Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis
Abstract
1. Introduction
2. Spatiotemporal-Specific Skeletal Stem Cells
3. Heterogeneity of BMSCs by scRNA-Seq
4. Cellular Dynamics of BMSCs
5. Role of BMSCs on Bone Regeneration
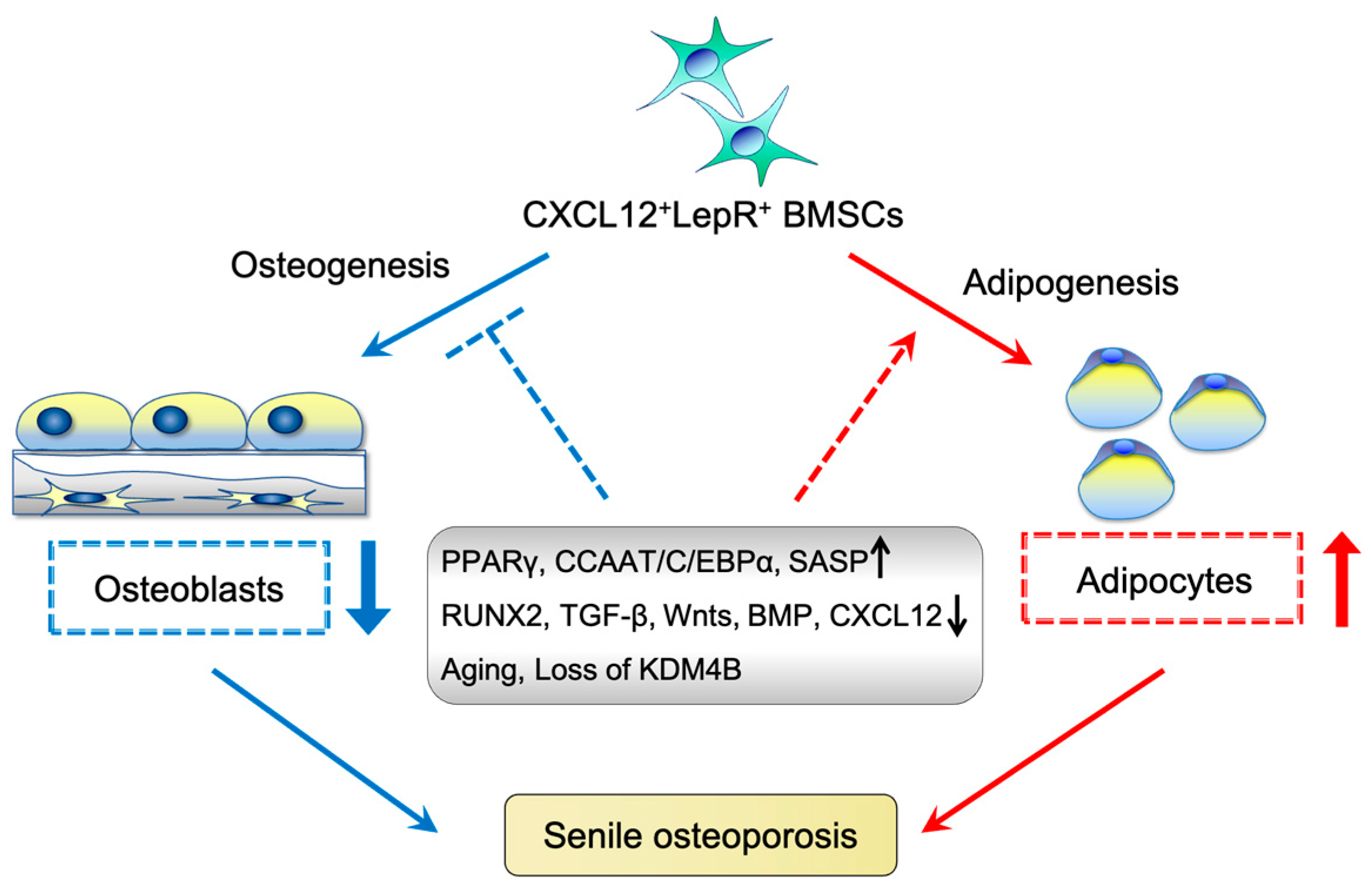
6. Characteristics of Aging BMSCs
7. Effect of BMSC Aging on Osteoporosis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fu, R.; Lv, W.C.; Xu, Y.; Gong, M.Y.; Chen, X.J.; Jiang, N.; Yao, Q.Q.; Di, L.; Lu, T.; Wang, L.M.; et al. Endothelial ZEB1 promotes angiogenesis-dependent bone formation and reverses osteoporosis. Nat. Commun. 2020, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Huo, L.; Liu, Y.; Deng, P.; Szymanski, J.; Li, J.; Luo, X.; Hong, C.; Lin, J.; Wang, C.Y. PGC-1alpha Controls Skeletal Stem Cell Fate and Bone-Fat Balance in Osteoporosis and Skeletal Aging by Inducing TAZ. Cell Stem Cell 2018, 23, 193–209.e5. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, Y.; Matsushita, Y. Bone regeneration: A message from clinical medicine and basic science. Clin. Anat. 2022, 35, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Rajawat, J. Skeletal Aging and Osteoporosis: Mechanisms and Therapeutics. Int. J. Mol. Sci. 2021, 22, 3553. [Google Scholar] [CrossRef]
- van Deursen, J.M. The role of senescent cells in ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef]
- Baker, D.J.; Weaver, R.L.; van Deursen, J.M. p21 both attenuates and drives senescence and aging in BubR1 progeroid mice. Cell Rep. 2013, 3, 1164–1174. [Google Scholar] [CrossRef]
- Farr, J.N.; Xu, M.; Weivoda, M.M.; Monroe, D.G.; Fraser, D.G.; Onken, J.L.; Negley, B.A.; Sfeir, J.G.; Ogrodnik, M.B.; Hachfeld, C.M.; et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017, 23, 1072–1079. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Piatetzky-Shapiro, I.I.; Petrakova, K.V. Osteogenesis in transplants of bone marrow cells. J. Embryol. Exp. Morphol. 1966, 16, 381–390. [Google Scholar] [CrossRef]
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.G.; Riminucci, M.; et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007, 131, 324–336. [Google Scholar] [CrossRef]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; Macarthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhyan, R.K.; Gerasimov, U.V. Bone marrow osteogenic stem cells: In vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet. 1987, 20, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Boyde, A. Confocal images of marrow stromal (Westen-Bainton) cells. Histochemistry 1993, 100, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Pinho, S.; Ahmed, J.; Kunisaki, Y.; Hanoun, M.; Mendelson, A.; Ono, N.; Kronenberg, H.M.; Frenette, P.S. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell 2014, 29, 340–349. [Google Scholar] [CrossRef]
- Shi, Y.; He, G.; Lee, W.C.; McKenzie, J.A.; Silva, M.J.; Long, F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017, 8, 2043. [Google Scholar] [CrossRef]
- Matsushita, Y.; Chu, A.K.Y.; Tsutsumi-Arai, C.; Orikasa, S.; Nagata, M.; Wong, S.Y.; Welch, J.D.; Ono, W.; Ono, N. The fate of early perichondrial cells in developing bones. Nat. Commun. 2022, 13, 7319. [Google Scholar] [CrossRef]
- Zhou, B.O.; Yue, R.; Murphy, M.M.; Peyer, J.G.; Morrison, S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 2014, 15, 154–168. [Google Scholar] [CrossRef]
- Seike, M.; Omatsu, Y.; Watanabe, H.; Kondoh, G.; Nagasawa, T. Stem cell niche-specific Ebf3 maintains the bone marrow cavity. Genes. Dev. 2018, 32, 359–372. [Google Scholar] [CrossRef]
- Tikhonova, A.N.; Dolgalev, I.; Hu, H.; Sivaraj, K.K.; Hoxha, E.; Cuesta-Domínguez, Á.; Pinho, S.; Akhmetzyanova, I.; Gao, J.; Witkowski, M.; et al. The bone marrow microenvironment at single-cell resolution. Nature 2019, 569, 222–228. [Google Scholar] [CrossRef]
- Greenblatt, M.B.; Ono, N.; Ayturk, U.M.; Debnath, S.; Lalani, S. The Unmixing Problem: A Guide to Applying Single-Cell RNA Sequencing to Bone. J. Bone Miner. Res. 2019, 34, 1207–1219. [Google Scholar] [CrossRef]
- Anaparthy, N.; Ho, Y.J.; Martelotto, L.; Hammell, M.; Hicks, J. Single-Cell Applications of Next-Generation Sequencing. Cold Spring Harb. Perspect. Med. 2019, 9, a026898. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Fan, C.; Yin, Z.; Wang, T.; Chen, X. Potential applications of deep learning in single-cell RNA sequencing analysis for cell therapy and regenerative medicine. Stem Cells 2021, 39, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, M.; Crosby, W.H. Transplantation of marrow to extramedullary sites. Science 1968, 161, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.; Seo, E.Y.; Chen, J.Y.; Lo, D.; McArdle, A.; Sinha, R.; Tevlin, R.; Seita, J.; Vincent-Tompkins, J.; Wearda, T.; et al. Identification and specification of the mouse skeletal stem cell. Cell 2015, 160, 285–298. [Google Scholar] [CrossRef]
- Morikawa, S.; Mabuchi, Y.; Kubota, Y.; Nagai, Y.; Niibe, K.; Hiratsu, E.; Suzuki, S.; Miyauchi-Hara, C.; Nagoshi, N.; Sunabori, T.; et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009, 206, 2483–2496. [Google Scholar] [CrossRef]
- Breitbach, M.; Kimura, K.; Luis, T.C.; Fuegemann, C.J.; Woll, P.S.; Hesse, M.; Facchini, R.; Rieck, S.; Jobin, K.; Reinhardt, J.; et al. In Vivo Labeling by CD73 Marks Multipotent Stromal Cells and Highlights Endothelial Heterogeneity in the Bone Marrow Niche. Cell Stem Cell 2018, 22, 262–276.e267. [Google Scholar] [CrossRef]
- Mizuhashi, K.; Ono, W.; Matsushita, Y.; Sakagami, N.; Takahashi, A.; Saunders, T.L.; Nagasawa, T.; Kronenberg, H.M.; Ono, N. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature 2018, 563, 254–258. [Google Scholar] [CrossRef]
- Debnath, S.; Yallowitz, A.R.; McCormick, J.; Lalani, S.; Zhang, T.; Xu, R.; Li, N.; Liu, Y.; Yang, Y.S.; Eiseman, M.; et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 2018, 562, 133–139. [Google Scholar] [CrossRef]
- Matsushita, Y.L.J.; Chu, A.K.Y.; Tsutsumi-Arai, C.; Nagata, M.; Arai, Y.; Ono, W.; Sakagami, N.Y.; Amamoto, K.; Saunders, T.L.; Welch, J.D.; et al. Bone marrow endosteal stem cells dictate active osteogenesis and aggressive tumorigenesis. Nat. Commun. 2023, 14, 1–23. [Google Scholar] [CrossRef]
- Krebsbach, P.H.; Kuznetsov, S.A.; Bianco, P.; Robey, P.G. Bone marrow stromal cells: Characterization and clinical application. Crit. Rev. Oral Biol. Med. 1999, 10, 165–181. [Google Scholar] [CrossRef]
- Kfoury, Y.; Scadden, D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell 2015, 16, 239–253. [Google Scholar] [CrossRef]
- Ara, T.; Itoi, M.; Kawabata, K.; Egawa, T.; Tokoyoda, K.; Sugiyama, T.; Fujii, N.; Amagai, T.; Nagasawa, T. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J. Immunol. 2003, 170, 4649–4655. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, A.; Hsu, Y.M.; Day, R.B.; Schuettpelz, L.G.; Christopher, M.J.; Borgerding, J.N.; Nagasawa, T.; Link, D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 2013, 495, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, Y.; Nagata, M.; Kozloff, K.M.; Welch, J.D.; Mizuhashi, K.; Tokavanich, N.; Hallett, S.A.; Link, D.C.; Nagasawa, T.; Ono, W.; et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat. Commun. 2020, 11, 332. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Saunders, T.L.; Enikolopov, G.; Morrison, S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012, 481, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Ara, T.; Tokoyoda, K.; Sugiyama, T.; Egawa, T.; Kawabata, K.; Nagasawa, T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity 2003, 19, 257–267. [Google Scholar] [CrossRef]
- Omatsu, Y.; Sugiyama, T.; Kohara, H.; Kondoh, G.; Fujii, N.; Kohno, K.; Nagasawa, T. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity 2010, 33, 387–399. [Google Scholar] [CrossRef]
- Matsushita, Y.; Chu, A.K.Y.; Ono, W.; Welch, J.D.; Ono, N. Intercellular Interactions of an Adipogenic CXCL12-Expressing Stromal Cell Subset in Murine Bone Marrow. J. Bone Miner. Res. 2021, 36, 1145–1158. [Google Scholar] [CrossRef]
- Zhou, B.O.; Yu, H.; Yue, R.; Zhao, Z.; Rios, J.J.; Naveiras, O.; Morrison, S.J. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 2017, 19, 891–903. [Google Scholar] [CrossRef]
- Zhong, L.; Yao, L.; Tower, R.J.; Wei, Y.; Miao, Z.; Park, J.; Shrestha, R.; Wang, L.; Yu, W.; Holdreith, N.; et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife 2020, 9, e54695. [Google Scholar] [CrossRef]
- Yue, R.; Zhou, B.O.; Shimada, I.S.; Zhao, Z.; Morrison, S.J. Leptin Receptor Promotes Adipogenesis and Reduces Osteogenesis by Regulating Mesenchymal Stromal Cells in Adult Bone Marrow. Cell Stem Cell 2016, 18, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.S.; Chung, N.C.; Chen, Y.R.; Huang, H.Y.; Chuang, W.P.; Lai, D.M. Imbalanced Osteogenesis and Adipogenesis in Mice Deficient in the Chemokine Cxcl12/Sdf1 in the Bone Mesenchymal Stem/Progenitor Cells. J. Bone Miner. Res. 2018, 33, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.S.; Liu, Y.L.; Tang, X.T.; Zhang, X.S.; Zhou, B.; Zou, W.; Zhou, B.O. Tracing the skeletal progenitor transition during postnatal bone formation. Cell Stem Cell 2021, 28, 2122–2136.e3. [Google Scholar] [CrossRef] [PubMed]
- Baryawno, N.; Przybylski, D.; Kowalczyk, M.S.; Kfoury, Y.; Severe, N.; Gustafsson, K.; Kokkaliaris, K.D.; Mercier, F.; Tabaka, M.; Hofree, M.; et al. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell 2019, 177, 1915–1932.e16. [Google Scholar] [CrossRef]
- Wolock, S.L.; Krishnan, I.; Tenen, D.E.; Matkins, V.; Camacho, V.; Patel, S.; Agarwal, P.; Bhatia, R.; Tenen, D.G.; Klein, A.M.; et al. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Rep. 2019, 28, 302–311.e5. [Google Scholar] [CrossRef]
- Mo, C.; Guo, J.; Qin, J.; Zhang, X.; Sun, Y.; Wei, H.; Cao, D.; Zhang, Y.; Zhao, C.; Xiong, Y.; et al. Single-cell transcriptomics of LepR-positive skeletal cells reveals heterogeneous stress-dependent stem and progenitor pools. EMBO J. 2022, 41, e108415. [Google Scholar] [CrossRef]
- Martin, J.F.; Olson, E.N. Identification of a prx1 limb enhancer. Genesis 2000, 26, 225–229. [Google Scholar] [CrossRef]
- Logan, M.; Martin, J.F.; Nagy, A.; Lobe, C.; Olson, E.N.; Tabin, C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 2002, 33, 77–80. [Google Scholar] [CrossRef]
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef]
- Falacho, R.I.; Palma, P.J.; Marques, J.A.; Figueiredo, M.H.; Caramelo, F.; Dias, I.; Viegas, C.; Guerra, F. Collagenated Porcine Heterologous Bone Grafts: Histomorphometric Evaluation of Bone Formation Using Different Physical Forms in a Rabbit Cancellous Bone Model. Molecules 2021, 26, 1339. [Google Scholar] [CrossRef]
- Brochado Martins, J.F.; Rodrigues, C.F.D.; Diogo, P.; Paulo, S.; Palma, P.J.; do Vale, F.F. Remodelling compartment in root cementum. Folia Morphol. 2021, 80, 972–979. [Google Scholar] [CrossRef]
- Ono, N.; Ono, W.; Nagasawa, T.; Kronenberg, H.M. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 2014, 16, 1157–1167. [Google Scholar] [CrossRef]
- Worthley, D.L.; Churchill, M.; Compton, J.T.; Tailor, Y.; Rao, M.; Si, Y.; Levin, D.; Schwartz, M.G.; Uygur, A.; Hayakawa, Y.; et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell 2015, 160, 269–284. [Google Scholar] [CrossRef]
- Long, J.T.; Leinroth, A.; Liao, Y.; Ren, Y.; Mirando, A.J.; Nguyen, T.; Guo, W.; Sharma, D.; Rouse, D.; Wu, C.; et al. Hypertrophic chondrocytes serve as a reservoir for marrow-associated skeletal stem and progenitor cells, osteoblasts, and adipocytes during skeletal development. eLife 2022, 11, e76932. [Google Scholar] [CrossRef]
- Sivaraj, K.K.; Jeong, H.W.; Dharmalingam, B.; Zeuschner, D.; Adams, S.; Potente, M.; Adams, R.H. Regional specialization and fate specification of bone stromal cells in skeletal development. Cell Rep. 2021, 36, 109352. [Google Scholar] [CrossRef]
- Duchamp de Lageneste, O.; Julien, A.; Abou-Khalil, R.; Frangi, G.; Carvalho, C.; Cagnard, N.; Cordier, C.; Conway, S.J.; Colnot, C. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun. 2018, 9, 773. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, E.C.; Mann, T.L.A.; Pool, J.A.; Zhao, Z.; Morrison, S.J. Bone marrow and periosteal skeletal stem/progenitor cells make distinct contributions to bone maintenance and repair. Cell Stem Cell 2022, 29, 1547–1561.e6. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Spencer, J.A.; Koh, B.I.; Kobayashi, T.; Fujisaki, J.; Clemens, T.L.; Lin, C.P.; Kronenberg, H.M.; Scadden, D.T. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 2012, 10, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, Y.; Sakamoto, K.; Tamamura, Y.; Shibata, Y.; Minamizato, T.; Kihara, T.; Ito, M.; Katsube, K.; Hiraoka, S.; Koseki, H.; et al. CCN3 protein participates in bone regeneration as an inhibitory factor. J. Biol. Chem. 2013, 288, 19973–19985. [Google Scholar] [CrossRef] [PubMed]
- Minear, S.; Leucht, P.; Jiang, J.; Liu, B.; Zeng, A.; Fuerer, C.; Nusse, R.; Helms, J.A. Wnt proteins promote bone regeneration. Sci. Transl. Med. 2010, 2, 29ra30. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Ii, M.; Matsumoto, T.; Kuroda, R.; Kuroda, T.; Kwon, S.M.; Kawamoto, A.; Akimaru, H.; Mifune, Y.; Shoji, T.; et al. SDF-1/CXCR4 axis in Tie2-lineage cells including endothelial progenitor cells contributes to bone fracture healing. J. Bone Miner. Res. 2015, 30, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, K.; Bandyopadhyay, A.; Harfe, B.D.; Cox, K.; Kakar, S.; Gerstenfeld, L.; Einhorn, T.; Tabin, C.J.; Rosen, V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat. Genet. 2006, 38, 1424–1429. [Google Scholar] [CrossRef]
- Marcellini, S.; Henriquez, J.P.; Bertin, A. Control of osteogenesis by the canonical Wnt and BMP pathways in vivo: Cooperation and antagonism between the canonical Wnt and BMP pathways as cells differentiate from osteochondroprogenitors to osteoblasts and osteocytes. Bioessays 2012, 34, 953–962. [Google Scholar] [CrossRef]
- Myers, T.J.; Longobardi, L.; Willcockson, H.; Temple, J.D.; Tagliafierro, L.; Ye, P.; Li, T.; Esposito, A.; Moats-Staats, B.M.; Spagnoli, A. BMP2 Regulation of CXCL12 Cellular, Temporal, and Spatial Expression is Essential During Fracture Repair. J. Bone Miner. Res. 2015, 30, 2014–2027. [Google Scholar] [CrossRef]
- Wang, C.; Inzana, J.A.; Mirando, A.J.; Ren, Y.; Liu, Z.; Shen, J.; O’Keefe, R.J.; Awad, H.A.; Hilton, M.J. NOTCH signaling in skeletal progenitors is critical for fracture repair. J. Clin. Investig. 2016, 126, 1471–1481. [Google Scholar] [CrossRef]
- Kimmel, J.C.; Penland, L.; Rubinstein, N.D.; Hendrickson, D.G.; Kelley, D.R.; Rosenthal, A.Z. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome. Res. 2019, 29, 2088–2103. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, T.H.; Marecic, O.; McArdle, A.; Sinha, R.; Gulati, G.S.; Tong, X.; Wang, Y.; Steininger, H.M.; Hoover, M.Y.; Koepke, L.S.; et al. Aged skeletal stem cells generate an inflammatory degenerative niche. Nature 2021, 597, 256–262. [Google Scholar] [CrossRef]
- Deng, P.; Yuan, Q.; Cheng, Y.; Li, J.; Liu, Z.; Liu, Y.; Li, Y.; Su, T.; Wang, J.; Salvo, M.E.; et al. Loss of KDM4B exacerbates bone-fat imbalance and mesenchymal stromal cell exhaustion in skeletal aging. Cell Stem Cell 2021, 28, 1057–1073.e7. [Google Scholar] [CrossRef]
- Ambrosi, T.H.; Scialdone, A.; Graja, A.; Gohlke, S.; Jank, A.M.; Bocian, C.; Woelk, L.; Fan, H.; Logan, D.W.; Schürmann, A.; et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 2017, 20, 771–784.e6. [Google Scholar] [CrossRef]
- Fan, Y.; Hanai, J.I.; Le, P.T.; Bi, R.; Maridas, D.; DeMambro, V.; Figueroa, C.A.; Kir, S.; Zhou, X.; Mannstadt, M.; et al. Parathyroid Hormone Directs Bone Marrow Mesenchymal Cell Fate. Cell Metab. 2017, 25, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes. Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ko, J. A novel PPARγ2 modulator sLZIP controls the balance between adipogenesis and osteogenesis during mesenchymal stem cell differentiation. Cell Death Differ. 2014, 21, 1642–1655. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Gao, Y.; Ueta, C.; Yamaguchi, A.; Komori, T. Multilineage differentiation of Cbfa1-deficient calvarial cells in vitro. Biochem. Biophys. Res. Commun. 2000, 273, 630–636. [Google Scholar] [CrossRef]
- Wu, X.; Pang, L.; Lei, W.; Lu, W.; Li, J.; Li, Z.; Frassica, F.J.; Chen, X.; Wan, M.; Cao, X. Inhibition of Sca-1-positive skeletal stem cell recruitment by alendronate blunts the anabolic effects of parathyroid hormone on bone remodeling. Cell Stem Cell 2010, 7, 571–580. [Google Scholar] [CrossRef]
- Clark, D.; Brazina, S.; Yang, F.; Hu, D.; Hsieh, C.L.; Niemi, E.C.; Miclau, T.; Nakamura, M.C.; Marcucio, R. Age-related changes to macrophages are detrimental to fracture healing in mice. Aging Cell 2020, 19, e13112. [Google Scholar] [CrossRef]
- Qadir, A.; Liang, S.; Wu, Z.; Chen, Z.; Hu, L.; Qian, A. Senile Osteoporosis: The Involvement of Differentiation and Senescence of Bone Marrow Stromal Cells. Int. J. Mol. Sci. 2020, 21, 349. [Google Scholar] [CrossRef]
- Fischer, J.; Koch, L.; Emmerling, C.; Vierkotten, J.; Peters, T.; Brüning, J.C.; Rüther, U. Inactivation of the Fto gene protects from obesity. Nature 2009, 458, 894–898. [Google Scholar] [CrossRef]
- Farr, J.N.; Rowsey, J.L.; Eckhardt, B.A.; Thicke, B.S.; Fraser, D.G.; Tchkonia, T.; Kirkland, J.L.; Monroe, D.G.; Khosla, S. Independent Roles of Estrogen Deficiency and Cellular Senescence in the Pathogenesis of Osteoporosis: Evidence in Young Adult Mice and Older Humans. J. Bone Miner. Res. 2019, 34, 1407–1418. [Google Scholar] [CrossRef]
- Nishida, S.; Endo, N.; Yamagiwa, H.; Tanizawa, T.; Takahashi, H.E. Number of osteoprogenitor cells in human bone marrow markedly decreases after skeletal maturation. J. Bone Miner. Metab. 1999, 17, 171–177. [Google Scholar] [CrossRef]
- Jilka, R.L.; Weinstein, R.S.; Takahashi, K.; Parfitt, A.M.; Manolagas, S.C. Linkage of decreased bone mass with impaired osteoblastogenesis in a murine model of accelerated senescence. J. Clin. Investig. 1996, 97, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.D.; Shi, S.; Xu, T.; Robey, P.G.; Young, M.F. Age-related osteoporosis in biglycan-deficient mice is related to defects in bone marrow stromal cells. J. Bone Miner. Res. 2002, 17, 331–340. [Google Scholar] [CrossRef]
- Xu, J.Z.; Zhou, Y.M.; Zhang, L.L.; Chen, X.J.; Yang, Y.Y.; Zhang, D.; Zhu, K.C.; Kong, X.K.; Sun, L.H.; Tao, B.; et al. BMP9 reduces age-related bone loss in mice by inhibiting osteoblast senescence through Smad1-Stat1-P21 axis. Cell Death Discov. 2022, 8, 254. [Google Scholar] [CrossRef] [PubMed]
- Farr, J.N.; Fraser, D.G.; Wang, H.; Jaehn, K.; Ogrodnik, M.B.; Weivoda, M.M.; Drake, M.T.; Tchkonia, T.; LeBrasseur, N.K.; Kirkland, J.L.; et al. Identification of Senescent Cells in the Bone Microenvironment. J. Bone Miner. Res. 2016, 31, 1920–1929. [Google Scholar] [CrossRef]
- Nelson, G.; Wordsworth, J.; Wang, C.; Jurk, D.; Lawless, C.; Martin-Ruiz, C.; von Zglinicki, T. A senescent cell bystander effect: Senescence-induced senescence. Aging Cell 2012, 11, 345–349. [Google Scholar] [CrossRef]
- Acosta, J.C.; Banito, A.; Wuestefeld, T.; Georgilis, A.; Janich, P.; Morton, J.P.; Athineos, D.; Kang, T.W.; Lasitschka, F.; Andrulis, M.; et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013, 15, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, B.M. Marrow adipocytes inhibit the differentiation of mesenchymal stem cells into osteoblasts via suppressing BMP-signaling. J. Biomed. Sci. 2017, 24, 11. [Google Scholar] [CrossRef]
- Muschler, G.F.; Nitto, H.; Boehm, C.A.; Easley, K.A. Age- and gender-related changes in the cellularity of human bone marrow and the prevalence of osteoblastic progenitors. J. Orthop. Res. 2001, 19, 117–125. [Google Scholar] [CrossRef]
- D’Ippolito, G.; Schiller, P.C.; Ricordi, C.; Roos, B.A.; Howard, G.A. Age-related osteogenic potential of mesenchymal stromal stem cells from human vertebral bone marrow. J. Bone Miner. Res. 1999, 14, 1115–1122. [Google Scholar] [CrossRef]
- Veldhuis-Vlug, A.G.; Rosen, C.J. Mechanisms of marrow adiposity and its implications for skeletal health. Metabolism 2017, 67, 106–114. [Google Scholar] [CrossRef]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.H.; Wang, C.Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Kim, H.N.; Han, L.; Zhou, D.; Thostenson, J.; Porter, R.M.; Ambrogini, E.; Manolagas, S.C.; Jilka, R.L. Increased marrow adipogenesis does not contribute to age-dependent appendicular bone loss in female mice. Aging Cell 2020, 19, e13247. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Guo, R.; Wang, Z.; Yan, H.; Lv, X.; Zhao, Q.; Jiang, X.; Zhang, C.; Zhang, D.; Yang, C.; et al. Circulating Monocytes Act as a Common Trigger for the Calcification Paradox of Osteoporosis and Carotid Atherosclerosis. Front. Endocrinol. 2022, 13, 944751. [Google Scholar] [CrossRef]
- Matsushita, Y.; Ono, W.; Ono, N. Bone regeneration via skeletal cell lineage plasticity: All hands mobilized for emergencies: Quiescent mature skeletal cells can be activated in response to injury and robustly participate in bone regeneration through cellular plasticity. Bioessays 2021, 43, e2000202. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.; Rodeheffer, M.S.; Rosen, C.J.; Horowitz, M.C. Adipose Tissue Residing Progenitors (Adipocyte Lineage Progenitors and Adipose Derived Stem Cells (ADSC). Curr. Mol. Biol. Rep. 2015, 1, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Rauch, A.; Haakonsson, A.K.; Madsen, J.G.S.; Larsen, M.; Forss, I.; Madsen, M.R.; Van Hauwaert, E.L.; Wiwie, C.; Jespersen, N.Z.; Tencerova, M.; et al. Osteogenesis depends on commissioning of a network of stem cell transcription factors that act as repressors of adipogenesis. Nat. Genet. 2019, 51, 716–727. [Google Scholar] [CrossRef]
- Kim, H.N.; Chang, J.; Shao, L.; Han, L.; Iyer, S.; Manolagas, S.C.; O’Brien, C.A.; Jilka, R.L.; Zhou, D.; Almeida, M. DNA damage and senescence in osteoprogenitors expressing Osx1 may cause their decrease with age. Aging Cell 2017, 16, 693–703. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Ohba, S.; Matsushita, Y. Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis. Int. J. Mol. Sci. 2023, 24, 9814. https://doi.org/10.3390/ijms24129814
Wu S, Ohba S, Matsushita Y. Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis. International Journal of Molecular Sciences. 2023; 24(12):9814. https://doi.org/10.3390/ijms24129814
Chicago/Turabian StyleWu, Sixun, Shinsuke Ohba, and Yuki Matsushita. 2023. "Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis" International Journal of Molecular Sciences 24, no. 12: 9814. https://doi.org/10.3390/ijms24129814
APA StyleWu, S., Ohba, S., & Matsushita, Y. (2023). Single-Cell RNA-Sequencing Reveals the Skeletal Cellular Dynamics in Bone Repair and Osteoporosis. International Journal of Molecular Sciences, 24(12), 9814. https://doi.org/10.3390/ijms24129814





