot sample preparation. Anal. Bioanal. Chem. 2018, 410, 3315–3323. [Google Scholar] [CrossRef] [PubMed]
- Carling, R.S.; Emmett, E.C.; Moat, S.J. Evaluation of volumetric collection devices for the measurement of phenylalanine and tyrosine to monitor patients with phenylketonuria. Clin. Chim. Acta 2022, 535, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Mazzarino, M.; Di Costanzo, L.; Comunità, F.; Stacchini, C.; de la Torre, X.; Botrè, F. UHPLC-HRMS method for the simultaneous screening of 235 drugs in capillary blood for doping control purpose: Comparative evaluation of volumetric and non-volumetric dried blood spotting devices. ACS Omega 2022, 7, 31845–31868. [Google Scholar] [CrossRef]
- Delahaye, L.; Veenhof, H.; Koch, B.C.P.; Alffenaar, J.C.; Linden, R.; Stove, C. Alternative sampling devices to collect dried blood microsamples: State-of-the-art. Ther. Drug Monit. 2021, 43, 310–321. [Google Scholar] [CrossRef]
- Velghe, S.; Stove, C.P. Evaluation of the Capitainer-B microfluidic device as a new hematocrit-independent alternative for dried blood spot collection. Anal. Chem. 2018, 90, 12893–12899. [Google Scholar] [CrossRef]
- Lavoie, P.; Boutin, M.; Auray-Blais, C. Multiplex analysis of novel urinary lyso-Gb3-related biomarkers for Fabry disease by tandem mass spectrometry. Anal. Chem. 2013, 85, 1743–1752. [Google Scholar] [CrossRef]
- Polo, G.; Burlina, A.P.; Ranieri, E.; Colucci, F.; Rubert, L.; Pascarella, A.; Duro, G.; Tummolo, A.; Padoan, A.; Plebani, M.; et al. Plasma and dried blood spot lysosphingolipids for the diagnosis of different sphingolipidoses: A comparative study. Clin. Chem. Lab. Med. 2019, 57, 1863–1874. [Google Scholar] [CrossRef]
| Whatman-GE 903 | Capitainer®B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sLQC | sMQC | sHQC | sLQC | sMQC | sHQC | |||||||
| nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | |
| Spiked concentration | 0.75 | na | 75.00 | na | 250.00 | na | 0.75 | na | 75.00 | na | 250.00 | na |
| Replicate 1 | 0.79 | 4.7 | 82.93 | 10.6 | 261.79 | 4.7 | 0.79 | 5.2 | 75.56 | 0.7 | 272.94 | 9.2 |
| Replicate 2 | 0.80 | 6.5 | 87.37 | 16.5 | 262.81 | 5.1 | 0.73 | −2.9 | 101.79 | 35.7 | 261.68 | 4.7 |
| Replicate 3 | 0.76 | 0.9 | 80.88 | 7.8 | 261.57 | 4.6 | 0.80 | 6.5 | 79.88 | 6.5 | 255.61 | 2.2 |
| Replicate 4 | 0.78 | 4.4 | 78.81 | 5.1 | 266.48 | 6.6 | 0.70 | −6.5 | 84.77 | 13.0 | 265.91 | 6.4 |
| Replicate 5 | 0.86 | 14.3 | 81.09 | 8.1 | 281.55 | 12.6 | 0.74 | −1.6 | 79.18 | 5.6 | 254.11 | 1.6 |
| Mean | 0.80 | 6.2 | 82.21 | 9.6 | 266.84 | 6.7 | 0.75 | 0.1 | 84.23 | 12.3 | 262.05 | 4.8 |
| SD | 0.04 | na | 3.23 | na | 8.46 | na | 0.04 | na | 10.35 | na | 7.71 | na |
| RSD% | 4.67 | na | 3.93 | na | 3.17 | na | 5.55 | na | 12.28 | na | 2.94 | na |
| Bias% min | na | 0.9 | na | 5.1 | na | 4.6 | na | −6.5 | na | 0.7 | na | 1.6 |
| Bias% max | na | 14.3 | na | 16.5 | na | 12.6 | na | 6.5 | na | 35.7 | na | 9.2 |
| Lyso-Gb3 Analogues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyso-Gb3 | −28 Da | −2 Da | +16 Da | +18 Da | +34 Da | ||||||||
| pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | ||
| nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | ||
| (A) Whatman- GE 903 | Replicate 1 | 47.93 | 126.95 | 2.24 | 7.34 | 9.27 | 27.09 | 3.33 | 11.68 | 2.73 | 9.89 | 4.34 | 16.67 |
| Replicate 2 | 45.75 | 120.97 | 2.38 | 7.10 | 8.52 | 25.63 | 3.81 | 11.43 | 2.42 | 10.11 | 3.99 | 15.84 | |
| Replicate 3 | 54.67 | 122.34 | 2.99 | 7.38 | 10.62 | 27.25 | 4.20 | 11.74 | 3.12 | 10.03 | 4.90 | 16.19 | |
| Replicate 4 | 49.73 | 114.15 | 2.33 | 6.83 | 9.16 | 23.91 | 3.65 | 10.41 | 2.64 | 9.49 | 4.49 | 15.04 | |
| Replicate 5 | 51.98 | 114.80 | 2.80 | 6.47 | 9.93 | 23.29 | 3.85 | 9.81 | 2.78 | 9.59 | 4.79 | 16.02 | |
| Mean | 50.01 | 119.84 | 2.55 | 7.02 | 9.50 | 25.43 | 3.77 | 11.01 | 2.74 | 9.82 | 4.50 | 15.95 | |
| SD | 3.47 | 5.38 | 0.33 | 0.38 | 0.80 | 1.80 | 0.32 | 0.86 | 0.25 | 0.27 | 0.36 | 0.60 | |
| RSD% | 6.9 | 4.5 | 12.8 | 5.4 | 8.4 | 7.1 | 8.4 | 7.8 | 9.3 | 2.8 | 8.0 | 3.7 | |
| (B) Capitainer®B | Replicate 1 | 67.70 | 173.59 | 4.54 | 11.04 | 15.80 | 40.05 | 6.87 | 18.24 | 5.31 | 16.04 | 8.66 | 24.76 |
| Replicate 2 | 70.01 | 189.64 | 4.36 | 11.41 | 14.74 | 40.74 | 6.15 | 18.04 | 5.96 | 16.95 | 8.70 | 25.05 | |
| Replicate 3 | 77.03 | 184.55 | 4.37 | 11.79 | 15.47 | 42.14 | 6.29 | 18.50 | 6.85 | 16.17 | 9.57 | 25.77 | |
| Replicate 4 | 73.57 | 190.14 | 4.13 | 11.40 | 14.95 | 42.13 | 6.17 | 18.81 | 5.85 | 19.35 | 9.34 | 28.32 | |
| Replicate 5 | 75.43 | 185.82 | 3.99 | 11.71 | 14.98 | 42.51 | 6.30 | 18.87 | 6.52 | 19.47 | 9.26 | 29.27 | |
| Mean | 72.75 | 184.75 | 4.28 | 11.47 | 15.19 | 41.51 | 6.36 | 18.49 | 6.10 | 17.59 | 9.10 | 26.63 | |
| SD | 3.85 | 6.68 | 0.22 | 0.30 | 0.43 | 1.06 | 0.29 | 0.36 | 0.60 | 1.69 | 0.41 | 2.03 | |
| RSD% | 5.3 | 3.6 | 5.1 | 2.6 | 2.9 | 2.6 | 4.6 | 1.9 | 9.9 | 9.6 | 4.5 | 7.6 | |
| Whatman-GE 903 | Capitainer®B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sLQC | sMQC | sHQC | sLQC | sMQC | sHQC | |||||||
| nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | nM | Bias% | |
| Spiked concentration | 0.75 | na | 75.00 | na | 250.00 | na | 0.75 | na | 75.00 | na | 250.00 | na |
| Day 1 | 0.80 | 6.2 | 82.21 | 9.6 | 266.84 | 6.7 | 0.75 | 0.1 | 84.23 | 12.3 | 262.05 | 4.8 |
| Day 2 | 0.79 | 5.1 | 84.00 | 12.0 | 272.55 | 9.0 | 0.80 | 6.4 | 73.06 | −2.6 | 260.73 | 4.3 |
| Day 3 | 0.76 | 0.9 | 77.69 | 3.6 | 270.24 | 8.1 | 0.80 | 6.3 | 80.39 | 7.2 | 271.74 | 8.7 |
| Day 4 | 0.76 | 1.1 | 77.38 | 3.2 | 239.97 | −4.0 | 0.68 | −9.9 | 72.95 | −2.7 | 262.98 | 5.2 |
| Day 5 | 0.80 | 6.2 | 78.29 | 4.4 | 254.29 | 1.7 | 0.73 | −2.7 | 82.95 | 10.6 | 256.45 | 2.6 |
| Mean | 0.78 | 3.9 | 79.91 | 6.6 | 260.78 | 4.3 | 0.75 | 0.0 | 78.72 | 5.0 | 262.79 | 5.1 |
| SD | 0.02 | na | 3.00 | na | 13.60 | na | 0.05 | na | 5.40 | na | 5.60 | na |
| RSD% | 2.6 | na | 3.8 | na | 5.2 | na | 6.8 | na | 6.9 | na | 2.1 | na |
| Bias% minimum | na | 0.9 | na | 3.2 | na | −4.0 | na | −9.9 | na | −2.7 | na | 2.6 |
| Bias% maximum | na | 6.2 | na | 12.0 | na | 9.0 | na | 6.4 | na | 12.3 | na | 8.7 |
| Lyso-Gb3 Analogues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyso-Gb3 | −28 Da | −2 Da | +16 Da | +18 Da | +34 Da | ||||||||
| pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | ||
| nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | nM | ||
| (A) Whatman- GE 903 | Day 1 | 50.01 | 119.84 | 2.55 | 7.02 | 9.50 | 25.43 | 3.77 | 11.01 | 2.74 | 9.82 | 4.50 | 15.95 |
| Day 2 | 44.51 | 122.23 | 2.38 | 7.06 | 8.35 | 25.50 | 3.55 | 11.11 | 2.83 | 8.50 | 4.09 | 13.75 | |
| Day 3 | 40.91 | 115.93 | 2.05 | 5.87 | 7.26 | 22.49 | 3.46 | 10.81 | 2.48 | 9.45 | 4.72 | 15.22 | |
| Day 4 | 40.53 | 113.24 | 2.04 | 6.20 | 7.60 | 24.10 | 3.96 | 13.34 | 2.85 | 7.51 | 5.04 | 14.98 | |
| Day 5 | 40.53 | 102.58 | 2.04 | 5.50 | 7.44 | 21.36 | 3.79 | 12.67 | 2.75 | 6.49 | 4.76 | 13.35 | |
| Mean | 43.30 | 114.76 | 2.21 | 6.33 | 8.03 | 23.78 | 3.71 | 11.79 | 2.73 | 8.35 | 4.62 | 14.65 | |
| SD | 4.11 | 7.64 | 0.24 | 0.69 | 0.92 | 1.82 | 0.20 | 1.14 | 0.15 | 1.37 | 0.35 | 1.07 | |
| RSD% | 9.5 | 6.7 | 10.7 | 11.0 | 11.5 | 7.7 | 5.4 | 9.7 | 5.4 | 16.4 | 7.6 | 7.3 | |
| (B) Capitainer®B | Day 1 | 72.75 | 184.75 | 4.28 | 11.47 | 15.19 | 41.51 | 6.36 | 18.49 | 6.10 | 17.59 | 9.10 | 26.63 |
| Day 2 | 66.06 | 194.13 | 3.66 | 10.37 | 13.50 | 38.14 | 5.59 | 15.69 | 5.89 | 19.45 | 8.39 | 26.80 | |
| Day 3 | 70.66 | 186.87 | 3.70 | 9.66 | 14.02 | 38.99 | 6.70 | 18.68 | 4.22 | 11.66 | 7.22 | 23.71 | |
| Day 4 | 75.98 | 197.24 | 3.80 | 9.62 | 15.09 | 38.51 | 7.71 | 19.81 | 4.57 | 13.21 | 7.69 | 22.67 | |
| Day 5 | 78.69 | 196.27 | 3.43 | 10.29 | 15.54 | 42.21 | 6.94 | 20.63 | 6.87 | 19.08 | 10.07 | 29.24 | |
| Mean | 72.83 | 191.85 | 3.77 | 10.28 | 14.67 | 39.87 | 6.66 | 18.66 | 5.53 | 16.20 | 8.49 | 25.81 | |
| SD | 4.87 | 5.68 | 0.31 | 0.75 | 0.87 | 1.86 | 0.78 | 1.88 | 1.10 | 3.55 | 1.13 | 2.63 | |
| RSD% | 6.7 | 3.0 | 8.3 | 7.3 | 5.9 | 4.7 | 11.7 | 10.1 | 20.0 | 21.9 | 13.3 | 10.2 | |
| LOD | LOQ | |||||
|---|---|---|---|---|---|---|
| Whatman-GE 903 | Capitainer®B | Plasma | Whatman-GE 903 | Capitainer®B | Plasma | |
| nM | nM | nM | nM | nM | nM | |
| Lyso-Gb3-13C6 | 0.21 | 0.40 | na | 0.70 | 1.34 | na |
| Lyso-Gb3 | 0.32 | 0.37 | 0.23 | 1.08 | 1.23 | 0.77 |
| Lyso-Gb3 (Analogue − 28 Da) | 0.11 | 0.18 | 0.06 | 0.36 | 0.59 | 0.21 |
| Lyso-Gb3 (Analogue −2 Da) | 0.22 | 0.15 | 0.29 | 0.73 | 0.55 | 0.97 |
| Lyso-Gb3 (Analogue +16 Da) | 0.10 | 0.39 | 0.22 | 0.32 | 1.29 | 0.72 |
| Lyso-Gb3 (Analogue +18 Da) | 0.13 | 0.23 | 0.14 | 0.42 | 0.77 | 0.47 |
| Lyso-Gb3 (Analogue +34 Da) | 0.13 | 0.19 | 0.24 | 0.43 | 0.64 | 0.79 |
| DBS Extraction Recovery (%) | SPE Recovery (%) | |||
|---|---|---|---|---|
| Whatman-GE 903 | Capitainer®B | Whatman-GE 903 | Capitainer®B | |
| sLQC | 69 | 68 | 71 | 72 |
| sMQC | 71 | 66 | 72 | 69 |
| sHQC | 66 | 68 | 74 | 74 |
| Whatman-GE 903 | Capitainer®B | |||||
|---|---|---|---|---|---|---|
| sLQC | sMQC | sHQC | sLQC | sMQC | sHQC | |
| Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | |
| Room temperature, 22 °C (7 days) | −9.6 | −1.3 | −4.2 | −8.3 | −1.9 | −4.3 |
| Room temperature, 22 °C (14 days) | −20.5 | −3.2 | −3.0 | −3.2 | −7.8 | 0.7 |
| Refrigerator, 4 °C (38 days) | −8.3 | −6.9 | −2.2 | 15.3 | 3.1 | −3.4 |
| Freezer, −20 °C (208 days) | −6.3 | −0.3 | −1.2 | 3.8 | 1.2 | −1.0 |
| Freeze/thaw cycles (n = 5) | −8.7 | 6.4 | 1.9 | 10.3 | −0.2 | 1.2 |
| UHPLC Autosampler, 4 °C (48 h) | 13.8 | −1.9 | −8.4 | −9.1 | −2.4 | −2.7 |
| Lyso-Gb3 Analogues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyso-Gb3 | −28 Da | −2 Da | +16 Da | +18 Da | +34 Da | ||||||||
| pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | pLQC | pHQC | ||
| Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | Bias% | ||
| (A) Whatman- GE 903 | RT, 22 °C (7 days) | 1.9 | −4.1 | 2.9 | −4.1 | 4.6 | 3.3 | −1.1 | 3.7 | 10.2 | −2.7 | 0.8 | −0.5 |
| RT, 22 °C (14 days) | −9.2 | 4.2 | −10.3 | 2.6 | −10.4 | 5.7 | −12.6 | 3.1 | 11.4 | 15.6 | −6.6 | 5.5 | |
| Refrigerator, 4 °C (38 days) | −5.0 | 10.8 | 2.2 | 12.0 | −1.1 | 10.9 | 10.5 | 7.5 | −12.7 | 12.0 | −3.8 | 13.7 | |
| Freezer, −20 °C (208 days) | −4.9 | −4.9 | −25.4 | −19.6 | −19.8 | −7.9 | −14.8 | −9.6 | 7.4 | −15.4 | 7.5 | −4.0 | |
| Freeze/thaw cycles (n = 5) | −16.0 | 6.8 | −18.9 | 5.2 | −17.5 | −1.4 | −23.4 | 0.9 | −7.6 | 15.0 | −19.6 | 5.2 | |
| UHPLC Autosampler, 4 °C (24 h) | −2.0 | −2.1 | 2.3 | −4.3 | −1.6 | −3.7 | −4.2 | −2.0 | 27.3 | 16.9 | 18.1 | 12.9 | |
| UHPLC Autosampler, 4 °C (48 h) | −9.4 | −16.9 | −13.8 | −14.4 | −14.8 | −16.7 | −13.1 | −14.4 | 8.2 | −1.7 | 2.6 | −2.6 | |
| (B) Capitainer®B | RT, 22 °C (7 days) | −4.2 | 0.8 | −0.4 | 3.6 | −0.2 | −0.6 | −1.7 | 6.6 | −7.1 | 4.5 | 2.3 | −0.7 |
| RT, 22 °C (14 days) | −2.7 | −5.2 | 1.6 | −6.6 | 0.3 | −2.3 | −8.2 | −10.0 | 7.0 | −0.9 | 6.6 | −0.5 | |
| Refrigerator, 4 °C (38 days) | −6.0 | −3.0 | −8.5 | −2.8 | −8.4 | −1.2 | 1.0 | −7.0 | −3.1 | 1.6 | −9.1 | −5.1 | |
| Freezer, −20 °C (208 days) | −1.3 | 0.0 | −18.7 | −19.5 | −1.6 | −5.5 | −7.1 | −12.7 | −7.7 | 1.0 | −4.8 | −6.2 | |
| Freeze/thaw cycles (n = 5) | −1.3 | 0.6 | 14.7 | −4.0 | 6.7 | 0.7 | 2.1 | −4.8 | 1.9 | 30.1 | 9.1 | 19.8 | |
| UHPLC Autosampler, 4 °C (24 h) | 0.2 | 1.1 | 4.5 | −0.1 | −2.2 | −1.3 | 0.1 | 0.2 | 11.0 | 15.5 | 23.3 | 16.7 | |
| UHPLC Autosampler, 4 °C (48 h) | −3.9 | −1.8 | 17.4 | 9.8 | 10.8 | 7.8 | 14.0 | 10.0 | 16.1 | 25.9 | 36.4 | 32.4 | |
| Biomarker | Blood Type | Collection Method | Mean | Min | Max | Median | 99th Percentile | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|---|
| Lyso-Gb3 (nM) | Venous | Whatman 903 | 3.96 | 2.25 | 6.25 | 3.86 | 6.14 | 100 | 92 |
| Capitainer®B | 4.60 | 3.16 | 8.43 | 4.18 | 8.19 | 85 | 92 | ||
| Capillary | Whatman 903 | 4.26 | 2.74 | 6.71 | 4.20 | 6.61 | 95 | 92 | |
| Capitainer®B | 4.15 | 2.95 | 5.93 | 4.19 | 5.92 | 100 | 100 | ||
| Plasma | EDTA K2 | 0.74 | 0.33 | 1.09 | 0.78 | 1.08 | 100 | 92 | |
| Lyso-Gb3 −28 Da (nM) | Venous | Whatman 903 | 0.25 | nd | 0.53 | 0.26 | 0.51 | 55 | 92 |
| Capitainer®B | nd | nd | 0.42 | nd | 0.40 | 85 | 92 | ||
| Capillary | Whatman 903 | 0.28 | nd | 0.58 | 0.27 | 0.56 | 60 | 92 | |
| Capitainer®B | nd | nd | 0.24 | nd | 0.24 | 100 | 92 | ||
| Plasma | EDTA K2 | nd | nd | nd | nd | nd | 100 | 92 | |
| Lyso-Gb3 −2 Da (nM) | Venous | Whatman 903 | 0.49 | 0.33 | 0.79 | 0.44 | 0.78 | 100 | 92 |
| Capitainer®B | 0.60 | 0.41 | 1.16 | 0.53 | 1.11 | 95 | 92 | ||
| Capillary | Whatman 903 | 0.56 | 0.35 | 1.03 | 0.53 | 1.00 | 100 | 92 | |
| Capitainer®B | 0.55 | 0.36 | 0.82 | 0.52 | 0.80 | 100 | 92 | ||
| Plasma | EDTA K2 | nd | nd | 0.31 | nd | 0.30 | 100 | 92 | |
| Lyso-Gb3 +16 Da (nM) | Venous | Whatman 903 | nd | nd | 0.21 | nd | 0.20 | 95 | 92 |
| Capitainer®B | nd | nd | nd | nd | nd | 80 | 100 | ||
| Capillary | Whatman 903 | nd | nd | nd | nd | nd | 100 | 100 | |
| Capitainer®B | nd | nd | nd | nd | nd | 84 | 100 | ||
| Plasma | EDTA K2 | nd | nd | nd | nd | nd | 95 | 100 | |
| Lyso-Gb3 +18 Da (nM) | Venous | Whatman 903 | nd | nd | nd | nd | nd | 100 | 100 |
| Capitainer®B | nd | nd | nd | nd | nd | 100 | 100 | ||
| Capillary | Whatman 903 | nd | nd | nd | nd | nd | 100 | 100 | |
| Capitainer®B | nd | nd | nd | nd | nd | 100 | 100 | ||
| Plasma | EDTA K2 | nd | nd | nd | nd | nd | 100 | 100 | |
| Lyso-Gb3 +34 Da (nM) | Venous | Whatman 903 | 0.28 | 0.17 | 0.39 | 0.25 | 0.38 | 90 | 92 |
| Capitainer®B | nd | nd | 0.25 | nd | 0.25 | 90 | 92 | ||
| Capillary | Whatman 903 | 0.30 | 0.21 | 0.35 | 0.30 | 0.35 | 85 | 100 | |
| Capitainer®B | nd | nd | 0.20 | nd | 0.20 | 95 | 92 | ||
| Plasma | EDTA K2 | nd | nd | nd | nd | nd | 100 | 92 |
| Biomarker | Slope | 95% CI (Slope) | Y-Intercept (nM) | 95% CI (Y-Intercept) | n |
|---|---|---|---|---|---|
| Lyso-Gb3 | 0.993 | 0.956–1.029 | 3.019 | 2.434–3.604 | 32 |
| Lyso-Gb3 −28 Da | 0.973 | 0.800–1.147 | 0.192 | 0.101–0.283 | 32 |
| Lyso-Gb3 −2 Da | 0.910 | 0.775–1.046 | 0.365 | 0.085–0.646 | 32 |
| Lyso-Gb3 +16 Da | 0.825 | 0.547–1.102 | 0.124 | −0.057–0.306 | 32 |
| Lyso-Gb3 +18 Da | 0.962 | 0.421–1.504 | −0.195 | −0.724–0.334 | 32 |
| Lyso-Gb3 +34 Da | 0.988 | 0.454–1.523 | −0.045 | −0.402–0.312 | 32 |
| Biomarker | Collection Method | Slope | 95% CI (Slope) | Y-Intercept (nM) | 95% CI (Y-Intercept) | n |
|---|---|---|---|---|---|---|
| Lyso-Gb3 | Whatman 903 | 1.243 | 0.650–1.837 | −2.576 | −9.022–3.869 | 32 |
| Capitainer®B | 1.134 | 0.867–1.401 | −1.678 | −4.638–1.282 | 31 | |
| Lyso-Gb3 −28 Da | Whatman 903 | 1.288 | 0.598–1.977 | −0.153 | −0.556–0.250 | 32 |
| Capitainer®B | 1.178 | 1.028–1.327 | −0.066 | −0.156–0.023 | 31 | |
| Lyso-Gb3 −2 Da | Whatman 903 | 1.301 | 0.564–2.037 | −0.643 | −2.203–0.917 | 32 |
| Capitainer®B | 1.119 | 0.933–1.305 | −0.262 | −0.670–0.147 | 31 | |
| Lyso-Gb3 +16 Da | Whatman 903 | 1.233 | 0.617–1.850 | −0.183 | −0.595–0.229 | 32 |
| Capitainer®B | 1.070 | 0.853–1.286 | −0.055 | −0.209–0.099 | 31 | |
| Lyso-Gb3 +18 Da | Whatman 903 | 1.193 | 0.856–1.531 | −0.170 | −0.437–0.098 | 32 |
| Capitainer®B | 0.979 | 0.762–1.197 | 0.007 | −0.184–0.199 | 31 | |
| Lyso-Gb3 +34 Da | Whatman 903 | 1.225 | 0.548–1.903 | −0.170 | −0.595–0.255 | 32 |
| Capitainer®B | 1.077 | 1.058–1.097 | −0.010 | −0.055–0.035 | 31 |
| Biomarker | Collection Method | Slope | 95% CI (Slope) | Y-Intercept (nM) | 95% CI (Y-Intercept) | r2 | p-Value | n |
|---|---|---|---|---|---|---|---|---|
| Lyso-Gb3 | Whatman 903 | 8.107 | −5.019–21.230 | −0.466 | −5.971–5.040 | 0.050 | 0.217 | 32 |
| Capitainer®B | −7.438 | −23.140–8.261 | 6.358 | −0.214–12.930 | 0.031 | 0.341 | 31 | |
| Lyso-Gb3 −28 Da | Whatman 903 | 1.272 | −0.217–2.762 | −0.358 | −0.983–0.266 | 0.092 | 0.091 | 32 |
| Capitainer®B | −0.439 | −1.538–0.660 | 0.309 | −0.149–0.767 | 0.023 | 0.420 | 30 | |
| Lyso-Gb3 −2 Da | Whatman 903 | −0.070 | −3.106–2.965 | 0.222 | −1.046–1.489 | 0.000 | 0.963 | 31 |
| Capitainer®B | −1.937 | −5.551–1.677 | 0.993 | −0.513–2.499 | 0.041 | 0.282 | 30 | |
| Lyso-Gb3 +16 Da | Whatman 903 | 0.130 | −0.967–1.226 | −0.041 | −0.499–0.417 | 0.002 | 0.811 | 31 |
| Capitainer®B | −1.605 | −2.853–(−0.357) | 0.702 | 0.182–1.222 | 0.199 | 0.014 | 30 | |
| Lyso-Gb3 +18 Da | Whatman 903 | 5.287 | 1.552–9.022 | −2.460 | −4.027–(−0.893) | 0.218 | 0.007 | 32 |
| Capitainer®B | 1.715 | −0.403–3.832 | −0.790 | −1.673–0.092 | 0.089 | 0.108 | 30 | |
| Lyso-Gb3 +34 Da | Whatman 903 | 1.434 | −0.608–3.475 | −0.610 | −1.468–0.247 | 0.066 | 0.162 | 31 |
| Capitainer®B | 0.981 | −0.805–2.767 | −0.465 | −1.210–0.279 | 0.043 | 0.270 | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boutin, M.; Lavoie, P.; Beaudon, M.; Kabala Ntumba, G.; Bichet, D.G.; Maranda, B.; Auray-Blais, C. Mass Spectrometry Analysis of Globotriaosylsphingosine and Its Analogues in Dried Blood Spots. Int. J. Mol. Sci. 2023, 24, 3223. https://doi.org/10.3390/ijms24043223
Boutin M, Lavoie P, Beaudon M, Kabala Ntumba G, Bichet DG, Maranda B, Auray-Blais C. Mass Spectrometry Analysis of Globotriaosylsphingosine and Its Analogues in Dried Blood Spots. International Journal of Molecular Sciences. 2023; 24(4):3223. https://doi.org/10.3390/ijms24043223
Chicago/Turabian StyleBoutin, Michel, Pamela Lavoie, Margot Beaudon, Georges Kabala Ntumba, Daniel G. Bichet, Bruno Maranda, and Christiane Auray-Blais. 2023. "Mass Spectrometry Analysis of Globotriaosylsphingosine and Its Analogues in Dried Blood Spots" International Journal of Molecular Sciences 24, no. 4: 3223. https://doi.org/10.3390/ijms24043223
APA StyleBoutin, M., Lavoie, P., Beaudon, M., Kabala Ntumba, G., Bichet, D. G., Maranda, B., & Auray-Blais, C. (2023). Mass Spectrometry Analysis of Globotriaosylsphingosine and Its Analogues in Dried Blood Spots. International Journal of Molecular Sciences, 24(4), 3223. https://doi.org/10.3390/ijms24043223