Abstract
Chitin deacetylase (CDA) can accelerate the conversion of chitin to chitosan, influencing the mechanical properties and permeability of the cuticle structures and the peritrophic membrane (PM) in insects. Putative Group V CDAs SeCDA6/7/8/9 (SeCDAs) were identified and characterized from beet armyworm Spodoptera exigua larvae. The cDNAs of SeCDAs contained open reading frames of 1164 bp, 1137 bp, 1158 bp and 1152 bp, respectively. The deduced protein sequences showed that SeCDAs are synthesized as preproteins of 387, 378, 385 and 383 amino acid residues, respectively. It was revealed via spatiotemporal expression analysis that SeCDAs were more abundant in the anterior region of the midgut. The SeCDAs were down-regulated after treatment with 20-hydroxyecdysone (20E). After treatment with a juvenile hormone analog (JHA), the expression of SeCDA6 and SeCDA8 was down-regulated; in contrast, the expression of SeCDA7 and SeCDA9 was up-regulated. After silencing SeCDAV (the conserved sequences of Group V CDAs) via RNA interference (RNAi), the layer of intestinal wall cells in the midgut became more compact and more evenly distributed. The vesicles in the midgut were small and more fragmented or disappeared after SeCDAs were silenced. Additionally, the PM structure was scarce, and the chitin microfilament structure was loose and chaotic. It was indicated in all of the above results that Group V CDAs are essential for the growth and structuring of the intestinal wall cell layer in the midgut of S. exigua. Additionally, the midgut tissue and the PM structure and composition were affected by Group V CDAs.
1. Introduction
Spodoptera exigua (Hübner) is a moth (Lepidoptera) of the Noctuidae family and a considerable polyphagous economic pest in the world [1,2], causing damage to a variety of crops. S. exigua is found in all provinces, regions and cities in China. Seventeen Chinese provinces have reported S. exigua outbreaks, resulting in significant losses in agricultural production [3,4]. Chemical pesticides are currently the primary means of controlling the reproduction of S. exigua. However, because of an over-reliance on chemical pesticides, S. exigua resistance has grown fast. In some regions, a high level of cross-resistance to various pesticides has been developed in field populations in recent years [5,6,7].
The hydrolysis of chitin N-acetylamino linkages is catalyzed via chitin deacetylase (CDA, EC3.5.1.41), a chitin hydrolase that results in the formation of chitosan [8]. CDA is an extracellular chitin modification enzyme that deacetylates β-(1,4)-linked N-acetylglucosamine homopolymer chitin to produce β-(1,4)-linked D-glucosamine residue polymer chitosan [9]. Chitin is hydrolyzed by chitinase and N-acetyl-β-D-glucosaminidase, whereas CDA enzymatically alter chitin through a deacetylating process [10]. Many CDAs have been isolated from fungi, bacteria, viruses, and insects.
Insect CDA proteins were classified into five homologous groups (Groups I–V) based on phylogeny and CDA-conserved motifs [9]. Group I CDA proteins include CDA1 and CDA2, while Group II CDA proteins only contain CDA3. Group I and II CDA proteins have three domains: the chitin-binding functional domain (ChBD), the low-density lipoprotein-receptor-binding domain (LDLa), and the chitin deacetylation catalytic domain (CDA). The CDA4 protein of Group III and the CDA5 protein of Group IV have ChBD and chitin deacetylation catalytic domains, with an extended intermediate region separating the ChBD and chitin deacetylation catalytic domains. The CDA6/7/8/9 protein of Group V only has the chitin-deacetylated catalytic domain [9]. Five conserved catalytic motifs exist in the chitin deacetylation domain of CE-4 family proteins in the insect catalytic domain [11]. During the development of insects, different CDA groups serve various purposes according to tissue and developmental stages. All nine CDAs have been found and characterized in Tribolium castaneum [12]. The knockdown of TcCDA1 or TcCDA2 via RNA interference (RNAi) caused a considerable molting failure phenomenon. However, after the knockdown of TcCDA3 to TcCDA9, no developmental problems were detected [12]. Four CDA genes were also discovered in Nilaparvata lugens using whole-genome sequencing [11]. Double-stranded RNA (dsRNA) injection for NlCDA3 resulted in no discernible morphological abnormalities, while molting failure and significant mortality were caused by the silencing of NlCDA1, NlCDA2 and NlCDA4 [11].
CDAs appear widespread in insects and have been found in various taxa. Guo et al. discovered an insect CDA protein as a novel peritrophic membrane (PM) protein, TnPM-P42, from the cabbage looper Trichoplusia ni, which exhibits chitin-binding activity [13]. Since then, many insect species from different groups, including Lepidoptera [10,14,15,16,17,18], Coleoptera [12,19,20,21], Orthoptera [22], Hemiptera [11], Hymenoptera [9], Diptera [23,24] and Isoptera [25], have been shown to contain CDA genes. Several CDAs in Bombyx mori and Mamestra configurata are primarily expressed in the PM and play significant roles in changing the PM’s physical characteristics [26,27]. The CDA protein in the PM of Helicoverpa armigera probably protects the gut from external infection and intercepts toxins, such as Bacillus thuringiensis (Bt) crystal proteins [28]. The silkworm midgut genes BmCDA6, BmCDA7 and BmCDA8 exhibit unique expressions in the anterior midgut. However, their temporal expression pattern is different [29]. The function of Group V CDAs and their role in the structure of the insect midgut was speculated upon in previous studies, but not in depth [26,27,28,29]. The functions of Group V CDAs SeCDA6/7/8/9 (SeCDAs) have not been clarified in S. exigua.
In this study, the genome database of S. exigua was used to identify and clone SeCDAs. The predicted amino acid sequences, protein domains and phylogenies of the genes were then investigated. The expression profiles of the SeCDAs in tissue and developmental expression were investigated using qRT-PCR. RNAi technology was used to silence SeCDAs to explore their biological role in S. exigua and to elucidate the mechanism of CDA action. These findings will facilitate our understanding of the functions of Group V CDAs and serve as the foundation for further research.
2. Results
2.1. Group V SeCDAs Sequence Analysis
Nucleotide sequences of the cDNAs for Group V SeCDAs were obtained from the S. exigua genome (Table 1). It was shown by the NCBI Blastp comparisons that SeCDAs only have a CDA domain (Figure 1). It was demonstrated by the deduced protein sequences that SeCDAs proteins are synthesized as preproteins with signal peptides using the SignalP 5.0 Server. The secreted SeCDAs contain multiple N-glycosylation sites and five conserved motifs of the CDA domain (Supplementary Figure S1).

Table 1.
Summary of Group V SeCDAs in S. exigua.
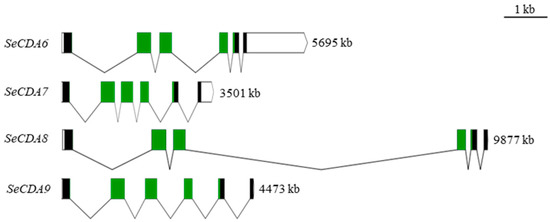
Figure 1.
Schematic diagram of exon–intron of SeCDAs gene and domain of SeCDAs amino acid. The structure of the genome sequence is plotted to scale. Exons are represented by solid rectangles, noncoding regions are represented by hollow rectangles, and the connecting line between two exons represents introns. The green rectangles represent the chitin deacetylation catalytic domain.
A phylogenetic analysis was performed based on the CDA amino acid residue sequences of several insect species and the SeCDAs amino acid sequence of S. exigua published in NCBI GenBank (Figure 2, Supplementary Figures S2 and S3). It was shown by the analysis that SeCDAs cluster among Group V CDA proteins. There was minimal genetic distance to the Lepidoptera species of Spodoptera litura, Spodoptera frugiperda and Helicoverpa armigera.

Figure 2.
Phylogenetic tree of the CDA amino acid residue sequence of S. exigua and other insects. SeCDAs of S. exigua are labeled with a solid red triangle. The origin of CDA species: Bm: Bombyx mori; Ha: Helicoverpa armigera; Hc: Hyphantria cunea; Mc: Mamestra configurata; Pm: Papilio machaon; Px: Papilio Xuthus; Sf: Spodoptera frugiperda; Sl: Spodoptera litura; Tc: Tribolium castaneum; Tn: Trichoplusia ni.
2.2. Group V SeCDAs Expression Analyses
It was shown via qRT-PCR analysis that Group V SeCDAs are expressed in different developmental stages of S. exigua. SeCDA6 gene expression was highest in eggs, followed by the fifth- and second-instar larvae. Expression of SeCDA7 and SeCDA9 genes was found in the fifth-instar larvae. The SeCDA8 gene was highly expressed in eggs and prepupae (Figure 3).
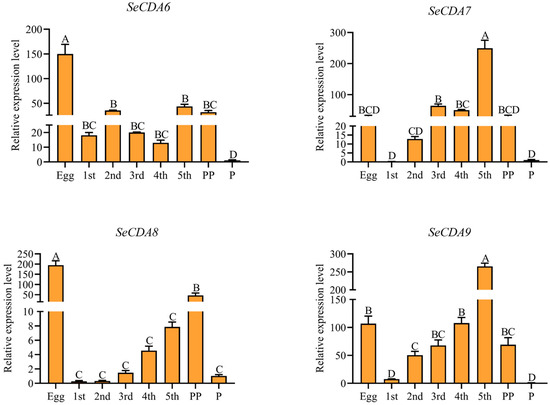
Figure 3.
Expression profiles of SeCDAs in different developmental stages. Developmental stages include the egg, first-to-fifth-instar larvae, prepupae and pupae. Data were calibrated by using β-actin and presented as means ± SE of three separate experiments. The group of the pupae data was used for normalization. Different letters above the bars represent the different significances of the samples (p < 0.01).
The expression of SeCDAs in different tissues of the fifth-instar day-2 larvae was shown via qRT-PCR analysis. SeCDAs were expressed in all tissues of the fifth-instar larvae and were highly expressed in the midgut (Figure 4A). The SeCDAs expression was more abundant in the anterior region of the midgut, followed by the middle and posterior regions of the midgut (Figure 4B).

Figure 4.
Detection of SeCDAs in various tissue samples and different midgut regions via qRT-PCR analysis. (A) IN: integument, MG: midgut, HG: hindgut, MT: malpighian tubule, FB: fat body. The MT data group was used for normalization. (B) AM: anterior part of the midgut, MM: middle part of the midgut, PM: posterior part of the midgut. The MM data group was used for normalization. Data were calibrated by using β-actin and presented as means ± SE of three separate experiments. Different letters above the bars represent the different significances of the samples (p < 0.01).
2.3. Expression of Group V SeCDAs in Response to 20E and JHA Signaling
Furthermore, qRT-PCR was used to analyze the effects of the injection of 20-hydroxyecdysone (20E) and juvenile hormone analog (JHA) on the SeCDAs of S. exigua. The injection resulted in the down-regulation of the transcriptional expression level of SeCDAs when treated with 20E (Figure 5A). The expression of SeCDA6 and SeCDA8 decreased after JHA treatment. However, the expression of SeCDA7 and SeCDA9 increased after JHA treatment (Figure 5B).
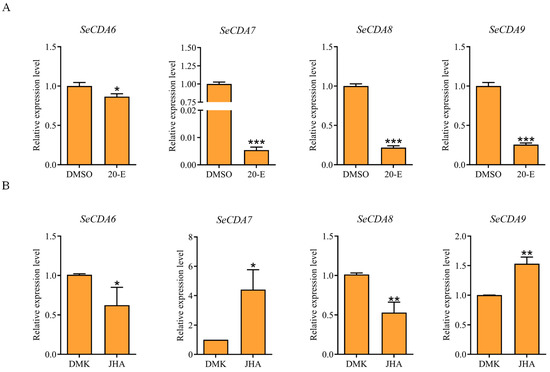
Figure 5.
Effect of two hormone analogs on the expression of SeCDAs. (A) Effect of 20E on the expression of SeCDAs. DMSO was used in the control group. (B) Effect of JHA on the expression of SeCDAs. DMK was used in the control group. Data are reported as means ± SE of three independent biological replications (*** p < 0.001; ** p < 0.01; * p < 0.05).
2.4. Functional Analysis of Group V SeCDAs
qRT-PCR analysis showed that the injection of dsSeCDAs can cause the silencing efficiency of SeCDAs to be 79.28%, 75.55%, 73.23% and 87.57%, respectively. The relative expression level of SeCDAs was reduced by 99.89%, 90.45%, 84.46% and 96.60% after the injection of dsSeCDAV (Figure 6). Group V SeCDAs were silenced in larvae injected with dsSeCDA6 and dsSeCDA8, respectively. SeCDA6 and SeCDA7 were silenced when dsSeCDA7 was injected into the fourth-instar larvae. Only the SeCDA9 gene was silenced when dsSeCDA9 was injected. This was indicated by the results that the silencing efficiency of SeCDAs can be up-regulated via dsSeCDAV injection. After the GFP and SeCDAs were silenced, the larvae were molted and pupated normally (Supplementary Figure S4).

Figure 6.
Relative expression levels of SeCDAs in S. exigua larvae injected with dsRNA-SeCDAs/GFP. Data are reported as means ± SE of three independent biological replications. (*** p < 0.001; ** p < 0.01; * p < 0.05).
2.5. Immunohistochemistry
The specific signal of SeCDAs was weak in the midgut after dsSeCDAV injection, which confirmed the specificity and silencing effect of the SeCDAs antibody. The immunohistochemistry method was used to explore the biological role of SeCDAs. Group V SeCDA protein was localized on the intestinal wall cells of the midgut of S. exigua. After silencing SeCDAs, the intestinal cell layer became thinner, and the intestinal cells were more closely arranged (Figure 7).

Figure 7.
Detection of SeCDAs using a specific antibody. The specific signal of SeCDAs was detected after dsGFP and dsSeCDAV injection into fourth-instar larvae. The negative control was with the pre-immune serum. The SeCDAs protein is represented by green and the cell nucleus by red. Cm: circular muscle; Pm: peritrophic membrane; Rc: regenerative cells; Dc: digestive cells; scale bar is 50 μm.
2.6. Scanning Electron Microscopy Analysis
Seventy-two hours after the injection of dsSeCDAV, the morphology of the midgut and PM were observed under a scanning electron microscope. The results showed that the vesicles on the midgut epithelia disappeared after the injection of dsSeCDAV compared with the control injected with dsGFP (Figure 8A). The PM treated with dsSeCDAV became rough with large folds and pores; in contrast, the chitin microfilament structure was loose and disordered (Figure 8B).

Figure 8.
The ultrastructure of S. exigua larvae injected with dsRNA-SeCDAV/GFP. (A) Midgut ultrastructure of S. exigua larvae injected with dsRNA-SeCDAV/GFP. Red arrowheads point to vesicles. (B) Peripheral membrane ultrastructure of S. exigua larvae injected with dsRNA-SeCDAV/GFP. Red arrowheads point to the pores. Mi: microvilli; Ve: vesicles. The scale bars were 50 μm, 25 μm, 5 μm, 2 μm, 500 nm, and 250 nm.
3. Discussion
According to the GenBank databases, five groups of CDA proteins have been classified according to the alignment of their predicted chitin deacetylase domain sequences. SeCDA6/7/8/9, which only contains a CDA catalytic domain, belongs to the Group V CDAs protein (Figure 1).
It was indicated by recent studies on CDA in insects that the abundance of these proteins was different in the tissue, developmental stages and species. The first CDA protein, TnPM-P42, a midgut-specific protein, was identified from T. ni in 2005 [13]. In subsequent studies, Group I CDA was mainly found in ectodermal tissues, such as the foregut, hindgut, integument and trachea. Group III CDA occurred primarily in specialized epidermal tissues, and Group V CDAs were mainly expressed in the midgut-specific proteins. BmCDA6/7/8 were specifically described in the anterior part of the midgut depending on the larval instar. The expression of BmCDA6 was concentrated primarily in the middle-molting stage. BmCDA7 and BmCDA8 were only highly expressed in the feeding stages. [29]. TcCDA6/7/8/9 were expressed specifically in the midgut during the larval feeding stage [12]. SeCDAs identified from S. exigua were more abundant in the most anterior region of the midgut, which was similar to Group V CDAs in B. mori [29] and T. castaneum [12]. This indicated that SeCDAs were primarily synthesized in the anterior part of the midgut. We speculate that SeCDA7 and SeCDA9 may be involved in modifying and maintaining midgut characteristics to stabilize the metabolism. Moreover, SeCDA6 and SeCDA8 proteins were more frequently detected in the eggs. We speculate that SeCDA6 and SeCDA8 may be involved in midgut formation during egg development and the modification of their structures.
In insect, the bioactive hormone 20E and juvenile hormone (JH) play a crucial role in development, metamorphosis and reproduction. Insect molting may be induced by this process [30] and regulate specific gene expression [31,32,33], such as five chitin synthesis pathway genes in S. exigua [34]. Insect egg maturation is triggered by JH, which also helps maintain the larval condition, adult sexual development regulation, and premature larval metamorphosis prevention [35]. The JHA was used instead of JH in the larvae. The treatment of 20E accounted for a significant increase in mRNA expression levels of BmCDA6; however, it decreased the expression levels of BmCDA7 and BmCDA8. The treatment of JHA up-regulated the expression of BmCDA7 and BmCDA8 [16]. Some results obtained after the treatment of 20E in other insects also regulated the CDA gene expression [16,36,37]. In this study, after the larvae of S. exigua were injected with 20E, the expression level of the SeCDA6/7/8/9 was decreased. After treatment with JHA, the expression of SeCDA6 and SeCDA8 decreased, while SeCDA7 and SeCDA9 expressions increased. These results indicate that both 20E and JHA can regulate the expression of CDA genes.
RNAi technology has been widely used to regulate post-transcriptional genes in fungi, nematodes, and insects. According to their functions, insect CDAs can be divided into intestinal-specific proteins (Group V CDA) and parenteral-specific proteins (Groups I, II, III and IV CDA). The TcCDA6/7/8/9 were only expressed in the midgut, which belongs to midgut-specific proteins. There was no aberrant phenotype in individuals treated with dsTcCDA6/7/8/9 [12]. RNAi was used to silence Mythimna seperata CDA1 to evaluate PM alterations [38]. It was shown in the investigation that in the treated specimens, the dsRNA concentration was raised, the PM holes were wide, and the chitin microfilament structure became looser [38]. In our study, the silencing effectiveness of the SeCDA6 gene following RNAi treatment was 79.28%. The SeCDA7 gene silencing effectiveness was 75.55%. SeCDA8 silencing efficacy was 73.23%, and SeCDA9 gene silencing effectiveness was 87.57%. After the injection of dsSeCDAV, the silencing efficacy of SeCDA6/7/8/9 was 99.89%, 90.45%, 84.46% and 96.60%, respectively. The silencing efficiency of all Group V CDAs was considerably better than that of any Group V CDA alone, and a new way to improve the silencing efficiency of Lepidoptera genes was provided. The expression of SeCDAs decreased in all larvae injected with dsSeCDA6 or dsSeCDA8. Additionally, the DNA sequence alignment identity of SeCDA6 and SeCDA8 was as high as 93.64%.
This result may be due to the high similarity of the gene sequences. The silence of the SeCDA7 gene led to decreased gene expression of SeCDA6 and SeCDA7. Only the SeCDA9 gene was silenced when dsSeCDA9 was injected into the larvae. The DNA sequence alignment similarity between SeCDA6 and SeCDA7 was 53.95%. The sequence alignment similarities between SeCDA9 and SeCDA6, SeCDA7, and SeCDA8 are 65.64%, 60.88%, and 65.49%, respectively. The sequence similarity between SeCDA7 and SeCDA8 was only 55.53%. It was suggested by these results that the desired precise targeting effect may not be produced by highly similar gene sequences or the dsRNA targeting the same domain. The reasons for these results are unclear, and more research will be studied in the future.
BmCDA8 was discovered in both the PM and the ectoperitrophic gap between the midgut epithelial cells and the PM. BmCDA7 was discovered in the PM of B. mori [27,39,40]. These midgut CDAs may have been involved in PM production in the anterior midgut, where PM synthesis begins [41,42]. The structure of the chitin matrix in many insects is impacted by the deacetylation process [24,43,44]. It has been noted that PM permeability in vitro may be boosted by recombinant HaCDA5a, a CDA from the moth H. armigera [45]. In this study, we examined the localization of SeCDAs using immunohistology to determine their biological function. SeCDAs are present in the intestinal wall cells of the midgut of S. exigua. When SeCDAs were silenced, the intestinal cell layer shrank, and the intestinal cells were more evenly distributed. Group V CDAs were demonstrated by these findings to be critical in developing and organizing the intestinal wall cell layer in the midgut of S. exigua.
The maintenance of PM mechanical strength in H. armigera is probably correlated with CDA [17]. A change in the peritrophic membrane might make insects more prone to infection, which may eventually be lethal [46]. It was shown via scanning electron microscopy (SEM) analysis of the surface structure of the S. exigua midgut after dsSeCDAV treatment that the microvilli on the epithelia of the larval midgut did not alter substantially. However, all vesicles disappeared after the injection of dsSeCDAV compared to the control. This demonstrates that CDAs do not damage the microvilli but are crucial to the development of vesicles. Compared with the control, the PM of the larvae injected with dsSeCDAV became coarse, with many folds, pores and gaps. The chitin microfilament structure became loose and disorderly, similar to the results of CDA gene silencing in M. separata [38]. Therefore, it is hypothesized that CDA is crucial for maintaining the structural integrity of the PM of S. exigua. Group V CDAs are essential in forming the midgut and PM of S. exigua. However, no phenotypic changes were observed during molting in S. exigua with silenced Group V CDAs. The silencing of Group V CDAs affected the structure of the midgut and the PM. It was demonstrated by all the above results that the PM structure was disrupted when the Group V CDAs were silenced.
4. Materials and Methods
4.1. Insect Larvae
Larvae of S. exigua were reared on an artificial diet by the Laboratory of Insect Molecular Biology at Hebei Agricultural University. The rearing temperature was 26.5 ± 1 °C, the relative humidity from the egg to the third-instar larvae was 75 ± 10%, the relative humidity from the third-instar larva to the pupal stage was 65 ± 15%, and they were reared under a 14 L:10 D photoperiod. Fifth-instar larvae were used for dissection to isolate the PMs and various tissues for analyses.
4.2. Sequence Analysis
The sequence of the gene encoding Group V CDA was analyzed using Clustal X 1.83 and GeneDoc. The gene structure was mapped using the Exon-Intron Graphic Maker (http://www.wormweb.org/exonintron (accessed on 17 May 2021)). The open reading frame, coded amino acid, isoelectric point (pI) and molecular protein weight were predicted using the DNAMAN 6.0 program (LynnonBiosoft, USA). The NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/ (accessed on 18 February 2022)) was used to predict the N-glycosylation sites. The TMHMM Server v.2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 (accessed on 19 February 2022)) was used to determine whether it belonged to a transmembrane protein, and SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0 (accessed on 19 February 2022)) was used to predict the presence or absence of signal peptides. The NCBI Conserved Domain Database (CCD, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 23 June 2021)) was used to predict the conservative protein structure domain of SeCDA. Clustal X 1.83 was used for multi-sequence alignment, MEGA 5.0 software was used to build a neighbor-joining (NJ) phylogenetic tree, and 1000 repetitions were calculated using the bootstrap method, and the model was the Poisson model.
4.3. Gene Expression Analyses
Five tissues (integument, midgut, hindgut, malpighian tubules and fat body) were dissected from fifth-instar larvae for total RNA extraction to determine the tissue-specific expression pattern of SeCDAs. The hindgut and malpighian tubules from 10 larvae and other tissues (integument, midgut and fat body) from three larvae were pooled as one treatment. Three biological replications were prepared (each with three technical replicates). Eggs, first-, second-, third-, fourth-, and fifth-instar larvae, prepupae, and pupae were collected to determine their expression profiles during development. Each sample contained 3–20 individuals. Samples from each developmental stage were analyzed in biological triplicate. Each replicate was given a sample of at least 100 mg of tissue or larvae. The samples were stored at −80 until all samples were collected for RNA extraction.
Total RNA from the frozen tissues was isolated using the RNAprep Pure Tissue Kit (TIANGEN, Co., Ltd., Beijing, China) and treated with DNase I. A spectrophotometer (BioSpectrometer fluorescence, Eppendorf, Germany) and 1.2% agarose gel electrophoresis were used to measure RNA concentration and quality. The entire experimental operation was conducted in an RNase-free environment. A total of 1 μg of total RNA per reaction was used to perform cDNA synthesis using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Beijing, China) with an Oligo dT Primer.
The primers for qRT-PCR were designed using DNAMAN 6.0 software and are listed in Table 2. The melting curve was determined for each sample to detect the gene-specific peak and verify the absence of primer dimers. qRT-PCR was carried out in a 20 μL reaction volume containing 1 μL of cDNA, 10 μM of each primer, 7 μL of H2O and 10 μL of TB Green® Premix Ex Taq™ II (Tli RNaseH Plus), ROX plus (TaKaRa, Beijing, China). PCR was conducted on a CFX-96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The amplifications were performed using an Eppendorf thermocycler. They comprised an initial denaturation step at 95 °C for 3 min, followed by 39 cycles at 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s, and a final extension step of 5 min at 72 °C. Relative transcript levels of target genes were calculated with the 2−ΔΔCt method. Three biological repeats, each with three technical replicates, were set for each cDNA, and β-actin was used as the internal reference gene to calibrate the total amount of RNA. β-actin proved to be a suitable housekeeping gene applied in the qRT-PCR of S. exigua in our previous study [46]. The amplification efficiencies of the target and reference were approximately equal (Supplementary Figure S5). The significance of differences between samples was assessed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA), and the data were presented as means and standard errors (SE).

Table 2.
Primers used in this study.
4.4. Preparation and Treatment with 20E and JHA
A total of 40 fourth-instar larvae of S. exigua with similar body weights and body shapes were selected for the experiment. 20E (Solarbio, Beijing, China) was dissolved in dimethyl sulfoxide (DMSO) at a working concentration. A total of 5 µL of a 100 ng/µL 20E solution was injected into the fourth-instar larvae on the second day, and DMSO was injected into the control group. JHA (Sigma-Aldrich, Shanghai, China) was dissolved in dimethyl ketone (DMK). JHA and DMK were injected into S. exigua as described for the 20E treatment and control. RNA was extracted from whole S. exigua larvae after 12 h of treatment. Reverse transcription was performed, and the expression of SeCDAs genes was analyzed using qRT-PCR.
4.5. Functional Analysis of Group V CDA Genes
RNAi was used to reveal the biological roles of SeCDA6, SeCDA7, SeCDA8 and SeCDA9 in the midgut and PM of S. exigua. The primers containing the T7 RNA polymerase promoter sequences were designed with DNAMAN (Table 2). PCR was conducted to prepare the cDNA template for the synthesis of the dsRNA of SeCDA6, SeCDA7, SeCDA8, SeCDA9, SeCDAV (the conserved sequences of Group V CDAs, SeCDA6: 325 bp-813 bp, SeCDA7: 304 bp-786 bp, SeCDA8: 325 bp-807 bp, SeCDA9: 316 bp-798 bp) and GFP (control). The PCR product was verified on a 0.8% agarose gel. Samples (2 μg) were collected for dsSeCDA6, dsSeCDA7, dsSeCDA8, dsSeCDA9, dsSeCDAV and dsGFP synthesis using the T7 RiboMAX Express RNAi System (Promega, Madison, WI, USA).
A total of 150 fourth-instar 1-day larvae were collected and randomly divided into five groups. Aliquots of 10 μg of dsSeCDA6, dsSeCDA7, dsSeCDA8, dsSeCDA9, dsSeCDAV and dsGFP were injected into the hemocoel of the abdomen between the second and third segments with a microinjector. The silencing efficiency of dsSeCDA6, dsSeCDA7, dsSeCDA8, dsSeCDA9 and dsSeCDAV was detected 72 h after dsRNA injection via qRT-PCR. Three biological replications (each with three technical replicates) were used to determine the silencing efficiency. The remaining larvae were maintained for phenotypical observation. The visible phenotypical changes were recorded daily until the larvae molted into adults.
4.6. Immunohistochemistry
Fourth-instar larvae were injected with dsGFP or dsSeCDAV separately to determine the specificity of the SeCDAs antibody. At 72 h after injection, the larvae were collected to prepare paraffin sections. Immunohistology was conducted as follows: SeCDAs were detected with the Group V SeCDAs rabbit antiserum (1:200) as primary antibodies and HRP Rabbit IgG/FITC (1:200) (Servicebio, Wuhan, China) as the secondary antibody for fluorescence detection. Propidium iodide (PI) is an analog of ethidium bromide that binds strongly to DNA and emits red fluorescence upon intercalation in double-stranded DNA, which stains the nucleus red. PI (Servicebio, Wuhan, China) was used for nuclear staining. Images were captured using a transmission electron microscope (HT7800/HT7700, Nikon, Japan) with excitation at 465–495 nm (HRP Rabbit IgG/FITC) and 330–380 nm (PI).
4.7. Scanning Electron Microscopy
The midgut and the PM of S. exigua larvae injected with dsGFP and dsSeCDAV were rinsed gently with PBS (Servicebio, Wuhan, China). The midgut and PM were quickly fixed using an electron microscope fixative (Servicebio, Wuhan, China) at room temperature for 2 h and transferred to 4° for storage. The midgut was washed, and PM was used three times for 15 min each in 0.1 M PBS. The tissues were placed in 1% OsO4 for 2 h at room temperature (Ted Pella Inc., Redding, CA, USA) in 0.1 M PBS. The tissues were washed three times for 15 min each in 0.1 M PBS. The tissues were sequentially added to 30%, 50%, 70%, 80%, 90%, 95% and 100% alcohol and isoamyl acetate for 15 min each time. The samples were dried with a Critical Point Dryer (K850, Quorum, UK) and sputter-coated with gold for 30 s with Lon Sputtering Apparatus (MC1000, Nikon, Japan). The tissues were observed under a scanning electron microscope (SU8100, Nikon, Japan), and the images were stored for analysis [38].
5. Conclusions
Group V SeCDAs (SeCDA6/7/8/9) were identified and characterized from the beet armyworm, S. exigua. The predicted proteins exhibited conserved structural features. SeCDAs were expressed at different developmental stages and in different larval tissues of S. exigua, and their expression was induced by injecting 20E or JHA. RNAi-mediated SeCDAs silencing considerably inhibited the expression of SeCDAs and caused structural changes in the midgut and PM. A reference for improving the silencing efficiency of Lepidoptera and enriching the biological function of Group V CDAs may be provided by the results of this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043076/s1.
Author Contributions
Conceptualization, W.G., D.Z. and H.W.; methodology, H.W. and D.Z.; investigation, X.-C.G. and Z.-R.L.; data curation, H.W. and D.Z.; software, H.W. and D.Z.; validation, W.G. and R.-J.L.; supervision, W.G. and X.-J.L.; writing—review and editing: H.W., W.G. and D.Z.; funding acquisition, W.G. and D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the earmarked fund for Modern Agro-industry Technology Research System (CARS-13), the National Natural Science Foundation of China (Grant No. 31471775), the Science and Technology Project of Hebei Education Department (QN2020119) and the Foundation of the Graduate School of the Chinese Academy of Agricultural Sciences (CAAS) (1610042022005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Azadeh, K.M.; Yaghoub, F.; Ali, A.T.; Mohammad, B. Life Table Parameters and Survivorship of Spodoptera exigua (Lepidoptera: Noctuidae) at Constant Temperatures. Environ. Entomol. 2014, 43, 795–803. [Google Scholar]
- Zhang, W.J.; Dai, H.Q. A quantitative study on development, fecundity and mortality of beet armyworm, Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae), infected by SeMNPV. Arthropods 2018, 7, 26–30. [Google Scholar]
- Zheng, X.L.; Cong, X.P.; Wang, X.P.; Lei, C.L. A Review of Geographic Distribution, Overwintering and Migration in Spodoptera exigua Hübner (Lepidoptera: Noctuidae). J. Entomol. Res. Soc. 2011, 13, 39–48. [Google Scholar]
- Yonow, T.; Kriticos, D.J.; Kirichenko, N.; Ota, N. Considering biology when inferring range-limiting stress mechanisms for agricultural pests: A case study of the beet armyworm. J. Pest Sci. 2018, 91, 523–538. [Google Scholar] [CrossRef]
- Zuo, Y.Y.; Huang, J.L.; Wang, J.; Feng, Y.; Han, T.T.; Wu, Y.D.; Yang, Y.H. Knockout of a P-glycoprotein gene increases susceptibility to abamectin and emamectin benzoate in Spodoptera exigua. Insect Mol. Biol. 2018, 27, 36–45. [Google Scholar] [CrossRef]
- Sun, X.X.; Li, H.Y.; Jiang, Y.J.; Zhang, J.X.; Gu, H.L.; Gao, B.; Ma, J.J.; Wang, F.; Zhou, J.C.; Tian, X.R.; et al. Resistance Risk Evaluated by Metaflumizone Selection and the Effects on Toxicities Over Other Insecticides in Spodoptera exigua (Lepidoptera: Noctuidae). J. Econ. Entomol. 2019, 112, 2354–2361. [Google Scholar] [CrossRef]
- Wang, X.G.; Xiang, X.; Yu, H.L.; Liu, S.H.; Yin, Y.; Cui, P.; Wu, Y.Q.; Yang, J.; Jiang, C.X.; Yang, Q.F. Monitoring and biochemical characterization of beta-cypermethrin resistance in Spodoptera exigua (Lepidoptera: Noctuidae) in Sichuan Province, China. Pestic. Biochem. Phys. 2018, 146, 71–79. [Google Scholar] [CrossRef]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef]
- Tetreau, G.; Cao, X.L.; Chen, Y.R.; Muthukrishnan, S.; Jiang, H.B.; Blissard, G.W.; Kanost, M.R.; Wang, P. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 2015, 62, 114–126. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Ye, Y.X.; Yu, B.; Zhang, C.X. Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 2014, 23, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Dixit, R.; Begum, K.; Park, Y.; Specht, C.A.; Merzendorfer, H.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2009, 39, 355–365. [Google Scholar] [CrossRef]
- Guo, W.; Li, G.; Pang, Y.; Wang, P. A novel chitin-binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 2005, 35, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Wu, H.; Yan, X.P.; Guo, W.; Lu, X.J.; Guo, X.C.; Liu, Z.R.; Li, Y.Z. Molecular cloning and functional characterization of two insect chitin deacetylases from the American white moth, Hyphantria cunea. J. Appl. Entomol. 2021, 145, 697–706. [Google Scholar] [CrossRef]
- Yan, X.P.; Zhao, D.; Zhang, Y.K.; Guo, W.; Wang, W.; Zhao, K.L.; Gao, Y.J.; Wang, X.Y. Identification and characterization of chitin deacetylase 2 from the American white moth, Hyphantria cunea (Drury). Gene 2018, 670, 98–105. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Yan, J.M.; Liu, Q.; Zhang, Y.H.; Gong, J.; Hou, Y. Genome-Wide Analysis and Hormone Regulation of Chitin Deacetylases in Silkworm. Int. J. Mol. Sci. 2019, 20, 1679. [Google Scholar] [CrossRef] [PubMed]
- Han, G.Y.; Li, X.M.; Zhang, T.; Zhu, X.T.; Li, J.G. Cloning and Tissue-Specific Expression of a Chitin Deacetylase Gene from Helicoverpa armigera (Lepidoptera: Noctuidae) and Its Response to Bacillus thuringiensis. J. Insect Sci. 2015, 15, 95. [Google Scholar] [CrossRef]
- Quan, G.; Ladd, T.; Duan, J.; Wen, F.; Doucet, D.; Cusson, M.; Krell, P.J. Characterization of a spruce budworm chitin deacetylase gene: Stage- and tissue-specific expression, and inhibition using RNA interference. Insect Biochem. Mol. Biol. 2013, 43, 683–691. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, K.K.; Yan, X.; Chen, C.X.; Cao, Y.; Meng, Y.L.; Li, C. Functional characterization of chitin deacetylase 1 gene disrupting larval–pupal transition in the drugstore beetle using RNA interference. Comp. Biochem. Physiol. B 2018, 219, 10–16. [Google Scholar] [CrossRef]
- Wu, J.J.; Chen, Z.C.; Wang, Y.W.; Fu, K.Y.; Guo, W.C.; Li, G.Q. Silencing chitin deacetylase 2 impairs larval–pupal and pupal-adult molts in Leptinotarsa decemlineata. Insect Mol. Biol. 2019, 28, 52–64. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, Z.R.; Wu, H.; Fu, C.R.; Li, Y.Z.; Lu, X.J.; Guo, W. RNA interference-mediated functional characterization of Group I chitin deacetylases in Holotrichia parallela Motschulsky. Pestic. Biochem. Physiol. 2021, 173, 104770. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.R.; Liu, W.M.; Zhao, X.M.; Zhang, M.; Li, D.Q.; Zuber, R.; Ma, E.B.; Zhu, K.Y.; Moussian, B.; Zhang, J.Z. LmCDA1 organizes the cuticle by chitin deacetylation in Locusta migratoria. Insect Mol. Biol. 2019, 28, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Li, H.F.; Yang, Y.; Yang, R.L.; Yang, W.J.; Jiang, H.B.; Dou, W.; Smagghe, G.; Wang, J.J. Genome-wide identification of chitinase and chitin deacetylase gene families in the oriental fruit fly, Bactrocera dorsalis (Hendel). Comp. Biochem. Physiol. D 2018, 27, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, S.; Batz, T.; Armbruster, K.; Krasnow, M.A. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef]
- Sandoval-Mojica, A.F.; Scharf, M.E. Gut genes associated with the peritrophic matrix in Reticulitermes flavipes (Blattodea: Rhinotermitidae): Identification and characterization. Arch. Insect Biochem. Physiol. 2016, 92, 127–142. [Google Scholar] [CrossRef]
- Toprak, U.; Baldwin, D.; Erlandson, M.; Gillott, C.; Hou, X.; Coutu, C.; Hegedus, D.D. A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol. Biol. 2008, 17, 573–585. [Google Scholar] [CrossRef]
- Zhong, X.W.; Wang, X.H.; Tan, X.; Xia, Q.Y.; Xiang, Z.H.; Zhao, P. Identification and molecular characterization of a chitin deacetylase from Bombyx mori peritrophic membrane. Int. J. Mol. Sci. 2014, 15, 1946–1961. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.M.; Cao, A.T.; Hines, E.R.; East, P.D.; Gordon, K.H.J. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. 2008, 38, 950–958. [Google Scholar] [CrossRef]
- Liu, L.; Qu, M.B.; Liu, T.; Chen, Q.; Guo, X.M.; Yang, J.; Yang, Q. Biochemical characterization of three midgut chitin deacetylases of the Lepidopteran insect Bombyx mori. J. Insect Physiol. 2019, 113, 42–48. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Rybczynski, R.; Warren, J.T. Control and Biochemical Nature of the Ecdysteroidogenic Pathway. Annu. Rev. Entomol. 2002, 47, 883–916. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.Z.; Zhang, K.; Li, C.Y.; Hu, X.; Kausar, S.; Gu, H.Y.; Yang, L.Q.; Cui, H.J. A hemocyte-specific cathepsin L-like cysteine protease is involved in response to 20-hydroxyecdysone and microbial pathogens stimulation in silkworm, Bombyx mori. Mol. Immunol. 2021, 131, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.L.; Wang, J.Y.; Wang, Y.Z.; Yang, J.J.; Wei, B.F.; Li, M.M.; Wang, B.; Li, X.Z.; Cao, Y.; Tian, L.; et al. Transcriptome Analysis Reveals Potential Function of Long Non-coding RNAs in 20-hydroxyecdysone Regulated Autophagy in Bombyx Mori. BMC Genomics 2021, 22, 374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Su, J.J.; Chen, S.Y.; Yu, S.; Tan, J.; Xu, M.; Liang, H.H.; Zhao, Y.Z.; Chao, H.J.; Yang, L.Q.; et al. Molecular cloning, characterization and expression analysis of cathepsin O in silkworm Bombyx mori related to bacterial response. Mol. Immunol. 2015, 66, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Zhang, D.W.; Tang, B.; Chen, J.; Chen, J.; Lu, L.; Zhang, W.Q. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway. PLoS ONE 2010, 5, e14058. [Google Scholar] [CrossRef]
- Cai, R.; Tao, G.; Zhao, P.; Xia, Q.Y.; He, H.W.; Wang, Y.J. POU-M2 promotes juvenile hormone biosynthesis by directly activating the transcription of juvenile hormone synthetic enzyme genes in Bombyx mori. Open Biol. 2022, 12, 220031. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.W.; Yao, X.G.; Yang, Q.F.; Wang, X.G.; Zhang, Y.M.; Wang, Y.M.; Shen, L.T. Fitness Costs of Chlorantraniliprole Resistance Related to the SeNPF Overexpression in the Spodoptera exigua (Lepidoptera: Noctuidae). Int. J. Mol. Sci. 2021, 22, 5027. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, K.K.; Yan, Y.; Li, C.; Jin, D.C. Role of Chitin Deacetylase 1 in the molting and metamorphosis of the Cigarette Beetle Lasioderma serricorne. Int. J. Mol. Sci. 2020, 21, 2449. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, J.; Yao, L.; Li, S.Y.; Chen, Y.R.; Yang, H.J.; Fan, D. Chitin deacetylase: A potential target for Mythimna separata (Walker) control. Arch. Insect. Biochem. Physiol. 2020, 104, e21666. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Xue, B.; Li, Y.Y.; Li, F.C.; Ni, M.; Shen, W.D.; Gu, Z.Y.; Li, B. Construction of silkworm midgut cDNA library for screen and sequence analysis of peritrophic membrane protein genes. Arch. Insect. Biochem. Physiol. 2016, 91, 3–16. [Google Scholar] [CrossRef]
- Liu, L.; Qu, M.; Yang, J.; Yang, Q. The physiological differentiation along the midgut of Bombyx mori–inspirations from proteomics and gene expression patterns of the secreted proteins in the ectoperitrophic space. Insect Mol. Biol. 2018, 27, 247–259. [Google Scholar] [CrossRef]
- Harper, M.S.; Hopkins, T.L. Peritrophic membrane structure and secretion in European corn borer larvae (Ostrinia nubilalis). Tissue Cell 1997, 29, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Toprak, U.; Hegedus, D.D.; Baldwin, D.; Coutu, C.; Erlandson, M. Spatial and temporal synthesis of Mamestra configurata peritrophic matrix through a larval stadium. Insect Biochem. Mol. Biol. 2014, 54, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jayaram, S.J.; Senti, K.; Tsarouhas, V.; Jin, H.; Samakovlis, C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006, 16, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.R.; Liu, W.M.; Li, D.Q.; Zhao, X.M.; Ding, G.W.; Zhang, M.; Ma, E.B.; Zhu, K.Y.; Li, S.; Moussian, B.; et al. Helicoidal Organization of Chitin in the Cuticle of the Migratory Locust Requires the Function of the Chitin Deacetylase2 Enzyme (LmCDA2). J. Biol. Chem. 2016, 291, 24352–24363. [Google Scholar] [CrossRef] [PubMed]
- Jakubowska, A.K.; Caccia, S.; Gordon, K.H.; Ferre, J.; Herrero, S. Downregulation of a chitin deacetylase-like protein in response to baculovirus infection and its application for improving baculovirus infectivity. J. Virol. 2010, 84, 2547–2555. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Zhao, D.; Wu, H.; Ji, Y.J.; Liu, Z.R.; Guo, X.C.; Guo, W.; Bi, Y. Bt GS57 interaction with gut microbiota accelerates Spodoptera exigua mortality. Front. Microbiol. 2022, 13, 835227. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).