The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats
Abstract
1. Introduction
2. Results
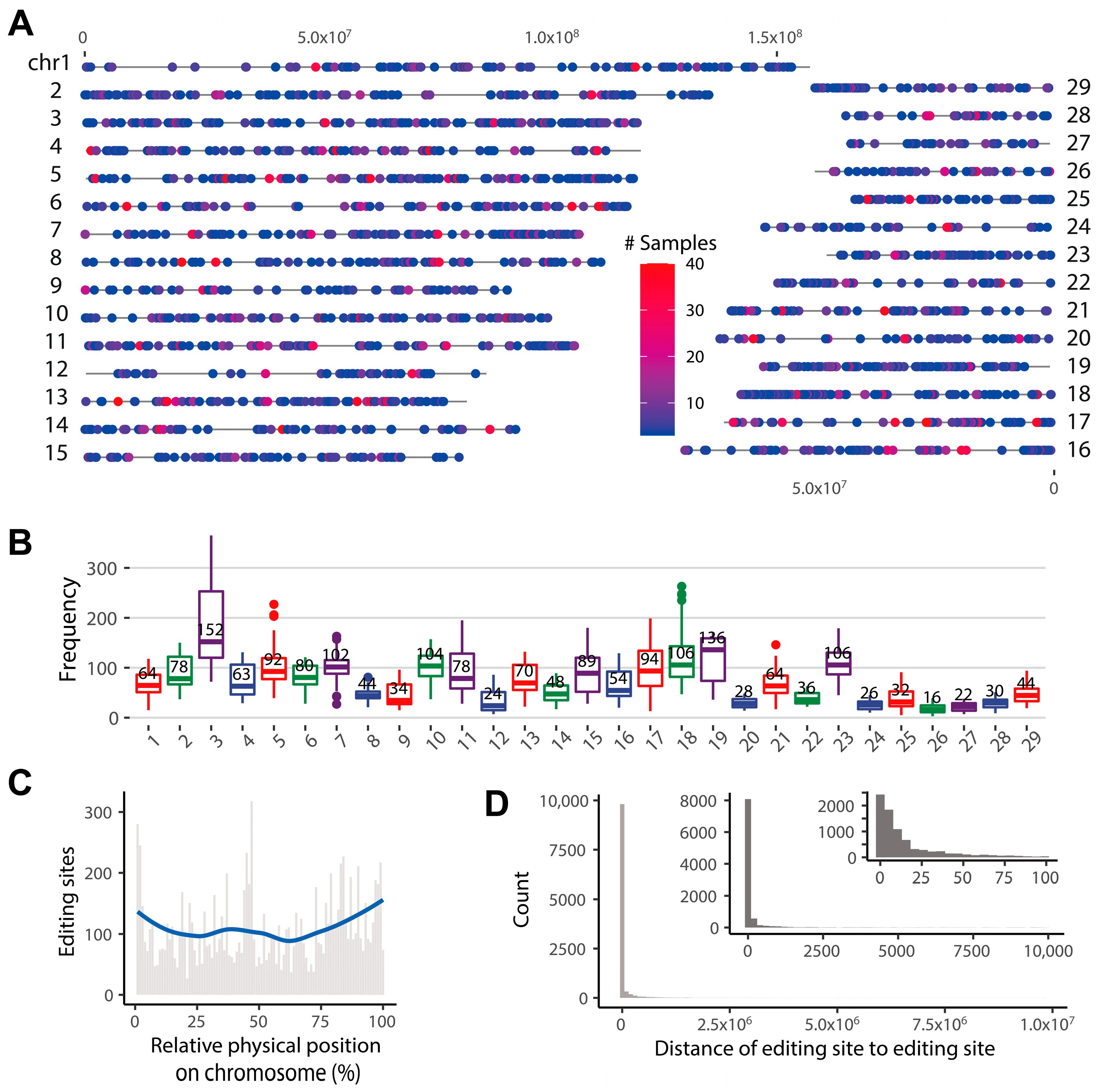
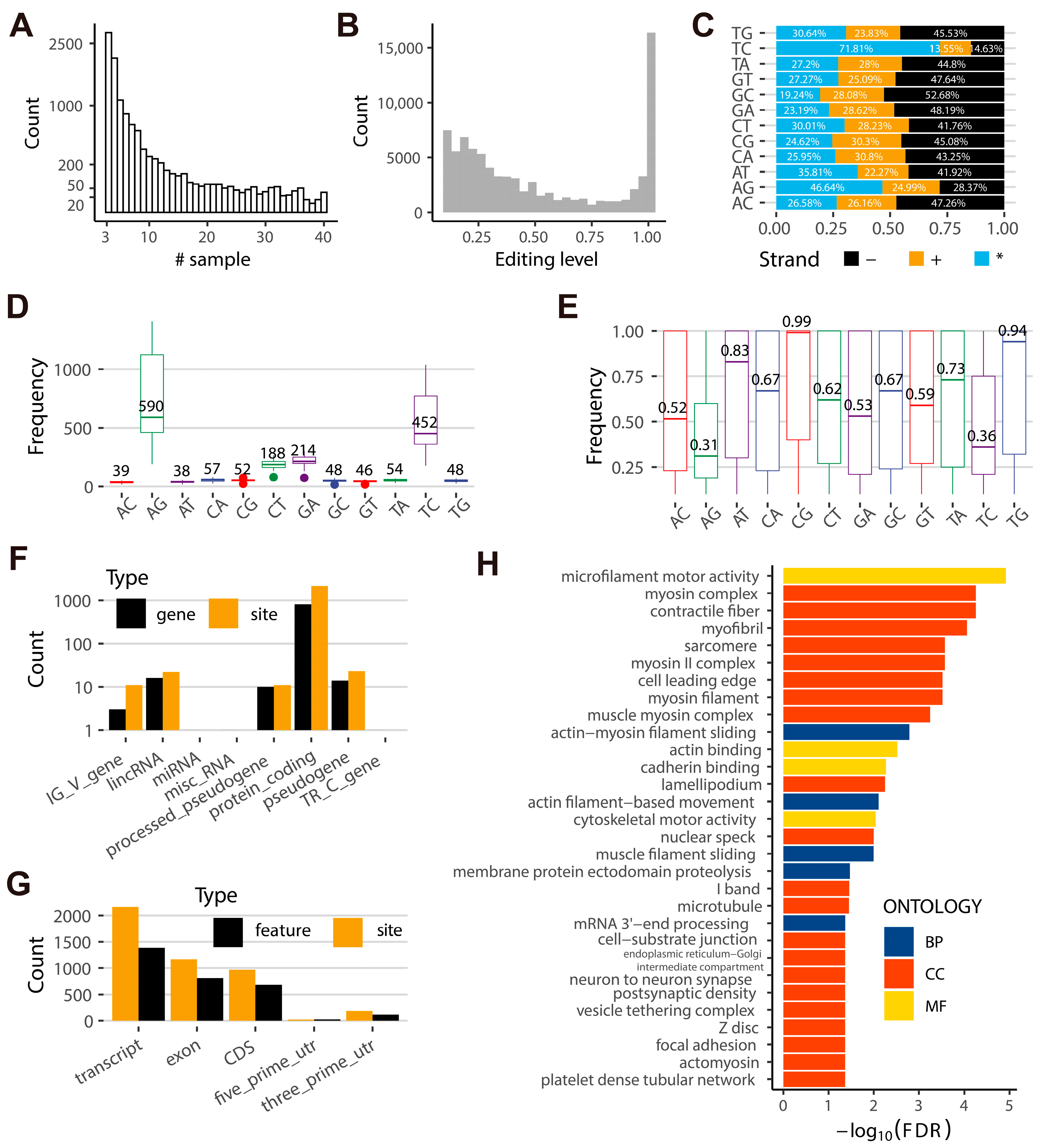
2.1. The Landscape of RNA Editome in Goats
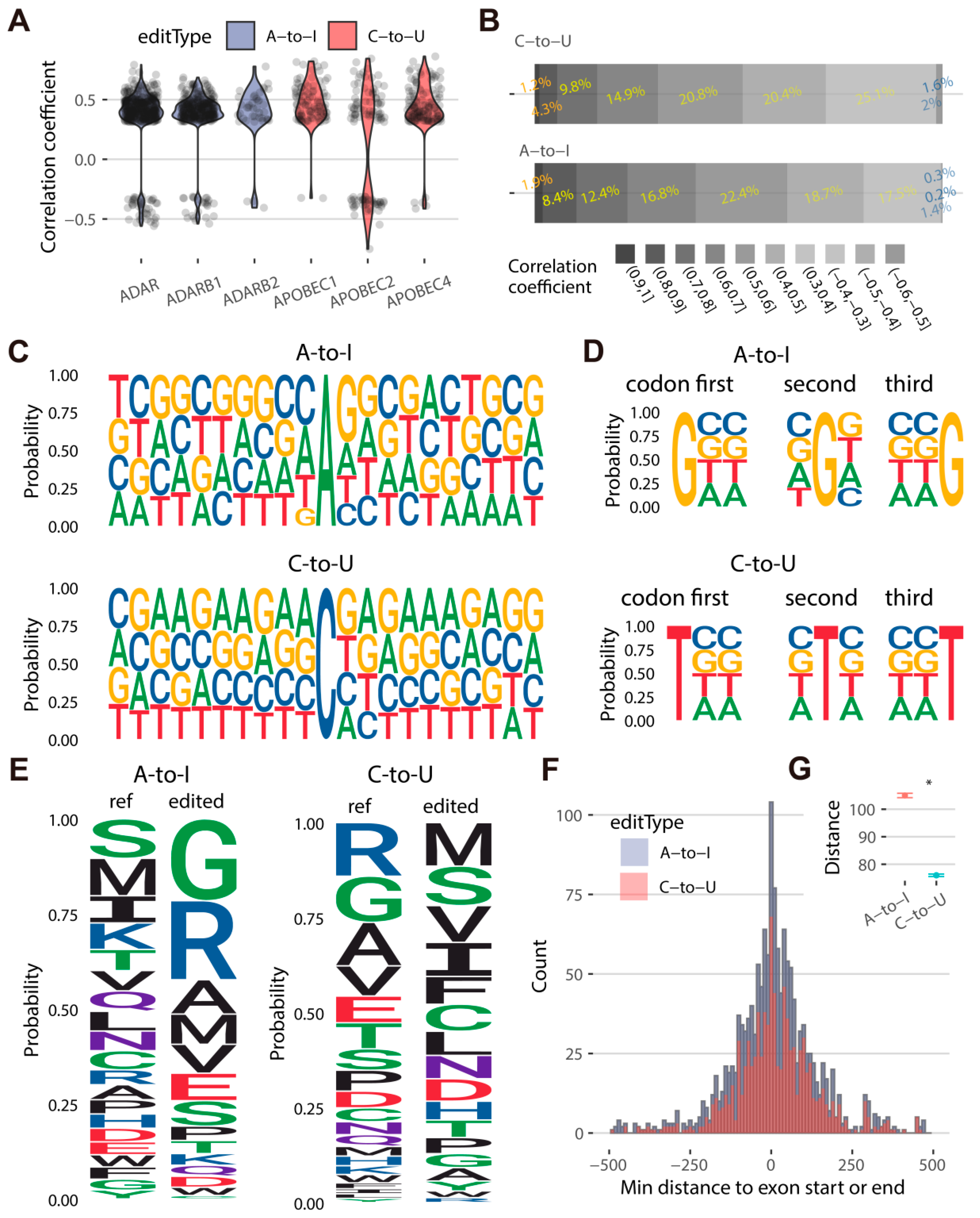
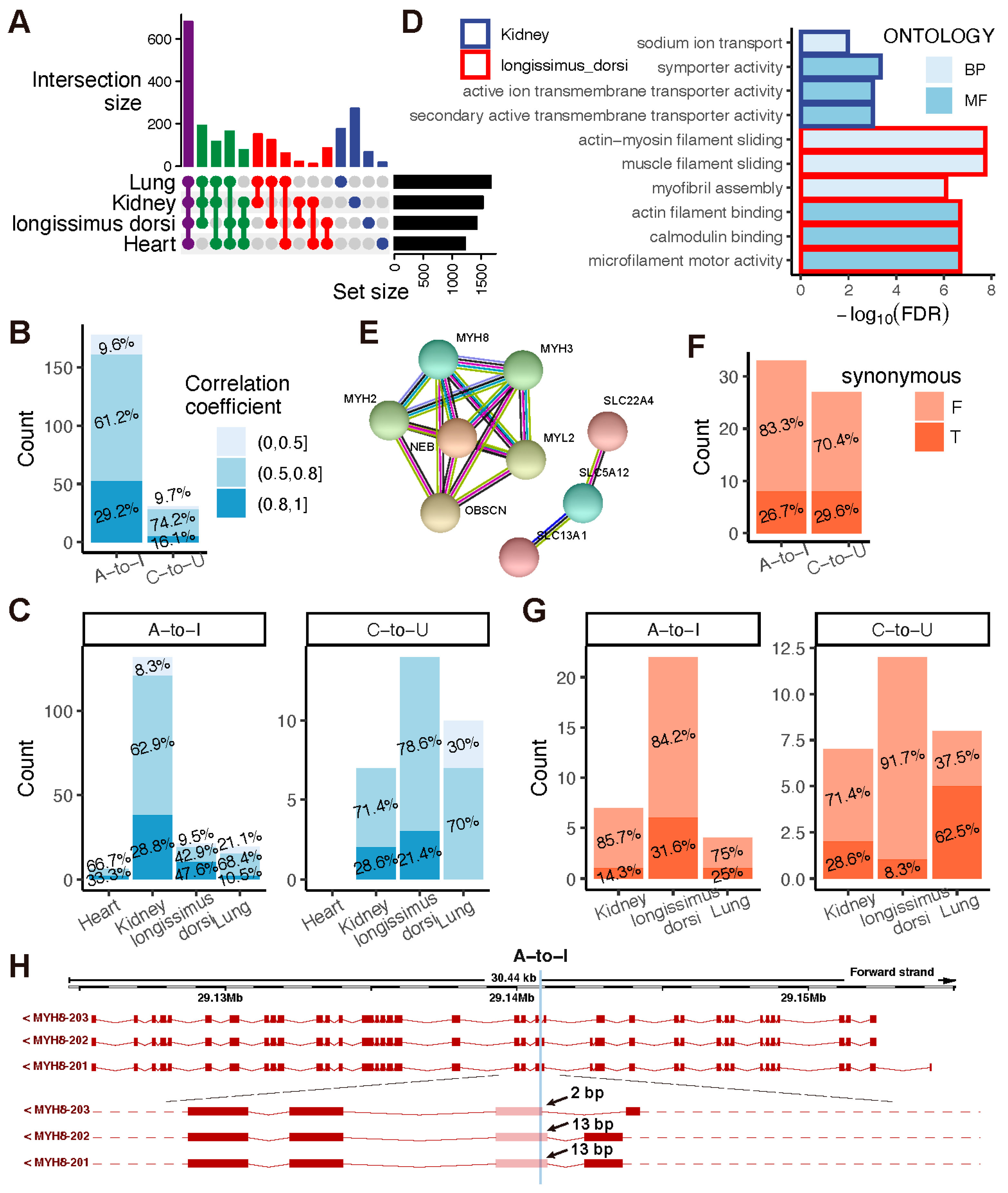
2.2. Different Flanking Sequences, Amino Acid Mutations, and Alternative Splicing Activities Are Observed at A-to-I and C-to-U Sites in Goats
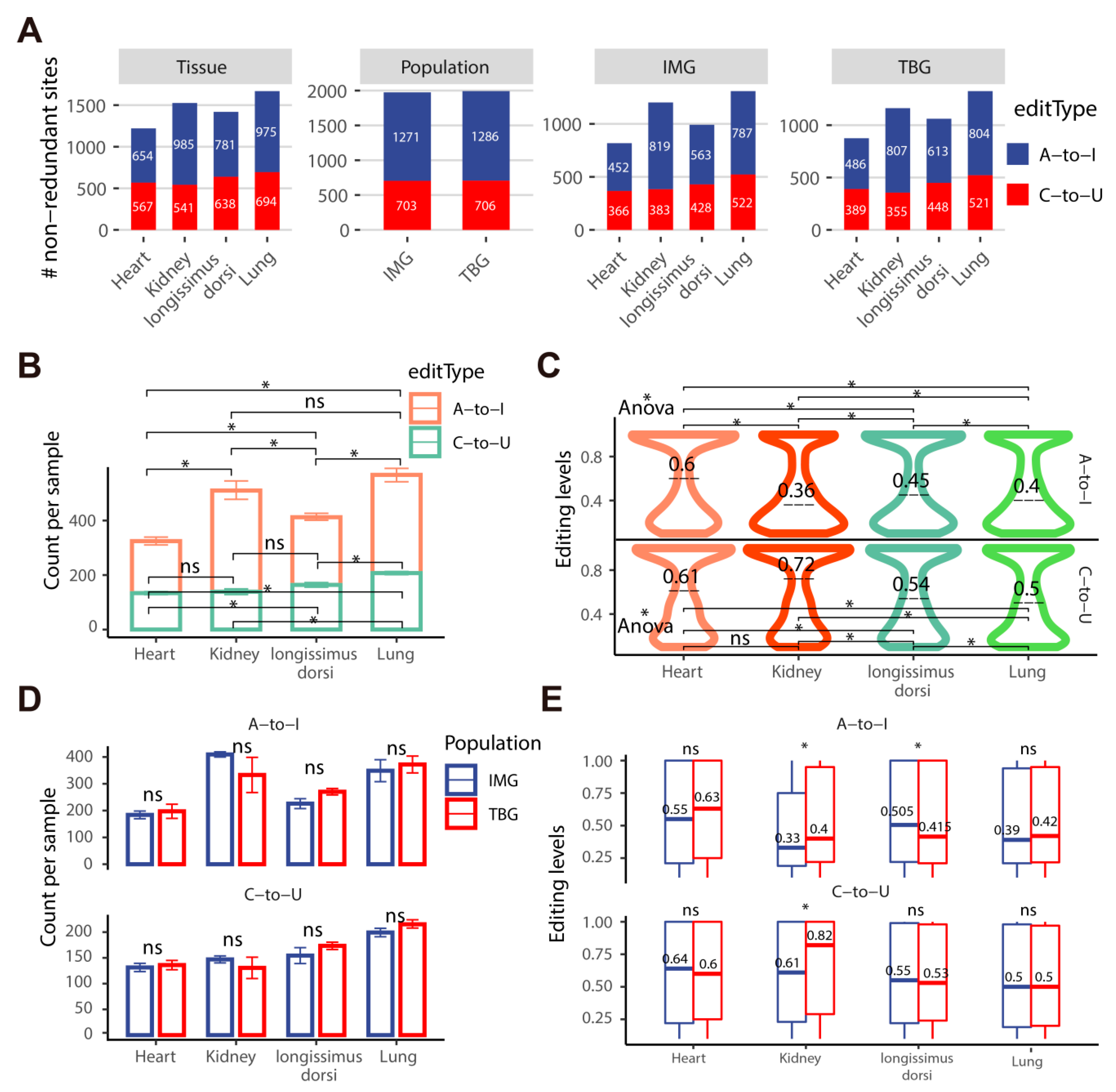
2.3. TBG Shows Higher A-to-I and C-to-U Editing Levels than IMG in the Kidney
2.4. The Kidney and Longissimus Dorsi Muscle Tissues Exhibit More Nonsynonymous A-to-I and C-to-U Sites in Goats
2.5. Population-Specific A-to-I and C-to-U RNA Editing Sites in TBG and IMG
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA and RNA Sequencing
4.3. Reads Alignment and Variant Calling
4.4. Identifying RESs
4.5. Gene Annotation and Enrichment Analysis
4.6. Sequence Preference
4.7. Population and Tissue Comparisons of RESs
4.8. Calculation of Gene Expression
4.9. Correlation Analysis between Gene Expression and RNA Editing Level
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IMG | Inner Mongolia cashmere goats |
| TBG | Tibetan cashmere goats |
| ADARs | Adenosine deaminase acting on the RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCGs | Protein coding genes |
| SNP | Single Nucleotide Polymorphism |
| CDS | Coding regions |
| UTR | Untranslated regions |
| tSESs | Tissue-specific editing sites |
| pSESs | Population-specific editing sites |
| pDESs | Population-differential editing sites |
References
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef]
- Sommer, B.; Kohler, M.; Sprengel, R.; Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E. Proteome Diversification by RNA Editing. Methods Mol. Biol. 2021, 2181, 229–251. [Google Scholar]
- Nakano, M.; Fukami, T.; Gotoh, S.; Nakajima, M. A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J. Biol. Chem. 2017, 292, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Williams, B.; Wold, B.J.; Mortazavi, A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012, 22, 1626–1633. [Google Scholar] [CrossRef]
- Hwang, T.; Park, C.K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.J.; Eterovic, A.K.; Yuan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, D.; Dong, X.; Wang, J.; Chen, J.; Yao, Y.; Darwish, H.Y.A.; Liu, W.; Deng, X. Genome-wide profiling of RNA editing sites in sheep. J. Anim. Sci. Biotechnol. 2019, 10, 31. [Google Scholar] [CrossRef]
- Li, I.C.; Chen, Y.C.; Wang, Y.Y.; Tzeng, B.W.; Ou, C.W.; Lau, Y.Y.; Wu, K.M.; Chan, T.M.; Lin, W.H.; Hwang, S.P.; et al. Zebrafish Adar2 Edits the Q/R site of AMPA receptor Subunit gria2alpha transcript to ensure normal development of nervous system and cranial neural crest cells. PLoS ONE 2014, 9, e97133. [Google Scholar]
- Hartner, J.C.; Schmittwolf, C.; Kispert, A.; Muller, A.M.; Higuchi, M.; Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004, 279, 4894–4902. [Google Scholar] [CrossRef]
- Roux, P.F.; Fresard, L.; Boutin, M.; Leroux, S.; Klopp, C.; Djari, A.; Esquerre, D.; Martin, P.G.; Zerjal, T.; Gourichon, D.; et al. The Extent of mRNA Editing Is Limited in Chicken Liver and Adipose, but Impacted by Tissular Context, Genotype, Age, and Feeding as Exemplified with a Conserved Edited Site in COG3. G3 2015, 6, 321–335. [Google Scholar] [CrossRef]
- Garrett, S.; Rosenthal, J.J. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 2012, 335, 848–851. [Google Scholar] [CrossRef]
- Bernard, A.; Khrestchatisky, M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J. Neurochem. 1994, 62, 2057–2060. [Google Scholar] [CrossRef]
- Yang, L.; Li, L.; Kyei, B.; Guo, J.; Zhan, S.; Zhao, W.; Song, Y.; Zhong, T.; Wang, L.; Xu, L.; et al. Systematic analyses reveal RNA editing events involved in skeletal muscle development of goat (Capra hircus). Funct. Integr. Genom. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Salehi, A.; Rivera, R.M. Genome-wide identification and analysis of A-to-I RNA editing events in bovine by transcriptome sequencing. PLoS ONE 2018, 13, e0193316. [Google Scholar]
- Yang, Y.; Zhu, M.; Fan, X.; Yao, Y.; Yan, J.; Tang, Y.; Liu, S.; Li, K.; Tang, Z. Developmental atlas of the RNA editome in Sus scrofa skeletal muscle. DNA Res. 2019, 26, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Montes, A.M.; Fernandez, A.; Perez-Montarelo, D.; Alves, E.; Benitez, R.M.; Nunez, Y.; Ovilo, C.; Ibanez-Escriche, N.; Folch, J.M.; Fernandez, A.I. Using RNA-Seq SNP data to reveal potential causal mutations related to pig production traits and RNA editing. Anim. Genet. 2017, 48, 151–165. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, X.; Tang, Z.; Li, S.C. Genome-Wide Investigation and Functional Analysis of Sus scrofa RNA Editing Sites across Eleven Tissues. Genes 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Yue, J.; Wei, X.; Wang, L.; Liu, X.; Gao, H.; Hou, X.; Zhao, F.; Yan, H.; et al. Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing. J. Anim. Sci. Biotechnol. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Darwin, C.R. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. Br. Foreign Med. Chir. Rev. 1860, 25, 367–404. [Google Scholar]
- Bailey, D.M.; Bartsch, P.; Knauth, M.; Baumgartner, R.W. Emerging concepts in acute mountain sickness and high-altitude cerebral edema: From the molecular to the morphological. Cell Mol. Life Sci. 2009, 66, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Brutsaert, T.D.; Soria, R.; Caceres, E.; Spielvogel, H.; Haas, J.D. Effect of developmental and ancestral high altitude exposure on chest morphology and pulmonary function in Andean and European/North American natives. Am. J. Hum. Biol. 1999, 11, 383–395. [Google Scholar] [CrossRef]
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.K.; Chen, L.; Ma, J.; et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef]
- Scott, G.R.; Schulte, P.M.; Egginton, S.; Scott, A.L.; Richards, J.G.; Milsom, W.K. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 2011, 28, 351–363. [Google Scholar] [CrossRef]
- Zaidan, H.; Ramaswami, G.; Golumbic, Y.N.; Sher, N.; Malik, A.; Barak, M.; Galiani, D.; Dekel, N.; Li, J.B.; Gaisler-Salomon, I. A-to-I RNA editing in the rat brain is age-dependent, region-specific and sensitive to environmental stress across generations. BMC Genom. 2018, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Neeman, Y.; Levanon, E.Y.; Jantsch, M.F.; Eisenberg, E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 2006, 12, 1802–1809. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Slotkin, W.; Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013, 5, 105. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Suzuki, T.; Tsuji, S.; Higuchi, M.; Seeburg, P.H.; Takahashi, R.; Misawa, H.; Kwak, S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010, 30, 11917–11925. [Google Scholar] [CrossRef] [PubMed]
- Galeano, F.; Tomaselli, S.; Locatelli, F.; Gallo, A. A-to-I RNA editing: The “ADAR” side of human cancer. Semin. Cell Dev. Biol. 2012, 23, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Perisic Matic, L.; John, D.; Lunella, F.F.; Jae, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Nakahama, T.; Yamasaki, R.; Costa Cruz, P.H.; Vongpipatana, T.; Inoue, M.; Kanou, N.; Xing, Y.; Todo, H.; Shibuya, T.; et al. RNA editing at a limited number of sites is sufficient to prevent MDA5 activation in the mouse brain. PLoS Genet. 2021, 17, e1009516. [Google Scholar] [CrossRef]
- Shafiei, H.; Bakhtiarizadeh, M.R.; Salehi, A. Large-scale potential RNA editing profiling in different adult chicken tissues. Anim. Genet. 2019, 50, 460–474. [Google Scholar] [CrossRef]
- Wei, C.; Lu, J.; Xu, L.; Liu, G.; Wang, Z.; Zhao, F.; Zhang, L.; Han, X.; Du, L.; Liu, C. Genetic structure of Chinese indigenous goats and the special geographical structure in the Southwest China as a geographic barrier driving the fragmentation of a large population. PLoS ONE 2014, 9, e94435. [Google Scholar] [CrossRef]
- Di, R.; Vahidi, S.M.; Ma, Y.H.; He, X.H.; Zhao, Q.J.; Han, J.L.; Guan, W.J.; Chu, M.X.; Sun, W.; Pu, Y.P. Microsatellite analysis revealed genetic diversity and population structure among Chinese cashmere goats. Anim. Genet. 2011, 42, 428–431. [Google Scholar] [CrossRef]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N.; et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genom. 2016, 17, 122. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, R.; Zhao, Z.; Xu, H.; Zhao, E.; Zhang, J. Genetic diversity and molecular phylogeography of Chinese domestic goats by large-scale mitochondrial DNA analysis. Mol. Biol. Rep. 2014, 41, 3695–3704. [Google Scholar] [CrossRef]
- Deng, J.; Feng, J.; Li, L.; Zhong, T.; Wang, L.; Guo, J.; Ba, G.; Song, T.; Zhang, H. Polymorphisms, differentiation, and phylogeny of 10 Tibetan goat populations inferred from mitochondrial D-loop sequences. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2018, 29, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chu, M.; Ma, X.; Pei, J.; Xiong, L.; Guo, X.; Liang, C.; Yan, P. Genome-Wide Identification of RNA Editing Sites Affecting Muscle Development in Yak. Front. Vet. Sci. 2022, 9, 871814. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Ben Hamidane, H.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.X.; So, E.; Devlin, J.L.; Zhao, Y.; Wu, M.; Cheung, V.G. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013, 5, 849–860. [Google Scholar] [CrossRef]
- Wang, Q.; Khillan, J.; Gadue, P.; Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 2000, 290, 1765–1768. [Google Scholar] [CrossRef]
- Anant, S.; Murmu, N.; Houchen, C.W.; Mukhopadhyay, D.; Riehl, T.E.; Young, S.G.; Morrison, A.R.; Stenson, W.F.; Davidson, N.O. Apobec-1 protects intestine from radiation injury through posttranscriptional regulation of cyclooxygenase-2 expression. Gastroenterology 2004, 127, 1139–1149. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Liu, H.; Huang, Y.; Kang, L.; Chen, H.W.; Chen, Y.T.; Wee, Y.R.; Chen, S.J.; Tan, B.C. ADAR1 deaminase contributes to scheduled skeletal myogenesis progression via stage-specific functions. Cell Death Differ. 2014, 21, 707–719. [Google Scholar] [CrossRef]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Marcucci, R.; Brindle, J.; Paro, S.; Casadio, A.; Hempel, S.; Morrice, N.; Bisso, A.; Keegan, L.P.; Del Sal, G.; O’Connell, M.A. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011, 30, 4211–4222. [Google Scholar] [CrossRef]
- Wahlstedt, H.; Daniel, C.; Enstero, M.; Ohman, M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009, 19, 978–986. [Google Scholar] [CrossRef]
- Soleymanjahi, S.; Blanc, V.; Davidson, N. APOBEC1 mediated C-to-U RNA editing: Target sequence and trans-acting factor contribution to 177 RNA editing events in 119 murine transcripts in-vivo. RNA 2021, 27, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Davidson, N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 594–602. [Google Scholar] [CrossRef]
- Weaver, T.E.; Conkright, J.J. Function of surfactant proteins B and C. Annu. Rev. Physiol. 2001, 63, 555–578. [Google Scholar] [CrossRef] [PubMed]
- Melton, K.R.; Nesslein, L.L.; Ikegami, M.; Tichelaar, J.W.; Clark, J.C.; Whitsett, J.A.; Weaver, T.E. SP-B deficiency causes respiratory failure in adult mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2003, 285, L543–L549. [Google Scholar] [CrossRef]
- Jin, S.; Kim, J.; Willert, T.; Klein-Rodewald, T.; Garcia-Dominguez, M.; Mosqueira, M.; Fink, R.; Esposito, I.; Hofbauer, L.C.; Charnay, P.; et al. Ebf factors and MyoD cooperate to regulate muscle relaxation via Atp2a1. Nat. Commun. 2014, 5, 3793. [Google Scholar] [CrossRef]
- Wang, K.; Cao, Y.; Rong, Y.; Ning, Q.; Jia, P.; Huang, Y.; Lan, X.; Dang, R.; Chen, H.; Lei, C. A Novel SNP in EIF2AK4 Gene Is Associated with Thermal Tolerance Traits in Chinese Cattle. Animals 2019, 9, 375. [Google Scholar] [CrossRef] [PubMed]
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; Horb, M.E.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Miura, A.; Itoh, K.; Takeshima, Y.; Nishio, H. RNA sequencing reveals abnormal LDB3 splicing in sudden cardiac death. Forensic Sci. Int. 2019, 302, 109906. [Google Scholar] [CrossRef]
- Markovich, D. Na+-sulfate cotransporter SLC13A1. Pflug. Arch. 2014, 466, 131–137. [Google Scholar] [CrossRef]
- Yoshino, J. ACMSD: A Novel Target for Modulating NAD(+) Homeostasis. Trends Endocrinol. Metab. 2019, 30, 229–232. [Google Scholar] [CrossRef]
- Cai, W.; Shi, L.; Cao, M.; Shen, D.; Li, J.; Zhang, S.; Song, J. Pan-RNA editing analysis of the bovine genome. RNA Biol. 2021, 18, 368–381. [Google Scholar] [CrossRef]
- Wu, X.; Ayalew, W.; Chu, M.; Pei, J.; Liang, C.; Bao, P.; Guo, X.; Yan, P. Characterization of RNA Editome in the Mammary Gland of Yaks during the Lactation and Dry Periods. Animals 2022, 12, 207. [Google Scholar] [CrossRef]
- Carmi, S.; Borukhov, I.; Levanon, E.Y. Identification of widespread ultra-edited human RNAs. PLoS Genet. 2011, 7, e1002317. [Google Scholar] [CrossRef]
- Porath, H.T.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 2017, 18, 185. [Google Scholar] [CrossRef]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Munday, J.S.; Perrott, M.; Wang, G.; Liu, X. Association of Age with the Expression of Hypoxia-Inducible Factors HIF-1α, HIF-2α, HIF-3α and VEGF in Lung and Heart of Tibetan Sheep. Animals 2019, 9, 673. [Google Scholar] [CrossRef] [PubMed]
- Frisancho, A.R. Developmental functional adaptation to high altitude: Review. Am. J. Hum. Biol. 2013, 25, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.G.; Smith, D.W.; Lee, C.J.; Ngo, J.P.; Gardiner, B.S. What Makes the Kidney Susceptible to Hypoxia? Anat. Rec. 2020, 303, 2544–2552. [Google Scholar] [CrossRef]
- Duan, Y.; Dou, S.; Porath, H.T.; Huang, J.; Eisenberg, E.; Lu, J. A-to-I RNA editing in honeybees shows signals of adaptation and convergent evolution. iScience 2021, 24, 101983. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, T.; Tong, P.; Li, Y.; Guo, H.; Wan, A.; Xia, L.; Liu, Y.; Li, Y.; Tian, Q.; et al. New ZNF644 mutations identified in patients with high myopia. Mol. Vis. 2014, 20, 939–946. [Google Scholar]
- Glembotski, C.C.; Arrieta, A.; Blackwood, E.A.; Stauffer, W.T. ATF6 as a Nodal Regulator of Proteostasis in the Heart. Front. Physiol. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, X.; Li, Z.; Feng, M.; Liu, X. ATF6 promotes liver fibrogenesis by regulating macrophage-derived interleukin-1alpha expression. Cell Immunol. 2021, 367, 104401. [Google Scholar] [CrossRef] [PubMed]
- Maurer, S.; Kopp, H.G.; Salih, H.R.; Kropp, K.N. Modulation of Immune Responses by Platelet-Derived ADAM10. Front. Immunol. 2020, 11, 44. [Google Scholar] [CrossRef]
- Wu, P.; Wang, K.; Zhou, J.; Chen, D.; Yang, X.; Jiang, A.; Shen, L.; Zhang, S.; Xiao, W.; Jiang, Y.; et al. Whole-genome sequencing association analysis reveals the genetic architecture of meat quality traits in Chinese Qingyu pigs. Genome 2020, 63, 503–515. [Google Scholar] [CrossRef]
- Ding, D.X.; Wang, Y.; Yan, W.; Fu, W.N. MYCT1 alters the glycogen shunt by regulating selective translation of RACK1-mediated enzymes. iScience 2022, 25, 103955. [Google Scholar] [CrossRef] [PubMed]
- Grogan, A.; Kontrogianni-Konstantopoulos, A. Unraveling obscurins in heart disease. Pflug. Arch. 2019, 471, 735–743. [Google Scholar] [CrossRef]
- Stoehr, A.; Kennedy, L.; Yang, Y.; Patel, S.; Lin, Y.; Linask, K.L.; Fergusson, M.; Zhu, J.; Gucek, M.; Zou, J.; et al. The ribosomal prolyl-hydroxylase OGFOD1 decreases during cardiac differentiation and modulates translation and splicing. JCI Insight 2019, 5, e128496. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.; Kling, E.; Schiller, E.; Zeh, R.; Schrewe, A.; Holter, S.M.; Mossbrugger, I.; Calzada-Wack, J.; Strecker, V.; Wittig, I.; et al. MTO1-deficient mouse model mirrors the human phenotype showing complex I defect and cardiomyopathy. PLoS ONE 2014, 9, e114918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; He, X.; Yao, S.; Lin, T.; Zhang, L.; Chen, D.; Chen, C.; Yang, Q.; Li, F.; Zhu, Y.M.; et al. Ablation of Mto1 in zebrafish exhibited hypertrophic cardiomyopathy manifested by mitochondrion RNA maturation deficiency. Nucleic Acids Res. 2021, 49, 4689–4704. [Google Scholar] [CrossRef]
- Luo, R.; Chen, P.W.; Kuo, J.C.; Jenkins, L.; Jian, X.; Waterman, C.M.; Randazzo, P.A. ARAP2 inhibits Akt independently of its effects on focal adhesions. Biol. Cell 2018, 110, 257–270. [Google Scholar] [CrossRef]
- Horvath, R.; Lewis-Smith, D.; Douroudis, K.; Duff, J.; Keogh, M.; Pyle, A.; Fletcher, N.; Chinnery, P.F. SCP2 mutations and neurodegeneration with brain iron accumulation. Neurology 2015, 85, 1909–1911. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.R.; Espenel, C.; Libanje, F.; Raingeaud, J.; Morgan, J.; Jaulin, F.; Kreitzer, G. KIF17 regulates RhoA-dependent actin remodeling at epithelial cell-cell adhesions. J. Cell Sci. 2016, 129, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Cadinu, D.; Henke, R.M.; Xin, X.; Dastidar, R.G.; Zhang, L. Deletion of a subgroup of ribosome-related genes minimizes hypoxia-induced changes and confers hypoxia tolerance. Physiol. Genom. 2011, 43, 855–872. [Google Scholar] [CrossRef]
- Zhou, G.; Dada, L.A.; Chandel, N.S.; Iwai, K.; Lecuona, E.; Ciechanover, A.; Sznajder, J.I. Hypoxia-mediated Na-K-ATPase degradation requires von Hippel Lindau protein. FASEB J. 2008, 22, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.L.; Lin, S.K.; Chao, L.H.; Hsiang-Hua Lai, E.; Chang, C.C.; Shun, C.T.; Lu, W.Y.; Wang, J.H.; Hsiao, M.; Hong, C.Y.; et al. Sirtuin 6 suppresses hypoxia-induced inflammatory response in human osteoblasts via inhibition of reactive oxygen species production and glycolysis-A therapeutic implication in inflammatory bone resorption. Biofactors 2017, 43, 170–180. [Google Scholar] [CrossRef]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Xu, X.; Xiao, M.; Huang, C.; Cao, J.; Zhan, S.; Guo, J.; Zhong, T.; Wang, L.; Yang, L.; et al. The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats. Int. J. Mol. Sci. 2023, 24, 3115. https://doi.org/10.3390/ijms24043115
Li L, Xu X, Xiao M, Huang C, Cao J, Zhan S, Guo J, Zhong T, Wang L, Yang L, et al. The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats. International Journal of Molecular Sciences. 2023; 24(4):3115. https://doi.org/10.3390/ijms24043115
Chicago/Turabian StyleLi, Li, Xiaoli Xu, Miao Xiao, Chunhua Huang, Jiaxue Cao, Siyuan Zhan, Jiazhong Guo, Tao Zhong, Linjie Wang, Liu Yang, and et al. 2023. "The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats" International Journal of Molecular Sciences 24, no. 4: 3115. https://doi.org/10.3390/ijms24043115
APA StyleLi, L., Xu, X., Xiao, M., Huang, C., Cao, J., Zhan, S., Guo, J., Zhong, T., Wang, L., Yang, L., & Zhang, H. (2023). The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats. International Journal of Molecular Sciences, 24(4), 3115. https://doi.org/10.3390/ijms24043115





