A Functional Polymorphism Downstream of Vitamin A Regulator Gene CYP26B1 Is Associated with Hand Osteoarthritis
Abstract
1. Introduction
2. Results
2.1. Overview of the Study Population
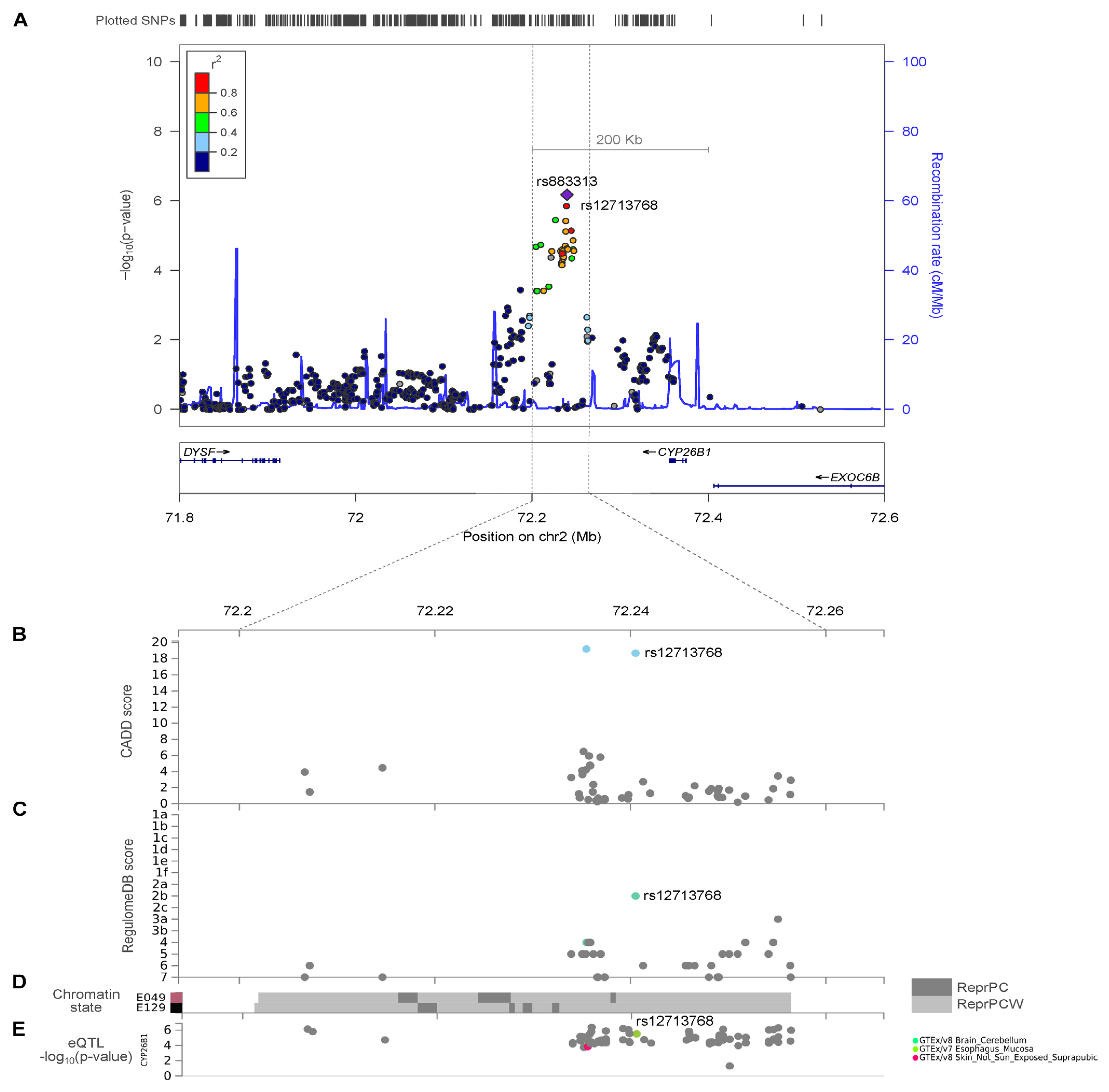
2.2. Hand OA Genome-Wide Association Analysis
2.3. CYP26B1/rs12713768 Is a Potential Causal Gene for Hand OA
2.4. Effect of SNP Variants rs12713768 and rs10208040 on the Predicted Enhancer Activity of CYP26B1 Gene
3. Discussion
4. Methods
4.1. Study Populations
4.2. Phenotype Definition for Hand OA
4.2.1. Inclusion Criteria
4.2.2. Exclusion Criteria
4.3. Phenotype Definition of Non-OA Control Subjects
4.3.1. Inclusion Criteria
4.3.2. Exclusion Criteria
4.4. Genotyping, Quality Control, and Imputation
4.5. Genome-Wide SNPs Cross-Platform Validation
4.6. Statistical and Bioinformatics Database Analysis
4.7. Construction of Luciferase Reporter Plasmids
4.8. Cell Cultures and Luciferase Reporter Assay
4.9. Statistics for Luciferase Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kloppenburg, M.; Kwok, W.-Y. Hand osteoarthritis—A heterogeneous disorder. Nat. Rev. Rheumatol. 2011, 8, 22–31. [Google Scholar] [CrossRef]
- Marshall, M.; Watt, F.E.; Vincent, T.L.; Dziedzic, K. Hand osteoarthritis: Clinical phenotypes, molecular mechanisms and disease management. Nat. Rev. Rheumatol. 2018, 14, 641–656. [Google Scholar] [CrossRef]
- Leung, G.J.; Rainsford, K.D.; Kean, W.F. Osteoarthritis of the hand I: Aetiology and pathogenesis, risk factors, investigation and diagnosis. J. Pharm. Pharmacol. 2014, 66, 339–346. [Google Scholar] [CrossRef]
- van Saase, J.L.; van Romunde, L.K.; Cats, A.; Vandenbroucke, J.P.; Valkenburg, H.A. Epidemiology of osteoarthritis: Zoetermeer survey. Comparison of radiological osteoarthritis in a Dutch population with that in 10 other populations. Ann. Rheum. Dis. 1989, 48, 271–280. [Google Scholar] [CrossRef]
- Carmona, L.; Ballina, J.; Gabriel, R.; Laffon, A.; EPISER Study Group. The burden of musculoskeletal diseases in the general population of Spain: Results from a national survey. Ann. Rheum. Dis. 2001, 60, 1040–1045. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, L.; Nevitt, M.C.; Niu, J.; Goggins, J.P.; Aliabadi, P.; Yu, W.; Lui, L.-Y.; Felson, D.T. Lower prevalence of hand osteoarthritis among Chinese subjects in Beijing compared with white subjects in the United States: The Beijing Osteoarthritis Study. Arthritis Rheum. 2003, 48, 1034–1040. [Google Scholar] [CrossRef]
- Kodama, R.; Muraki, S.; Oka, H.; Iidaka, T.; Teraguchi, M.; Kagotani, R.; Asai, Y.; Yoshida, M.; Morizaki, Y.; Tanaka, S.; et al. Prevalence of hand osteoarthritis and its relationship to hand pain and grip strength in Japan: The third survey of the ROAD study. Mod. Rheumatol. 2016, 26, 767–773. [Google Scholar] [CrossRef]
- Sowers, M.; Lachance, L.; Hochberg, M.; Jamadar, D. Radiographically defined osteoarthritis of the hand and knee in young and middle-aged African American and Caucasian women. Osteoarthr. Cartil. 2000, 8, 69–77. [Google Scholar] [CrossRef]
- Toba, N.; Sakai, A.; Aoyagi, K.; Yoshida, S.; Honda, S.; Nakamura, T. Prevalence and involvement patterns of radiographic hand osteoarthritis in Japanese women: The Hizen-Oshima Study. J. Bone Miner. Metab. 2006, 24, 344–348. [Google Scholar] [CrossRef]
- Bang, S.-Y.; Son, C.-N.; Sung, Y.-K.; Choi, B.K.; Joo, K.-B.; Jun, J.-B. Joint-specific prevalence and radiographic pattern of hand osteoarthritis in Korean. Rheumatol. Int. 2011, 31, 361–364. [Google Scholar] [CrossRef]
- Zhang, R.; Yao, J.; Xu, P.; Ji, B.; Luck, J.V.; Chin, B.; Lu, S.; Kelsoe, J.R.; Ma, J. A comprehensive meta-analysis of association between genetic variants of GDF5 and osteoarthritis of the knee, hip and hand. Inflamm. Res. 2015, 64, 405–414. [Google Scholar] [CrossRef]
- Chapman, K.; Takahashi, A.; Meulenbelt, I.; Watson, C.; Rodriguez-Lopez, J.; Egli, R.; Tsezou, A.; Malizos, K.N.; Kloppenburg, M.; Shi, D.; et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5’ UTR of GDF5 with osteoarthritis susceptibility. Hum. Mol. Genet. 2008, 17, 1497–1504. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Mabuchi, A.; Shi, D.; Kubo, T.; Takatori, Y.; Saito, S.; Fujioka, M.; Sudo, A.; Uchida, A.; Yamamoto, S.; et al. A functional polymorphism in the 5’ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007, 39, 529–533. [Google Scholar] [CrossRef]
- Stecher, R.M. Heberden’s nodes: The incidence of hypertrophic arthritis of the fingers. N. Engl. J. Med. 1940, 222, 300–308. [Google Scholar] [CrossRef]
- Spector, T.D.; Cicuttini, F.; Baker, J.; Loughlin, J.; Hart, D. Genetic influences on osteoarthritis in women: A twin study. BMJ 1996, 312, 940–943. [Google Scholar] [CrossRef]
- Horton, W.E.; Lethbridge-Cejku, M.; Hochberg, M.C.; Balakir, R.; Precht, P.; Plato, C.C.; Tobin, J.D.; Meek, L.; Doege, K. An association between an aggrecan polymorphic allele and bilateral hand osteoarthritis in elderly white men: Data from the Baltimore Longitudinal Study of Aging (BLSA). Osteoarthr. Cartil. 1998, 6, 245–251. [Google Scholar] [CrossRef]
- Kirk, K.M.; Doege, K.J.; Hecht, J.; Bellamy, N.; Martin, N.G. Osteoarthritis of the hands, hips and knees in an Australian twin sample--evidence of association with the aggrecan VNTR polymorphism. Twin Res. 2003, 6, 62–66. [Google Scholar]
- Kämäräinen, O.P.; Solovieva, S.; Vehmas, T.; Luoma, K.; Leino-Arjas, P.; Riihimäki, H.; Ala-Kokko, L.; Männikkö, M. Aggrecan core protein of a certain length is protective against hand osteoarthritis. Osteoarthr. Cartil. 2006, 14, 1075–1080. [Google Scholar] [CrossRef]
- Ross, J.M.; Kowalchuk, R.M.; Shaulinsky, J.; Ross, L.; Ryan, D.; Phatak, P.D. Association of heterozygous hemochromatosis C282Y gene mutation with hand osteoarthritis. J. Rheumatol. 2003, 30, 121–125. [Google Scholar]
- Carroll, G.J. HFE gene mutations are associated with osteoarthritis in the index or middle finger metacarpophalangeal joints. J. Rheumatol. 2006, 33, 741–743. [Google Scholar]
- Zhai, G.; van Meurs, J.B.J.; Livshits, G.; Meulenbelt, I.; Valdes, A.M.; Soranzo, N.; Hart, D.; Zhang, F.; Kato, B.S.; Richards, J.B.; et al. A genome-wide association study suggests that a locus within the ataxin 2 binding protein 1 gene is associated with hand osteoarthritis: The Treat-OA consortium. J. Med. Genet. 2009, 46, 614–616. [Google Scholar] [CrossRef]
- Styrkarsdottir, U.; Thorleifsson, G.; Helgadottir, H.T.; Bomer, N.; Metrustry, S.; Bierma-Zeinstra, S.; Strijbosch, A.M.; Evangelou, E.; Hart, D.; Beekman, M.; et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat. Genet. 2014, 46, 498–502. [Google Scholar] [CrossRef]
- den Hollander, W.; Boer, C.G.; Hart, D.J.; Yau, M.S.; Ramos, Y.F.M.; Metrustry, S.; Broer, L.; Deelen, J.; Cupples, L.A.; Rivadeneira, F.; et al. Genome-wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Ann. Rheum. Dis. 2017, 76, 2046–2053. [Google Scholar] [CrossRef]
- Boer, C.G.; Yau, M.S.; Rice, S.J.; Coutinho de Almeida, R.; Cheung, K.; Styrkarsdottir, U.; Southam, L.; Broer, L.; Wilkinson, J.M.; Uitterlinden, A.G.; et al. Genome-wide association of phenotypes based on clustering patterns of hand osteoarthritis identify WNT9A as novel osteoarthritis gene. Ann. Rheum. Dis. 2020, 3, 367–375. [Google Scholar] [CrossRef]
- Boer, C.G.; Hatzikotoulas, K.; Southam, L.; Stefánsdóttir, L.; Zhang, Y.; Coutinho de Almeida, R.; Wu, T.T.; Zheng, J.; Hartley, A.; Teder-Laving, M.; et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 2021, 184, 4784–4818. [Google Scholar] [CrossRef]
- Reynard, L.N.; Barter, M.J. Osteoarthritis year in review 2019: Genetics, genomics and epigenetics. Osteoarthr. Cartil. 2020, 28, 275–284. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, J.-C.; Hon, C.-C.; Fukui, N.; Tanaka, N.; Zhang, Z.; Lee, M.T.M.; Minoda, A. Chromatin accessibility landscape of articular knee cartilage reveals aberrant enhancer regulation in osteoarthritis. Sci. Rep. 2018, 8, 15499. [Google Scholar] [CrossRef]
- White, J.A.; Ramshaw, H.; Taimi, M.; Stangle, W.; Zhang, A.; Everingham, S.; Creighton, S.; Tam, S.P.; Jones, G.; Petkovich, M. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc. Natl. Acad. Sci. USA 2000, 97, 6403–6408. [Google Scholar] [CrossRef]
- MacLean, G.; Abu-Abed, S.; Dollé, P.; Tahayato, A.; Chambon, P.; Petkovich, M. Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech. Dev. 2001, 107, 195–201. [Google Scholar] [CrossRef]
- Williams, J.A.; Kane, M.; Okabe, T.; Enomoto-Iwamoto, M.; Napoli, J.L.; Pacifici, M.; Iwamoto, M. Endogenous retinoids in mammalian growth plate cartilage: Analysis and roles in matrix homeostasis and turnover. J. Biol. Chem. 2010, 285, 36674–36681. [Google Scholar] [CrossRef]
- Yashiro, K.; Zhao, X.; Uehara, M.; Yamashita, K.; Nishijima, M.; Nishino, J.; Saijoh, Y.; Sakai, Y.; Hamada, H. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev. Cell 2004, 6, 411–422. [Google Scholar] [CrossRef]
- Maclean, G.; Dollé, P.; Petkovich, M. Genetic disruption of CYP26B1 severely affects development of neural crest derived head structures, but does not compromise hindbrain patterning. Dev. Dyn. 2009, 238, 732–745. [Google Scholar] [CrossRef]
- Carvalho, J.E.; Theodosiou, M.; Chen, J.; Chevret, P.; Alvarez, S.; De Lera, A.R.; Laudet, V.; Croce, J.C.; Schubert, M. Lineage-specific duplication of amphioxus retinoic acid degrading enzymes (CYP26) resulted in sub-functionalization of patterning and homeostatic roles. BMC Evol. Biol. 2017, 17, 24. [Google Scholar] [CrossRef]
- Shepherd, C.; Zhu, D.; Skelton, A.J.; Combe, J.; Threadgold, H.; Zhu, L.; Vincent, T.L.; Stuart, P.; Reynard, L.N.; Loughlin, J. Functional Characterization of the Osteoarthritis Genetic Risk Residing at ALDH1A2 Identifies rs12915901 as a Key Target Variant. Arthritis Rheumatol. 2018, 70, 1577–1587. [Google Scholar] [CrossRef]
- Davies, M.R.; Ribeiro, L.R.; Downey-Jones, M.; Needham, M.R.C.; Oakley, C.; Wardale, J. Ligands for retinoic acid receptors are elevated in osteoarthritis and may contribute to pathologic processes in the osteoarthritic joint. Arthritis Rheum. 2009, 60, 1722–1732. [Google Scholar] [CrossRef]
- Schaefer, L.F.; McAlindon, T.E.; Eaton, C.B.; Roberts, M.B.; Haugen, I.K.; Smith, S.E.; Duryea, J.; Driban, J.B. The associations between radiographic hand osteoarthritis definitions and hand pain: Data from the osteoarthritis initiative. Rheumatol. Int. 2018, 38, 403–413. [Google Scholar] [CrossRef]
- McDougall, J.J.; Albacete, S.; Schuelert, N.; Mitchell, P.G.; Lin, C.; Oskins, J.L.; Bui, H.H.; Chambers, M.G. Lysophosphatidic acid provides a missing link between osteoarthritis and joint neuropathic pain. Osteoarthr. Cartil. 2017, 25, 926–934. [Google Scholar] [CrossRef]
- Cho, C.H.; Lho, Y.M.; Ha, E.; Hwang, I.; Song, K.S.; Min, B.W.; Bae, K.C.; Kim, D.H. Up-regulation of acid-sensing ion channels in the capsule of the joint in frozen shoulder. Bone Jt. J. 2015, 97-B, 824–829. [Google Scholar] [CrossRef]
- Kanaya, K.; Iba, K.; Dohke, T.; Okazaki, S.; Yamashita, T. TRPV1, ASICs and P2X2/3 expressed in bone cells simultaneously regulate bone metabolic markers in ovariectomized mice. J. Musculoskelet. Neuronal Interact. 2016, 16, 145–151. [Google Scholar]
- Hanaka, M.; Iba, K.; Dohke, T.; Kanaya, K.; Okazaki, S.; Yamashita, T. Antagonists to TRPV1, ASICs and P2X have a potential role to prevent the triggering of regional bone metabolic disorder and pain-like behavior in tail-suspended mice. Bone 2018, 110, 284–294. [Google Scholar] [CrossRef]
- Suzuki, A.; Minamide, M.; Iwaya, C.; Ogata, K.; Iwata, J. Role of metabolism in bone development and homeostasis. Int. J. Mol. Sci. 2020, 21, 8992. [Google Scholar] [CrossRef]
- Jahr, H.; van Driel, M.; van Osch, G.J.V.M.; Weinans, H.; van Leeuwen, J.P.T.M. Identification of acid-sensing ion channels in bone. Biochem. Biophys. Res. Commun. 2005, 337, 349–354. [Google Scholar] [CrossRef]
- Haringman, J.J.; Smeets, T.J.M.; Reinders-Blankert, P.; Tak, P.P. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann. Rheum. Dis. 2006, 65, 294–300. [Google Scholar] [CrossRef]
- Votta, B.J.; White, J.R.; Dodds, R.A.; James, I.E.; Connor, J.R.; Lee-Rykaczewski, E.; Eichman, C.F.; Kumar, S.; Lark, M.W.; Gowen, M. CKbeta-8 [CCL23], a novel CC chemokine, is chemotactic for human osteoclast precursors and is expressed in bone tissues. J. Cell Physiol. 2000, 183, 196–207. [Google Scholar] [CrossRef]
- Gao, K.; Zhu, W.; Li, H.; Ma, D.; Liu, W.; Yu, W.; Wang, L.; Cao, Y.; Jiang, Y. Association between cytokines and exosomes in synovial fluid of individuals with knee osteoarthritis. Mod. Rheumatol. 2020, 30, 758–764. [Google Scholar] [CrossRef]
- Vergunst, C.E.; van de Sande, M.G.H.; Lebre, M.C.; Tak, P.P. The role of chemokines in rheumatoid arthritis and osteoarthritis. Scand. J. Rheumatol. 2005, 34, 415–425. [Google Scholar] [CrossRef]
- Goldring, M.B.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 471–478. [Google Scholar] [CrossRef]
- Carey, D.J.; Fetterolf, S.N.; Davis, F.D.; Faucett, W.A.; Kirchner, H.L.; Mirshahi, U.; Murray, M.F.; Smelser, D.T.; Gerhard, G.S.; Ledbetter, D.H. The Geisinger MyCode community health initiative: An electronic health record-linked biobank for precision medicine research. Genet. Med. 2016, 18, 906–913. [Google Scholar] [CrossRef]
- Kraus, V.B.; Jordan, J.M.; Doherty, M.; Wilson, A.G.; Moskowitz, R.; Hochberg, M.; Loeser, R.; Hooper, M.; Renner, J.B.; Crane, M.M.; et al. The Genetics of Generalized Osteoarthritis (GOGO) study: Study design and evaluation of osteoarthritis phenotypes. Osteoarthr. Cartil. 2007, 15, 120–127. [Google Scholar] [CrossRef]
- Kellgren, J.H.; Lawrence, J.S. Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 1957, 16, 494–502. [Google Scholar] [CrossRef]
- American Rheumatism Association Glossary Committee. Dictionary of the rheumatic diseases. Signs Symptoms 1982, 1, 1–80. [Google Scholar]
- Devlin, B.; Roeder, K.; Wasserman, L. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol. 2001, 60, 155–166. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated tool for Genome-Wide Association Study. BioRxiv 2020, 19, 619–628. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Nomura, A.; Tada, H.; Teramoto, R.; Konno, T.; Hodatsu, A.; Won, H.-H.; Kathiresan, S.; Ino, H.; Fujino, N.; Yamagishi, M.; et al. Whole exome sequencing combined with integrated variant annotation prediction identifies a causative myosin essential light chain variant in hypertrophic cardiomyopathy. J. Cardiol. 2016, 67, 133–139. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2—Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-0-387-98140-6. [Google Scholar]




| Characteristic | Affected | Unaffected Control |
|---|---|---|
| N | 420 | 716 |
| F/M (n, %) | 365 (87)/55 (13) | 547 (76)/169 (24) |
| Age at sampling, years ± SD a (min–max) | 64.7 ± 10.4 (32–94) | 64.1 ± 9.7 (34–93) |
| Height, cm ± SD b (min–max) | 155.81 ± 6.9 (137–184) | 157.58 ± 6.9 (138.5–180) |
| Weight, kg ± SD (min–max) | 59.87 ± 9.21 (36–100) | 61.32 ± 9.91 (35–106) |
| BMI, kg/m2 ± SD b (min–max) | 24.64 ± 3.34 (15.51–39.11) | 24.67 ± 3.53 (13.01–36.16) |
| BMI ≤ 24.9 kg/m2 | 244 (58.10%) | 414 (57.90%) |
| 25.0 > BMI < 29.9 kg/m2 | 155 (36.90%) | 246 (34.41%) |
| BMI ≥ 30.9 kg/m2 | 21 (5%) | 55 (7.69%) |
| Chr | SNP | Position (bp) | Allele Format | Risk Allele | Risk Allele OR (95% CI) | RAF Controls | RAF Cases | p-Value | Nearest Gene a | |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs1517396 | 72,228,008 | CT | T | 1.588 (1.308–1.928) | 0.6568 | 0.7525 | 3.43 × 10−6 | CYP26B1 | Imputed |
| 2 | rs883313 | 72,239,692 | TC | C | 1.755 (1.409–2.185) | 0.7468 | 0.8381 | 6.76 × 10−7 | CYP26B1 | Imputed |
| 2 | rs883312 | 72,239,762 | CT | T | 1.581 (1.305–1.916) | 0.6620 | 0.7560 | 3.65 × 10−6 | CYP26B1 | |
| 2 | rs883311 | 72,239,809 | GA | A | 1.560 (1.287–1.891) | 0.6636 | 0.7548 | 7.31 × 10−6 | CYP26B1 | Imputed |
| 2 | rs12713768 | 72,240,527 | AC | C | 1.737 (1.393–2.164) | 0.7521 | 0.8405 | 1.36 × 10−6 | CYP26B1 | |
| 2 | rs191066740 | 72,245,915 | TG | G | 1.721 (1.364–2.173) | 0.7746 | 0.8554 | 6.97 × 10−6 | CYP26B1 | Imputed |
| 6 | rs1614699 | 38,738,748 | AT | A | 1.809 (1.399–2.338) | 0.0949 | 0.1595 | 5.30 × 10−6 | DNAH8, ZRF1PS | Imputed |
| 6 | rs1678657 | 38,742,793 | GT | G | 1.809 (1.399–2.338) | 0.0949 | 0.1595 | 5.30 × 10−6 | DNAH8, ZRF1PS | Imputed |
| 6 | rs1738263 | 38,743,252 | CT | C | 1.809 (1.399–2.338) | 0.0949 | 0.1595 | 5.30 × 10−6 | DNAH8, ZRF1PS | Imputed |
| 6 | rs12197517 | 161,666,190 | CG | G | 2.327 (1.577–3.432) | 0.9088 | 0.9586 | 8.50 × 10−6 | AGPAT4 | Imputed |
| 6 | rs62437572 | 161,666,765 | CT | T | 2.360 (1.601–3.479) | 0.9076 | 0.9586 | 5.62 × 10−6 | AGPAT4 | Imputed |
| 6 | rs10945720 | 161,668,745 | AG | G | 2.366 (1.605–3.488) | 0.9076 | 0.9587 | 5.25 × 10−6 | AGPAT4 | Imputed |
| 6 | rs12190239 | 161,669,231 | GC | C | 2.362 (1.602–3.482) | 0.9077 | 0.9587 | 5.48 × 10−6 | AGPAT4 | Imputed |
| 6 | rs112790908 | 161,670,764 | AAT | AT | 2.358 (1.600–3.477) | 0.9079 | 0.9587 | 5.72 × 10−6 | AGPAT4 | Imputed |
| 6 | rs12207205 | 161,671,403 | AG | G | 2.367 (1.606–3.489) | 0.9075 | 0.9587 | 5.05 × 10−6 | AGPAT4 | Imputed |
| 6 | rs73019329 | 161,678,741 | CT | T | 2.353 (1.596–3.469) | 0.9083 | 0.9588 | 6.06 × 10−6 | AGPAT4 | Imputed |
| 12 | rs11108612 | 97,041,461 | AG | G | 1.484 (1.244–1.771) | 0.5380 | 0.6335 | 9.70 × 10−6 | CFAP54 | Imputed |
| 12 | rs11108617 | 97,047,776 | AG | G | 1.499 (1.253–1.792) | 0.5359 | 0.6338 | 7.45 × 10−6 | CFAP54 | Imputed |
| 17 | rs76055737 | 31,565,197 | TC | C | 1.947 (1.442–2.629) | 0.8632 | 0.9248 | 9.89 × 10−6 | ASIC2 | Imputed |
| 17 | rs79186365 | 31,565,269 | AG | G | 1.953 (1.447–2.638) | 0.8629 | 0.9248 | 8.90 × 10−6 | ASIC2 | Imputed |
| 17 | rs111761296 | 34,309,769 | CCCCTT | C | 2.124 (1.552–2.905) | 0.0579 | 0.1156 | 2.56 × 10−6 | LYZL6, CCL14, CCL15, CCL16, CCL23 | Imputed |
| 17 | rs76190126 | 34,310,256 | AC | A | 2.024 (1.485–2.758) | 0.0601 | 0.1147 | 8.39 × 10−6 | LYZL6, CCL14, CCL15, CCL16, CCL23 | Imputed |
| 17 | rs10491118 | 34,314,886 | TG | T | 1.971 (1.475–2.636) | 0.0677 | 0.1253 | 5.99 × 10−6 | LYZL6, CCL14, CCL15, CCL16, CCL23 | Imputed |
| 17 | rs12051658 | 35,988,597 | GA | G | 1.988 (1.488–2.656) | 0.0677 | 0.1262 | 4.35 × 10−6 | LYZL6, CCL14, CCL15, CCL16, CCL23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khosasih, V.; Liu, K.-M.; Huang, C.-M.; Liou, L.-B.; Hsieh, M.-S.; Lee, C.-H.; Tsai, C.-Y.; Kuo, S.-Y.; Hwa, S.-Y.; Yu, C.-L.; et al. A Functional Polymorphism Downstream of Vitamin A Regulator Gene CYP26B1 Is Associated with Hand Osteoarthritis. Int. J. Mol. Sci. 2023, 24, 3021. https://doi.org/10.3390/ijms24033021
Khosasih V, Liu K-M, Huang C-M, Liou L-B, Hsieh M-S, Lee C-H, Tsai C-Y, Kuo S-Y, Hwa S-Y, Yu C-L, et al. A Functional Polymorphism Downstream of Vitamin A Regulator Gene CYP26B1 Is Associated with Hand Osteoarthritis. International Journal of Molecular Sciences. 2023; 24(3):3021. https://doi.org/10.3390/ijms24033021
Chicago/Turabian StyleKhosasih, Vivia, Kai-Ming Liu, Chung-Ming Huang, Lieh-Bang Liou, Ming-Shium Hsieh, Chian-Her Lee, Chang-Youh Tsai, San-Yuan Kuo, Su-Yang Hwa, Chia-Li Yu, and et al. 2023. "A Functional Polymorphism Downstream of Vitamin A Regulator Gene CYP26B1 Is Associated with Hand Osteoarthritis" International Journal of Molecular Sciences 24, no. 3: 3021. https://doi.org/10.3390/ijms24033021
APA StyleKhosasih, V., Liu, K.-M., Huang, C.-M., Liou, L.-B., Hsieh, M.-S., Lee, C.-H., Tsai, C.-Y., Kuo, S.-Y., Hwa, S.-Y., Yu, C.-L., Chang, C.-H., Lin, C.-J., Hsieh, S.-C., Cheng, C.-Y., Chen, W.-M., Chen, L.-K., Chuang, H.-P., Chen, Y.-T., Tsai, P.-C., ... Wu, J.-Y. (2023). A Functional Polymorphism Downstream of Vitamin A Regulator Gene CYP26B1 Is Associated with Hand Osteoarthritis. International Journal of Molecular Sciences, 24(3), 3021. https://doi.org/10.3390/ijms24033021





