Applying Next-Generation Sequencing and Multi-Omics in Chronic Obstructive Pulmonary Disease
Abstract
1. Introduction
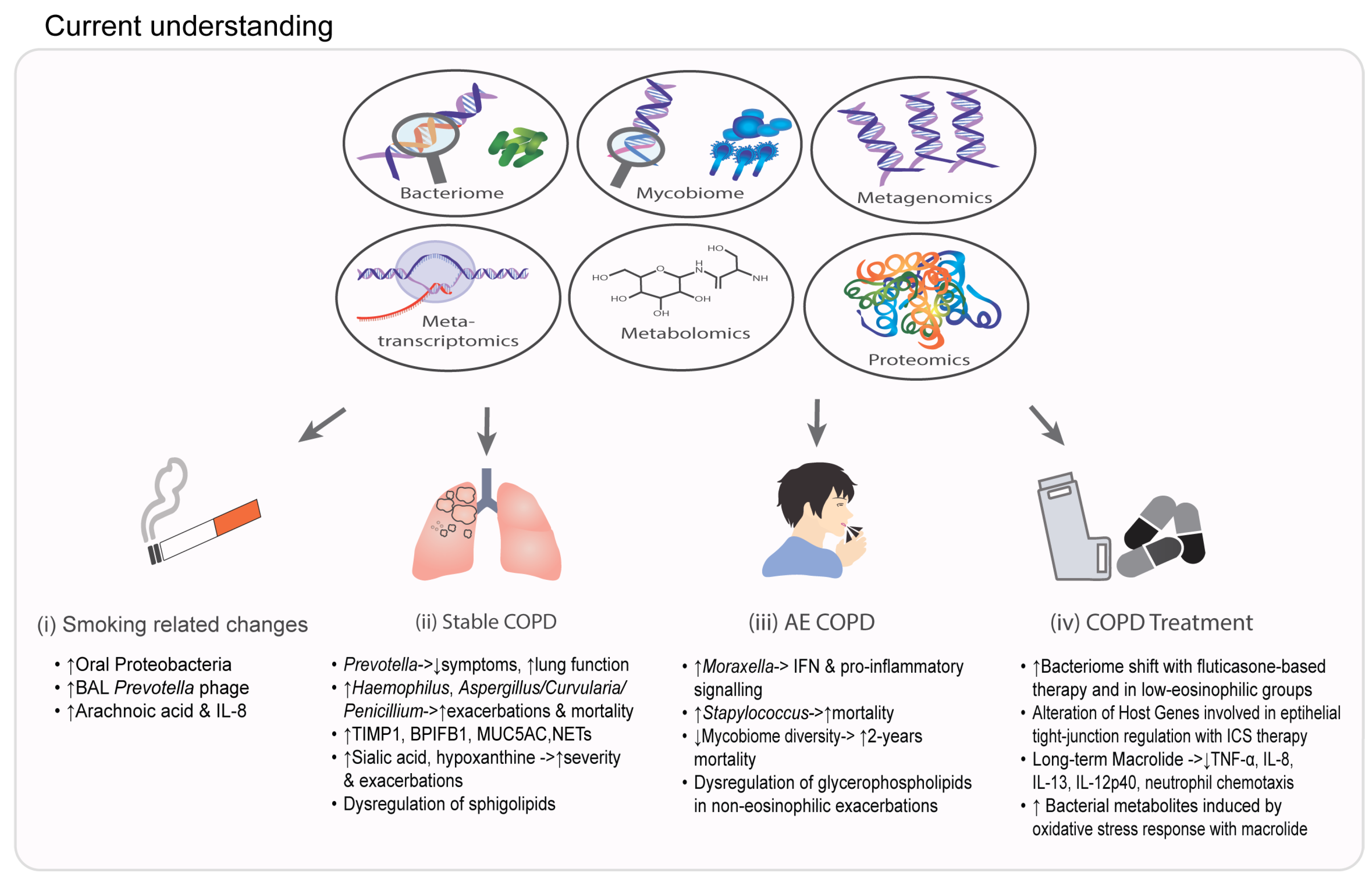
2. NGS in Endophenotyping and Prognostication of COPD
3. The Application of Proteomics and Metabolomics in COPD
4. Understanding COPD Therapeutics Using NGS
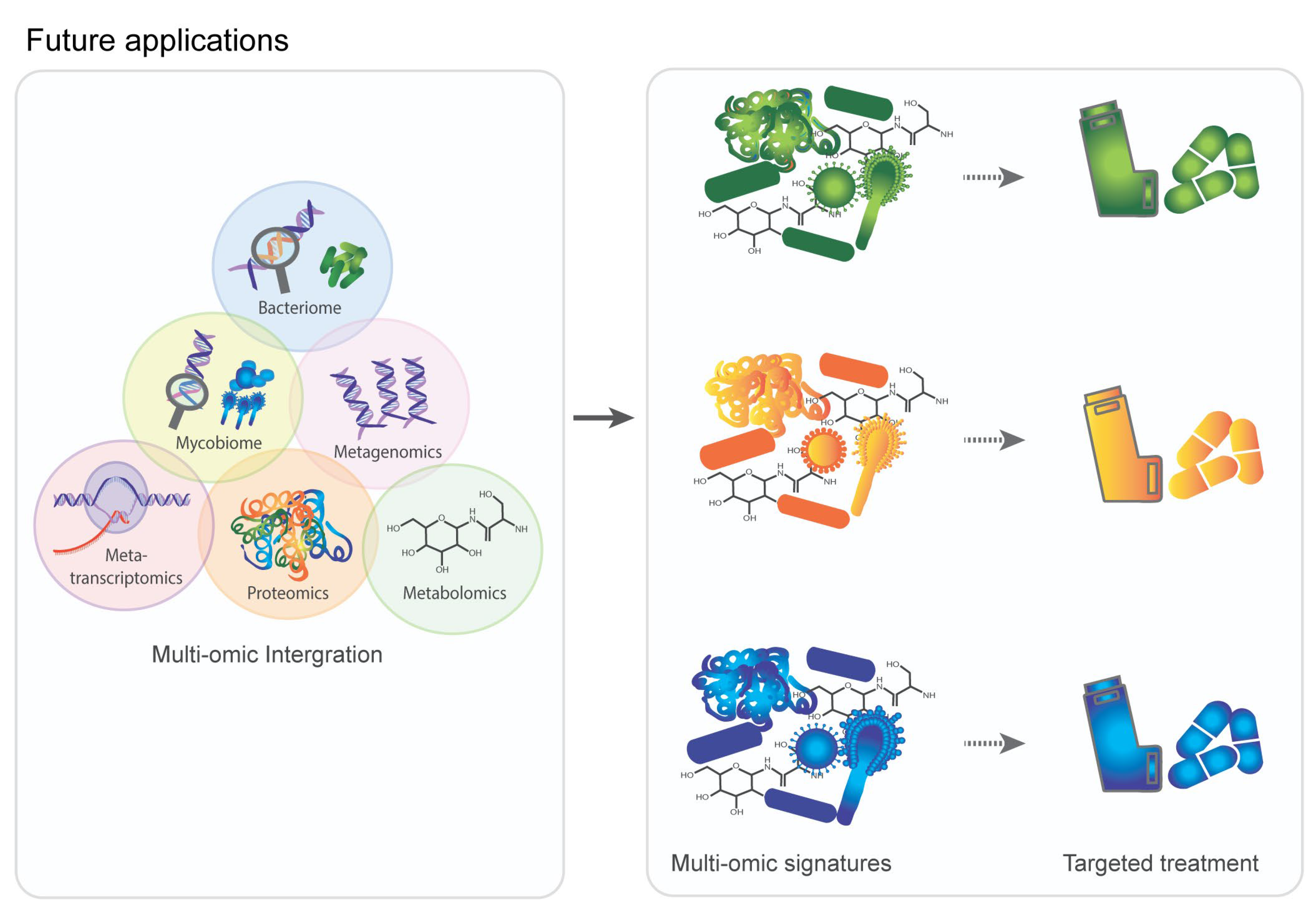
5. Multi-Omic Data Integration in COPD
6. Clinical Application and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.H.; Adcock, I.M.; Chotirmall, S.H.; Chung, K.F.; et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019, 7, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Chotirmall, S.H.; Gellatly, S.L.; Budden, K.F.; Mac Aogain, M.; Shukla, S.D.; Wood, D.L.; Hugenholtz, P.; Pethe, K.; Hansbro, P.M. Microbiomes in respiratory health and disease: An Asia-Pacific perspective. Respirology 2017, 22, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Aogain, M.M.; Jaggi, T.K.; Chotirmall, S.H. The Airway Microbiome: Present and Future Applications. Arch. Bronconeumol. 2022, 58, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.M.; Tiew, P.Y.; Mac Aogain, M.; Budden, K.F.; Yong, V.F.; Thomas, S.S.; Pethe, K.; Hansbro, P.M.; Chotirmall, S.H. The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 2017, 22, 634–650. [Google Scholar] [CrossRef]
- Ali, N.A.B.M.; Mac Aogain, M.; Morales, R.F.; Tiew, P.Y.; Chotirmall, S.H. Optimisation and Benchmarking of Targeted Amplicon Sequencing for Mycobiome Analysis of Respiratory Specimens. Int. J. Mol. Sci. 2019, 20, 4991. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Jaggi, T.K.; Chan, L.L.Y.; Chotirmall, S.H. The airway microbiome in COPD, bronchiectasis and bronchiectasis-COPD overlap. Clin. Respir. J. 2021, 15, 123–133. [Google Scholar] [CrossRef]
- Ditz, B.; Christenson, S.; Rossen, J.; Brightling, C.; Kerstjens, H.A.M.; van den Berge, M.; Faiz, A. Sputum microbiome profiling in COPD: Beyond singular pathogen detection. Thorax 2020, 75, 338–344. [Google Scholar] [CrossRef]
- Godbole, S.; Bowler, R.P. Metabolome Features of COPD: A Scoping Review. Metabolites 2022, 12, 621. [Google Scholar] [CrossRef]
- D’Amato, M.; Iadarola, P.; Viglio, S. Proteomic Analysis of Human Sputum for the Diagnosis of Lung Disorders: Where Are We Today? Int. J. Mol. Sci. 2022, 23, 5692. [Google Scholar] [CrossRef]
- Narayana, J.K.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Microbiomics-focused Data Integration: A Fresh Solve for the Rubik’s Cube of Endophenotyping? Am. J. Respir. Crit. Care Med. 2022, 206, 365–368. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogain, M.; Ali, N.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Similarity network fusion for the integration of multi-omics and microbiomes in respiratory disease. Eur. Respir. J. 2021, 58, 2101016. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Al Bataineh, M.T.; Dash, N.R.; Elkhazendar, M.; Alnusairat, D.M.H.; Darwish, I.M.I.; Al-Hajjaj, M.S.; Hamid, Q. Revealing oral microbiota composition and functionality associated with heavy cigarette smoking. J. Transl. Med. 2020, 18, 421. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Sullivan, M.B.; Segal, L.N.; Keller, B.C. Smoking is associated with quantifiable differences in the human lung DNA virome and metabolome. Respir. Res. 2018, 19, 174. [Google Scholar] [CrossRef]
- Ramsheh, M.Y.; Haldar, K.; Esteve-Codina, A.; Purser, L.F.; Richardson, M.; Muller-Quernheim, J.; Greulich, T.; Nowinski, A.; Barta, I.; Stendardo, M.; et al. Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: A bacterial 16S rRNA gene sequencing and host transcriptomic analysis. Lancet Microbe 2021, 2, e300–e310. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Dicker, A.J.; Keir, H.R.; Poh, M.E.; Pang, S.L.; Mac Aogain, M.; Chua, B.Q.Y.; Tan, J.L.; Xu, H.; Koh, M.S.; et al. A high-risk airway mycobiome is associated with frequent exacerbation and mortality in COPD. Eur. Respir. J. 2021, 57, 2002050. [Google Scholar] [CrossRef]
- Keir, H.R.; Dicker, A.; Lonergan, M.; Crichton, M.; Miller, B.E.; Tal-Singer, R.; Chalmers, J.D. Clinical endotypes of exacerbation are associated with differences in microbial composition and diversity in COPD. Eur. Respir. J. 2020, 56, 2000391. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Thng, K.X.; Chotirmall, S.H. Clinical Aspergillus Signatures in COPD and Bronchiectasis. J. Fungi 2022, 8, 480. [Google Scholar] [CrossRef]
- Yang, M.; Kohler, M.; Heyder, T.; Forsslund, H.; Garberg, H.K.; Karimi, R.; Grunewald, J.; Berven, F.S.; Magnus Skold, C.; Wheelock, A.M. Long-term smoking alters abundance of over half of the proteome in bronchoalveolar lavage cell in smokers with normal spirometry, with effects on molecular pathways associated with COPD. Respir. Res. 2018, 19, 40. [Google Scholar] [CrossRef]
- Britto, C.J.; Cohn, L. Bactericidal/Permeability-increasing protein fold-containing family member A1 in airway host protection and respiratory disease. Am. J. Respir. Cell Mol. Biol. 2015, 52, 525–534. [Google Scholar] [CrossRef]
- Esther, C.R., Jr.; O’Neal, W.K.; Anderson, W.H.; Kesimer, M.; Ceppe, A.; Doerschuk, C.M.; Alexis, N.E.; Hastie, A.T.; Barr, R.G.; Bowler, R.P.; et al. Identification of Sputum Biomarkers Predictive of Pulmonary Exacerbations in COPD. Chest 2022, 161, 1239–1249. [Google Scholar] [CrossRef]
- Halper-Stromberg, E.; Gillenwater, L.; Cruickshank-Quinn, C.; O’Neal, W.K.; Reisdorph, N.; Petrache, I.; Zhuang, Y.; Labaki, W.W.; Curtis, J.L.; Wells, J.; et al. Bronchoalveolar Lavage Fluid from COPD Patients Reveals More Compounds Associated with Disease than Matched Plasma. Metabolites 2019, 9, 157. [Google Scholar] [CrossRef]
- Mayhew, D.; Devos, N.; Lambert, C.; Brown, J.R.; Clarke, S.C.; Kim, V.L.; Magid-Slav, M.; Miller, B.E.; Ostridge, K.K.; Patel, R.; et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018, 73, 422–430. [Google Scholar] [CrossRef]
- Martinsen, E.M.H.; Eagan, T.M.L.; Leiten, E.O.; Haaland, I.; Husebo, G.R.; Knudsen, K.S.; Drengenes, C.; Sanseverino, W.; Paytuvi-Gallart, A.; Nielsen, R. The pulmonary mycobiome-A study of subjects with and without chronic obstructive pulmonary disease. PLoS ONE 2021, 16, e0248967. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Q.; Liu, C.; Pang, R.; Yin, Y. Plasma Metabolomics and Lipidomics Reveal Perturbed Metabolites in Different Disease Stages of Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstr. Pulm. Dis. 2020, 15, 553–565. [Google Scholar] [CrossRef]
- Prokic, I.; Lahousse, L.; de Vries, M.; Liu, J.; Kalaoja, M.; Vonk, J.M.; van der Plaat, D.A.; van Diemen, C.C.; van der Spek, A.; Zhernakova, A.; et al. A cross-omics integrative study of metabolic signatures of chronic obstructive pulmonary disease. BMC Pulm. Med. 2020, 20, 193. [Google Scholar] [CrossRef]
- Gillenwater, L.A.; Pratte, K.A.; Hobbs, B.D.; Cho, M.H.; Zhuang, Y.; Halper-Stromberg, E.; Cruickshank-Quinn, C.; Reisdorph, N.; Petrache, I.; Labaki, W.W.; et al. Plasma Metabolomic Signatures of Chronic Obstructive Pulmonary Disease and the Impact of Genetic Variants on Phenotype-Driven Modules. Netw. Syst. Med. 2020, 3, 159–181. [Google Scholar] [CrossRef]
- Albert, R.K.; Connett, J.; Bailey, W.C.; Casaburi, R.; Cooper, J.A., Jr.; Criner, G.J.; Curtis, J.L.; Dransfield, M.T.; Han, M.K.; Lazarus, S.C.; et al. Azithromycin for prevention of exacerbations of COPD. N. Engl. J. Med. 2011, 365, 689–698. [Google Scholar] [CrossRef]
- Wu, J.; Peters, B.A.; Dominianni, C.; Zhang, Y.; Pei, Z.; Yang, L.; Ma, Y.; Purdue, M.P.; Jacobs, E.J.; Gapstur, S.M.; et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016, 10, 2435–2446. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, W.; Cai, Q.Y.; Shrubsole, M.J.; Pei, Z.; Brucker, R.; Steinwandel, M.D.; Bordenstein, S.R.; Li, Z.; Blot, W.J.; et al. Cigarette smoking and oral microbiota in low-income and African-American populations. J. Epidemiol. Community Health 2019, 73, 1108–1115. [Google Scholar] [CrossRef]
- Charlson, E.S.; Chen, J.; Custers-Allen, R.; Bittinger, K.; Li, H.; Sinha, R.; Hwang, J.; Bushman, F.D.; Collman, R.G. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS ONE 2010, 5, e15216. [Google Scholar] [CrossRef]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B.; et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011, 6, e16384. [Google Scholar] [CrossRef]
- Haldar, K.; George, L.; Wang, Z.; Mistry, V.; Ramsheh, M.Y.; Free, R.C.; John, C.; Reeve, N.F.; Miller, B.E.; Tal-Singer, R.; et al. The sputum microbiome is distinct between COPD and health, independent of smoking history. Respir. Res. 2020, 21, 183. [Google Scholar] [CrossRef]
- Einarsson, G.G.; Comer, D.M.; McIlreavey, L.; Parkhill, J.; Ennis, M.; Tunney, M.M.; Elborn, J.S. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016, 71, 795–803. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Chotirmall, S.H. The current understanding and future directions for sputum microbiome profiling in chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2022, 28, 121–133. [Google Scholar] [CrossRef]
- Opron, K.; Begley, L.A.; Erb-Downward, J.R.; Freeman, C.; Madapoosi, S.; Alexis, N.E.; Barjaktarevic, I.; Graham Barr, R.; Bleecker, E.R.; Bowler, R.P.; et al. Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort. NPJ Biofilms Microbiomes 2021, 7, 14. [Google Scholar] [CrossRef]
- Madapoosi, S.S.; Cruickshank-Quinn, C.; Opron, K.; Erb-Downward, J.R.; Begley, L.A.; Li, G.; Barjaktarevic, I.; Barr, R.G.; Comellas, A.P.; Couper, D.; et al. Lung Microbiota and Metabolites Collectively Associate with Clinical Outcomes in Milder Stage COPD. Am. J. Respir. Crit. Care Med. 2022, 206, 427–439. [Google Scholar] [CrossRef]
- Tarran, R. Regulation of airway surface liquid volume and mucus transport by active ion transport. Proc. Am. Thorac. Soc. 2004, 1, 42–46. [Google Scholar] [CrossRef]
- Garcia-Nunez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Perez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monso, E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef]
- Galiana, A.; Aguirre, E.; Rodriguez, J.C.; Mira, A.; Santibanez, M.; Candela, I.; Llavero, J.; Garcinuno, P.; Lopez, F.; Ruiz, M.; et al. Sputum microbiota in moderate versus severe patients with COPD. Eur. Respir. J. 2014, 43, 1787–1790. [Google Scholar] [CrossRef]
- Leitao Filho, F.S.; Alotaibi, N.M.; Ngan, D.; Tam, S.; Yang, J.; Hollander, Z.; Chen, V.; FitzGerald, J.M.; Nislow, C.; Leung, J.M.; et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am. J. Respir. Crit. Care Med. 2019, 199, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Millares, L.; Pascual, S.; Monton, C.; Garcia-Nunez, M.; Lalmolda, C.; Faner, R.; Casadevall, C.; Seto, L.; Capilla, S.; Moreno, A.; et al. Relationship between the respiratory microbiome and the severity of airflow limitation, history of exacerbations and circulating eosinophils in COPD patients. BMC Pulm. Med. 2019, 19, 112. [Google Scholar] [CrossRef]
- Bouquet, J.; Tabor, D.E.; Silver, J.S.; Nair, V.; Tovchigrechko, A.; Griffin, M.P.; Esser, M.T.; Sellman, B.R.; Jin, H. Microbial burden and viral exacerbations in a longitudinal multicenter COPD cohort. Respir. Res. 2020, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bafadhel, M.; Haldar, K.; Spivak, A.; Mayhew, D.; Miller, B.E.; Tal-Singer, R.; Johnston, S.L.; Ramsheh, M.Y.; Barer, M.R.; et al. Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 2016, 47, 1082–1092. [Google Scholar] [CrossRef] [PubMed]
- Beech, A.S.; Lea, S.; Kolsum, U.; Wang, Z.; Miller, B.E.; Donaldson, G.C.; Wedzicha, J.A.; Brightling, C.E.; Singh, D. Bacteria and sputum inflammatory cell counts; a COPD cohort analysis. Respir. Res. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Winslow, S.; Odqvist, L.; Diver, S.; Riise, R.; Abdillahi, S.; Wingren, C.; Lindmark, H.; Wellner, A.; Lundin, S.; Yrlid, L.; et al. Multi-omics links IL-6 trans-signalling with neutrophil extracellular trap formation and Haemophilus infection in COPD. Eur. Respir. J. 2021, 58, 2003312. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Li, S.W.; Chin, C.Y.; Hsu, C.W.; Lee, C.C.; Yeh, Y.M.; Wu, K.A. Association of exacerbation phenotype with the sputum microbiome in chronic obstructive pulmonary disease patients during the clinically stable state. J. Transl. Med. 2021, 19, 121. [Google Scholar] [CrossRef]
- Dicker, A.J.; Crichton, M.L.; Pumphrey, E.G.; Cassidy, A.J.; Suarez-Cuartin, G.; Sibila, O.; Furrie, E.; Fong, C.J.; Ibrahim, W.; Brady, G.; et al. Neutrophil extracellular traps are associated with disease severity and microbiota diversity in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2018, 141, 117–127. [Google Scholar] [CrossRef]
- Dicker, A.J.; Huang, J.T.J.; Lonergan, M.; Keir, H.R.; Fong, C.J.; Tan, B.; Cassidy, A.J.; Finch, S.; Mullerova, H.; Miller, B.E.; et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2021, 147, 158–167. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Ter, S.K.; Aliberti, S.; Chalmers, J.D.; Chotirmall, S.H. Respiratory Mycoses in COPD and Bronchiectasis. Mycopathologia 2021, 186, 623–638. [Google Scholar] [CrossRef]
- Ali, N.; Ivan, F.X.; Mac Aogain, M.; Narayana, J.K.; Lee, S.Y.; Lim, C.L.; Chotirmall, S.H. The Healthy Airway Mycobiome in Individuals of Asian Descent. Chest 2021, 159, 544–548. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Ko, F.W.S.; Narayana, J.K.; Poh, M.E.; Xu, H.; Neo, H.Y.; Loh, L.C.; Ong, C.K.; Mac Aogain, M.; Tan, J.H.Y.; et al. “High-Risk” Clinical and Inflammatory Clusters in COPD of Chinese Descent. Chest 2020, 158, 145–156. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; O’Donoghue, E.; Bennett, K.; Gunaratnam, C.; O’Neill, S.J.; McElvaney, N.G. Sputum Candida albicans presages FEV(1) decline and hospital-treated exacerbations in cystic fibrosis. Chest 2010, 138, 1186–1195. [Google Scholar] [CrossRef]
- Wang, Z.; Locantore, N.; Haldar, K.; Ramsheh, M.Y.; Beech, A.S.; Ma, W.; Brown, J.R.; Tal-Singer, R.; Barer, M.R.; Bafadhel, M.; et al. Inflammatory Endotype-associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 1488–1502. [Google Scholar] [CrossRef]
- Wang, Z.; Maschera, B.; Lea, S.; Kolsum, U.; Michalovich, D.; Van Horn, S.; Traini, C.; Brown, J.R.; Hessel, E.M.; Singh, D. Airway host-microbiome interactions in chronic obstructive pulmonary disease. Respir. Res. 2019, 20, 113. [Google Scholar] [CrossRef]
- Wang, Z.; Singh, R.; Miller, B.E.; Tal-Singer, R.; Van Horn, S.; Tomsho, L.; Mackay, A.; Allinson, J.P.; Webb, A.J.; Brookes, A.J.; et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 2018, 73, 331–338. [Google Scholar] [CrossRef]
- Baraniuk, J.N.; Casado, B.; Pannell, L.K.; McGarvey, P.B.; Boschetto, P.; Luisetti, M.; Iadarola, P. Protein networks in induced sputum from smokers and COPD patients. Int. J. Chron. Obstr. Pulm. Dis. 2015, 10, 1957–1975. [Google Scholar] [CrossRef]
- Titz, B.; Sewer, A.; Schneider, T.; Elamin, A.; Martin, F.; Dijon, S.; Luettich, K.; Guedj, E.; Vuillaume, G.; Ivanov, N.V.; et al. Alterations in the sputum proteome and transcriptome in smokers and early-stage COPD subjects. J. Proteom. 2015, 128, 306–320. [Google Scholar] [CrossRef]
- Gao, J.; Ohlmeier, S.; Nieminen, P.; Toljamo, T.; Tiitinen, S.; Kanerva, T.; Bingle, L.; Araujo, B.; Ronty, M.; Hoyhtya, M.; et al. Elevated sputum BPIFB1 levels in smokers with chronic obstructive pulmonary disease: A longitudinal study. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L17–L26. [Google Scholar] [CrossRef]
- Reidel, B.; Radicioni, G.; Clapp, P.W.; Ford, A.A.; Abdelwahab, S.; Rebuli, M.E.; Haridass, P.; Alexis, N.E.; Jaspers, I.; Kesimer, M. E-Cigarette Use Causes a Unique Innate Immune Response in the Lung, Involving Increased Neutrophilic Activation and Altered Mucin Secretion. Am. J. Respir. Crit. Care Med. 2018, 197, 492–501. [Google Scholar] [CrossRef]
- Meldrum, O.W.; Chotirmall, S.H. Mucus, Microbiomes and Pulmonary Disease. Biomedicines 2021, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- Gulcev, M.; Reilly, C.; Griffin, T.J.; Broeckling, C.D.; Sandri, B.J.; Witthuhn, B.A.; Hodgson, S.W.; Woodruff, P.G.; Wendt, C.H. Tryptophan catabolism in acute exacerbations of chronic obstructive pulmonary disease. Int. J. Chron. Obstr. Pulm. Dis. 2016, 11, 2435–2446. [Google Scholar] [CrossRef]
- Petrache, I.; Natarajan, V.; Zhen, L.; Medler, T.R.; Richter, A.T.; Cho, C.; Hubbard, W.C.; Berdyshev, E.V.; Tuder, R.M. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat. Med. 2005, 11, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Berdyshev, E.V.; Serban, K.A.; Schweitzer, K.S.; Bronova, I.A.; Mikosz, A.; Petrache, I. Ceramide and sphingosine-1 phosphate in COPD lungs. Thorax 2021, 76, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Telenga, E.D.; Hoffmann, R.F.; Ruben, t.K.; Hoonhorst, S.J.; Willemse, B.W.; van Oosterhout, A.J.; Heijink, I.H.; van den Berge, M.; Jorge, L.; Sandra, P.; et al. Untargeted lipidomic analysis in chronic obstructive pulmonary disease. Uncovering sphingolipids. Am. J. Respir. Crit. Care Med. 2014, 190, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Guo, C.; Zhang, L.; Zhang, L.; Abulikemu, M.; Wang, J.; Zhou, Q.; Chen, Y.; Sun, Y.; Chang, C. Serum Glycerophospholipid Profile in Acute Exacerbation of Chronic Obstructive Pulmonary Disease. Front. Physiol. 2021, 12, 646010. [Google Scholar] [CrossRef] [PubMed]
- Bowler, R.P.; Jacobson, S.; Cruickshank, C.; Hughes, G.J.; Siska, C.; Ory, D.S.; Petrache, I.; Schaffer, J.E.; Reisdorph, N.; Kechris, K. Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. Am. J. Respir. Crit. Care Med. 2015, 191, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Suresh, B.; Lim, M.N.; Hong, S.H.; Kim, K.S.; Song, H.E.; Lee, H.Y.; Yoo, H.J.; Kim, W.J. Metabolomics Reveals Dysregulated Sphingolipid and Amino Acid Metabolism Associated with Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstr. Pulm. Dis. 2022, 17, 2343–2353. [Google Scholar] [CrossRef]
- Liu, D.; Meister, M.; Zhang, S.; Vong, C.I.; Wang, S.; Fang, R.; Li, L.; Wang, P.G.; Massion, P.; Ji, X. Identification of lipid biomarker from serum in patients with chronic obstructive pulmonary disease. Respir. Res. 2020, 21, 242. [Google Scholar] [CrossRef]
- Cruickshank-Quinn, C.I.; Jacobson, S.; Hughes, G.; Powell, R.L.; Petrache, I.; Kechris, K.; Bowler, R.; Reisdorph, N. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci. Rep. 2018, 8, 17132. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Flexeder, C.; McGarrah, R.W., III; Wyss, A.; Morrison, A.C.; North, K.E.; Boerwinkle, E.; Kastenmuller, G.; Gieger, C.; Suhre, K.; et al. Metabolomics Identifies Novel Blood Biomarkers of Pulmonary Function and COPD in the General Population. Metabolites 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Luettich, K.; Leroy, P.; Boue, S.; Vuillaume, G.; Vihervaara, T.; Ekroos, K.; Martin, F.; Peitsch, M.C.; Hoeng, J. Alterations in Serum Polyunsaturated Fatty Acids and Eicosanoids in Patients with Mild to Moderate Chronic Obstructive Pulmonary Disease (COPD). Int. J. Mol. Sci. 2016, 17, 1583. [Google Scholar] [CrossRef] [PubMed]
- Contoli, M.; Pauletti, A.; Rossi, M.R.; Spanevello, A.; Casolari, P.; Marcellini, A.; Forini, G.; Gnesini, G.; Marku, B.; Barnes, N.; et al. Long-term effects of inhaled corticosteroids on sputum bacterial and viral loads in COPD. Eur. Respir. J. 2017, 50, 1700451. [Google Scholar] [CrossRef]
- Singanayagam, A.; Glanville, N.; Cuthbertson, L.; Bartlett, N.W.; Finney, L.J.; Turek, E.; Bakhsoliani, E.; Calderazzo, M.A.; Trujillo-Torralbo, M.B.; Footitt, J.; et al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci. Transl. Med. 2019, 11, eaav3879. [Google Scholar] [CrossRef] [PubMed]
- Begley, L.A.; Opron, K.; Bian, G.; Kozik, A.J.; Liu, C.; Felton, J.; Wen, B.; Sun, D.; Huang, Y.J. Effects of Fluticasone Propionate on Klebsiella pneumoniae and Gram-Negative Bacteria Associated with Chronic Airway Disease. mSphere 2022, 7, e0037722. [Google Scholar] [CrossRef]
- Patterson, C.M.; Morrison, R.L.; D’Souza, A.; Teng, X.S.; Happel, K.I. Inhaled fluticasone propionate impairs pulmonary clearance of Klebsiella pneumoniae in mice. Respir. Res. 2012, 13, 40. [Google Scholar] [CrossRef]
- Leitao Filho, F.S.; Takiguchi, H.; Akata, K.; Ra, S.W.; Moon, J.Y.; Kim, H.K.; Cho, Y.; Yamasaki, K.; Milne, S.; Yang, J.; et al. Effects of Inhaled Corticosteroid/Long-Acting beta2-Agonist Combination on the Airway Microbiome of Patients with Chronic Obstructive Pulmonary Disease: A Randomized Controlled Clinical Trial (DISARM). Am. J. Respir. Crit. Care Med. 2021, 204, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Yip, W.; Li, X.; Koelwyn, G.J.; Milne, S.; Leitao Filho, F.S.; Yang, C.X.; Hernandez Cordero, A.I.; Yang, J.; Yang, C.W.T.; Shaipanich, T.; et al. Inhaled Corticosteroids Selectively Alter the Microbiome and Host Transcriptome in the Small Airways of Patients with Chronic Obstructive Pulmonary Disease. Biomedicines 2022, 10, 1110. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Ali, N.; Thng, K.X.; Goh, K.; Lau, K.J.X.; Chotirmall, S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia 2020, 185, 207–231. [Google Scholar] [CrossRef]
- Yi, X.; Li, Y.; Liu, H.; Liu, X.; Yang, J.; Gao, J.; Yang, Y.; Liang, Z.; Wang, F.; Chen, D.; et al. Inflammatory Endotype-Associated Airway Resistome in Chronic Obstructive Pulmonary Disease. Microbiol. Spectr. 2022, 10, e0259321. [Google Scholar] [CrossRef]
- Mac Aogain, M.; Lau, K.J.X.; Cai, Z.; Kumar Narayana, J.; Purbojati, R.W.; Drautz-Moses, D.I.; Gaultier, N.E.; Jaggi, T.K.; Tiew, P.Y.; Ong, T.H.; et al. Metagenomics Reveals a Core Macrolide Resistome Related to Microbiota in Chronic Respiratory Disease. Am. J. Respir. Crit. Care Med. 2020, 202, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Yamaya, M.; Azuma, A.; Takizawa, H.; Kadota, J.; Tamaoki, J.; Kudoh, S. Macrolide effects on the prevention of COPD exacerbations. Eur. Respir. J. 2012, 40, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Brill, S.E.; Law, M.; El-Emir, E.; Allinson, J.P.; James, P.; Maddox, V.; Donaldson, G.C.; McHugh, T.D.; Cookson, W.O.; Moffatt, M.F.; et al. Effects of different antibiotic classes on airway bacteria in stable COPD using culture and molecular techniques: A randomised controlled trial. Thorax 2015, 70, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Segal, L.N.; Clemente, J.C.; Wu, B.G.; Wikoff, W.R.; Gao, Z.; Li, Y.; Ko, J.P.; Rom, W.N.; Blaser, M.J.; Weiden, M.D. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 2017, 72, 13–22. [Google Scholar] [CrossRef]
- Huang, Y.J.; Sethi, S.; Murphy, T.; Nariya, S.; Boushey, H.A.; Lynch, S.V. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 2813–2823. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, D.; Lin, Y.; Liu, Z.; Liang, Z.; Su, J.; Chen, R.; Zhou, H.; Wang, Z. Association of sputum microbiome with clinical outcome of initial antibiotic treatment in hospitalized patients with acute exacerbations of COPD. Pharm. Res. 2020, 160, 105095. [Google Scholar] [CrossRef]
- Mac Aogain, M.; Narayana, J.K.; Tiew, P.Y.; Ali, N.; Yong, V.F.L.; Jaggi, T.K.; Lim, A.Y.H.; Keir, H.R.; Dicker, A.J.; Thng, K.X.; et al. Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 2021, 27, 688–699. [Google Scholar] [CrossRef]
- Murray, M.A.; Chotirmall, S.H. The Impact of Immunosenescence on Pulmonary Disease. Mediat. Inflamm. 2015, 2015, 692546. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Hoopmann, M.R.; Castaldi, P.J.; Simonsen, K.A.; Midha, M.K.; Cho, M.H.; Criner, G.J.; Bueno, R.; Liu, J.; Moritz, R.L.; et al. Lung proteomic biomarkers associated with chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 321, L1119–L1130. [Google Scholar] [CrossRef]
- Ubhi, B.K.; Cheng, K.K.; Dong, J.; Janowitz, T.; Jodrell, D.; Tal-Singer, R.; MacNee, W.; Lomas, D.A.; Riley, J.H.; Griffin, J.L.; et al. Targeted metabolomics identifies perturbations in amino acid metabolism that sub-classify patients with COPD. Mol. Biosyst. 2012, 8, 3125–3133. [Google Scholar] [CrossRef]
- Stockley, R.A.; Edgar, R.G.; Pillai, A.; Turner, A.M. Individualized lung function trends in alpha-1-antitrypsin deficiency: A need for patience in order to provide patient centered management? Int. J. Chron. Obstr. Pulm. Dis. 2016, 11, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Cant, E.; Keir, H.R.; Barton, A.K.; Kuzmanova, E.; Shuttleworth, M.; Pollock, J.; Finch, S.; Polverino, E.; Bottier, M.; et al. Endotyping Chronic Obstructive Pulmonary Disease, Bronchiectasis, and the “Chronic Obstructive Pulmonary Disease-Bronchiectasis Association”. Am. J. Respir. Crit. Care Med. 2022, 206, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Tiew, P.Y.; Ko, F.W.S.; Pang, S.L.; Matta, S.A.; Sio, Y.Y.; Poh, M.E.; Lau, K.J.X.; Mac Aogain, M.; Jaggi, T.K.; Ivan, F.X.; et al. Environmental fungal sensitisation associates with poorer clinical outcomes in COPD. Eur. Respir. J. 2020, 56, 2000418. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Narayana, J.K.; Quek, M.S.L.; Ang, Y.Y.; Ko, F.W.S.; Poh, M.E.; Jaggi, T.K.; Xu, H.; Thng, K.X.; Koh, M.S.; et al. Sensitisation to recombinant Aspergillus fumigatus allergens and clinical outcomes in COPD. Eur. Respir. J. 2023, 61, 2200507. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Chen, B.; Yang, Y.; Yi, X.; Wei, M.; Ecklu-Mensah, G.; Buschmann, M.M.; Liu, H.; Gao, J.; Liang, W.; et al. Multi-omics analyses of airway host-microbe interactions in chronic obstructive pulmonary disease identify potential therapeutic interventions. Nat. Microbiol. 2022, 7, 1361–1375. [Google Scholar] [CrossRef]
- Whelan, F.J.; Waddell, B.; Syed, S.A.; Shekarriz, S.; Rabin, H.R.; Parkins, M.D.; Surette, M.G. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat. Microbiol. 2020, 5, 379–390. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogain, M.; Goh, W.W.B.; Xia, K.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Mathematical-based microbiome analytics for clinical translation. Comput. Struct. Biotechnol. J. 2021, 19, 6272–6281. [Google Scholar] [CrossRef]
- Singh, S.; Natalini, J.G.; Segal, L.N. Lung microbial-host interface through the lens of multi-omics. Mucosal Immunol. 2022, 15, 837–845. [Google Scholar] [CrossRef]
- Li, C.X.; Wheelock, C.E.; Skold, C.M.; Wheelock, A.M. Integration of multi-omics datasets enables molecular classification of COPD. Eur. Respir. J. 2018, 51, 1701930. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Licursi, V.; Morrow, J.; Hersh, C.; Cho, M.; Castaldi, P.; Glass, K.; Silverman, E.K.; et al. Integrated transcriptomic correlation network analysis identifies COPD molecular determinants. Sci. Rep. 2020, 10, 3361. [Google Scholar] [CrossRef]
- Hornung, B.V.H.; Zwittink, R.D.; Kuijper, E.J. Issues and current standards of controls in microbiome research. FEMS Microbiol. Ecol. 2019, 95, fiz045. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wang, J.; Ji, X.B.; Wang, Z.Y.; Wang, Y.; Zhang, L.Y.; Li, H.P.; Zhang, Z.M.; Li, Q.Y. Transcriptomics Analysis Identifies the Presence of Upregulated Ribosomal Housekeeping Genes in the Alveolar Macrophages of Patients with Smoking-Induced Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstr. Pulm. Dis. 2021, 16, 2653–2664. [Google Scholar] [CrossRef] [PubMed]
- Noell, G.; Cosio, B.G.; Faner, R.; Monso, E.; Peces-Barba, G.; de Diego, A.; Esteban, C.; Gea, J.; Rodriguez-Roisin, R.; Garcia-Nunez, M.; et al. Multi-level differential network analysis of COPD exacerbations. Eur. Respir. J. 2017, 50, 1700075. [Google Scholar] [CrossRef] [PubMed]
- Burgel, P.R.; Paillasseur, J.L.; Caillaud, D.; Tillie-Leblond, I.; Chanez, P.; Escamilla, R.; Court-Fortune, I.; Perez, T.; Carre, P.; Roche, N.; et al. Clinical COPD phenotypes: A novel approach using principal component and cluster analyses. Eur. Respir. J. 2010, 36, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, C.; Klinger, K.; Rajpal, D.K.; Zhu, C. An integrative network-based approach for drug target indication expansion. PLoS ONE 2021, 16, e0253614. [Google Scholar] [CrossRef]
- Erler, J.T.; Linding, R. Network medicine strikes a blow against breast cancer. Cell 2012, 149, 731–733. [Google Scholar] [CrossRef]
- Narayana, J.K.; Aliberti, S.; Mac Aogain, M.; Jaggi, T.K.; Binte Mohamed Ali, N.A.; Xaverius Ivan, F.; Cheng, H.S.; Yip, Y.S.; Gerard Vos, M.I.; Low, Z.S.; et al. Microbial Dysregulation of the Gut-Lung Axis in Bronchiectasis. Am. J. Respir. Crit. Care Med. 2022. accepted. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Bogaert, D.; Chalmers, J.D.; Cox, M.J.; Hansbro, P.M.; Huang, Y.J.; Molyneaux, P.L.; O’Dwyer, D.N.; Pragman, A.A.; Rogers, G.B.; et al. Therapeutic Targeting of the Respiratory Microbiome. Am. J. Respir. Crit. Care Med. 2022, 206, 535–544. [Google Scholar] [CrossRef]
- Juul, F.E.; Garborg, K.; Bretthauer, M.; Skudal, H.; Oines, M.N.; Wiig, H.; Rose, O.; Seip, B.; Lamont, J.T.; Midtvedt, T.; et al. Fecal Microbiota Transplantation for Primary Clostridium difficile Infection. N. Engl. J. Med. 2018, 378, 2535–2536. [Google Scholar] [CrossRef]
- Guo, Q.; Goldenberg, J.Z.; Humphrey, C.; El Dib, R.; Johnston, B.C. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 2019, 4, CD004827. [Google Scholar] [CrossRef]
- Tano, K.; Grahn Hakansson, E.; Holm, S.E.; Hellstrom, S. A nasal spray with alpha-haemolytic streptococci as long term prophylaxis against recurrent otitis media. Int. J. Pediatr. Otorhinolaryngol. 2002, 62, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Hakansson, E.G.; Holm, S. Effect of recolonisation with “interfering” alpha streptococci on recurrences of acute and secretory otitis media in children: Randomised placebo controlled trial. BMJ 2001, 322, 210–212. [Google Scholar] [CrossRef] [PubMed]
- McHeick, H.; Saleh, L.; Ajami, H.; Mili, H. Context Relevant Prediction Model for COPD Domain Using Bayesian Belief Network. Sensors 2017, 17, 1486. [Google Scholar] [CrossRef] [PubMed]
- Adeloye, D.; Song, P.; Zhu, Y.; Campbell, H.; Sheikh, A.; Rudan, I.; NIHR RESPIRE Global Respiratory Health Unit. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: A systematic review and modelling analysis. Lancet Respir. Med. 2022, 10, 447–458. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiew, P.Y.; Meldrum, O.W.; Chotirmall, S.H. Applying Next-Generation Sequencing and Multi-Omics in Chronic Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2023, 24, 2955. https://doi.org/10.3390/ijms24032955
Tiew PY, Meldrum OW, Chotirmall SH. Applying Next-Generation Sequencing and Multi-Omics in Chronic Obstructive Pulmonary Disease. International Journal of Molecular Sciences. 2023; 24(3):2955. https://doi.org/10.3390/ijms24032955
Chicago/Turabian StyleTiew, Pei Yee, Oliver W. Meldrum, and Sanjay H. Chotirmall. 2023. "Applying Next-Generation Sequencing and Multi-Omics in Chronic Obstructive Pulmonary Disease" International Journal of Molecular Sciences 24, no. 3: 2955. https://doi.org/10.3390/ijms24032955
APA StyleTiew, P. Y., Meldrum, O. W., & Chotirmall, S. H. (2023). Applying Next-Generation Sequencing and Multi-Omics in Chronic Obstructive Pulmonary Disease. International Journal of Molecular Sciences, 24(3), 2955. https://doi.org/10.3390/ijms24032955





