Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy
Abstract
1. Introduction
2. Results
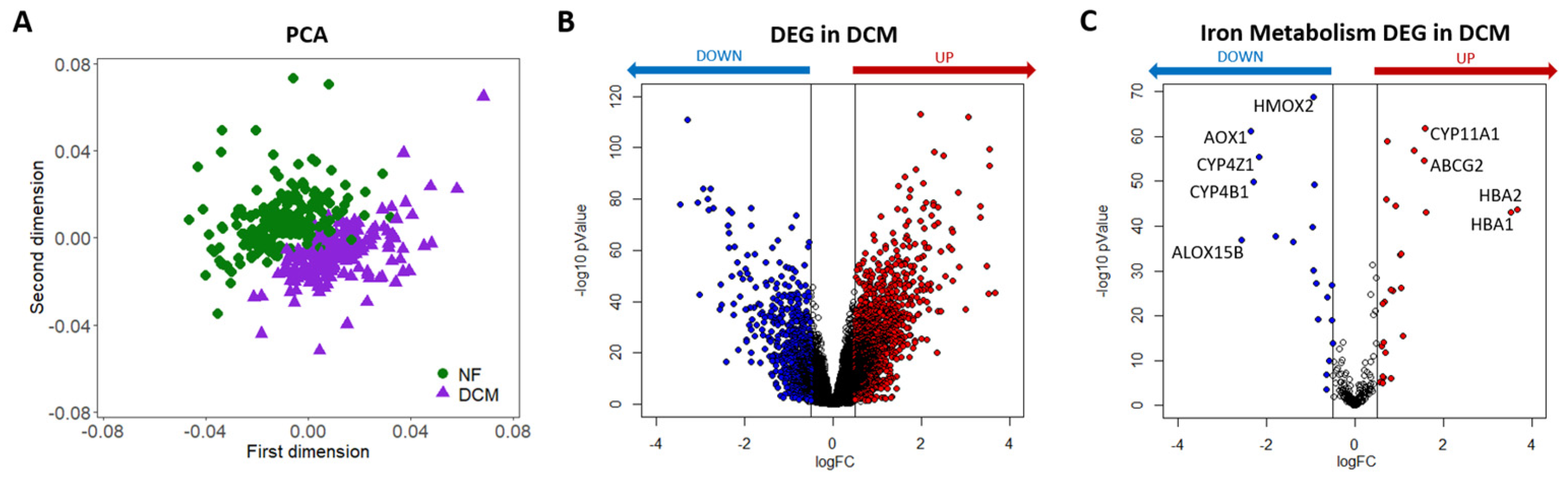
2.1. Bulk RNA-Seq Analysis in DCM
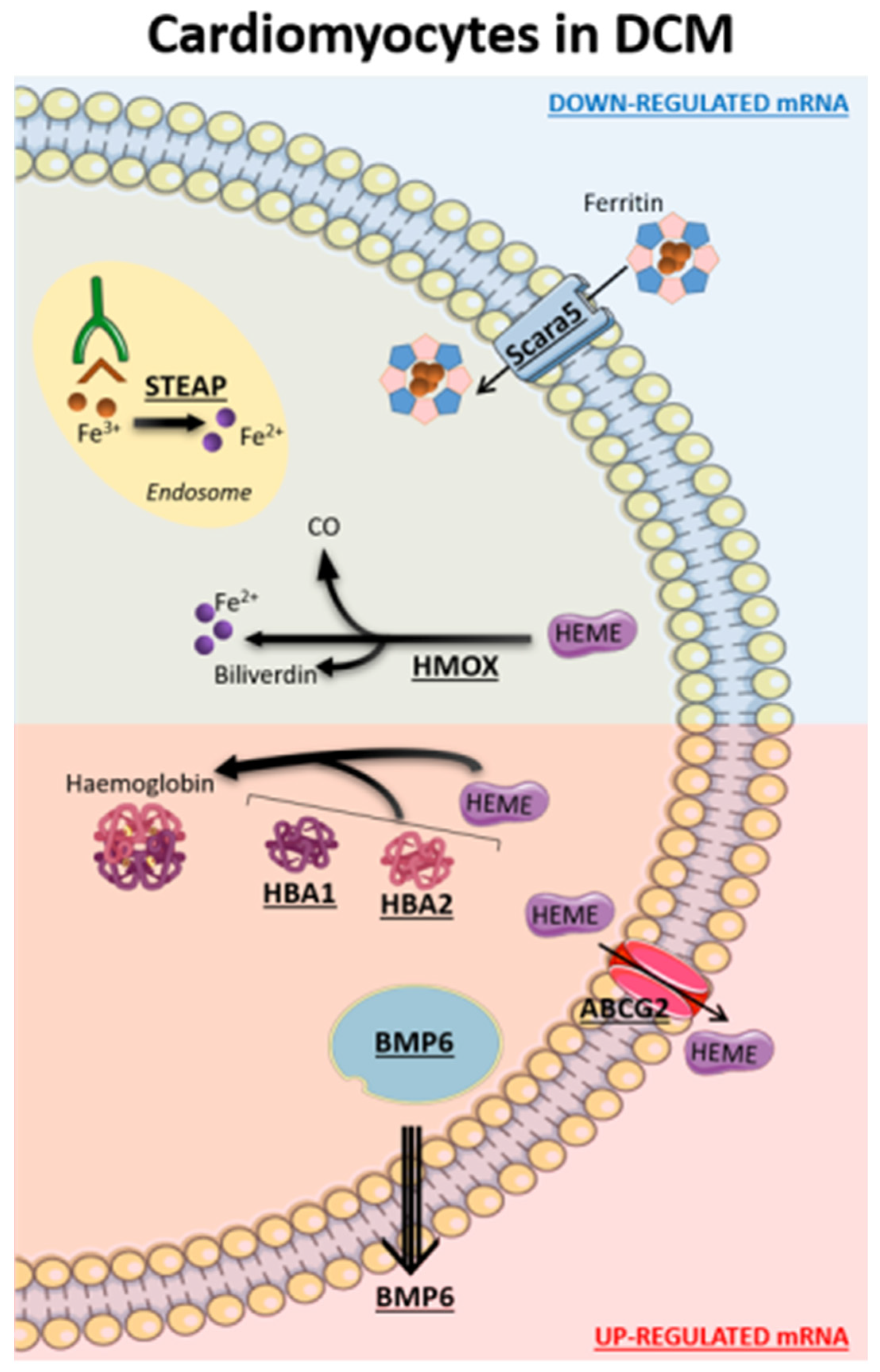
2.2. Implication of Iron Metabolism at Cardiac Tissue Level in DCM (Bulk RNA-Seq Data)
2.3. Implication of Iron Metabolism at Cardiac Cell Level in DCM (snRNA-Seq Data)
3. Discussion
4. Materials and Methods
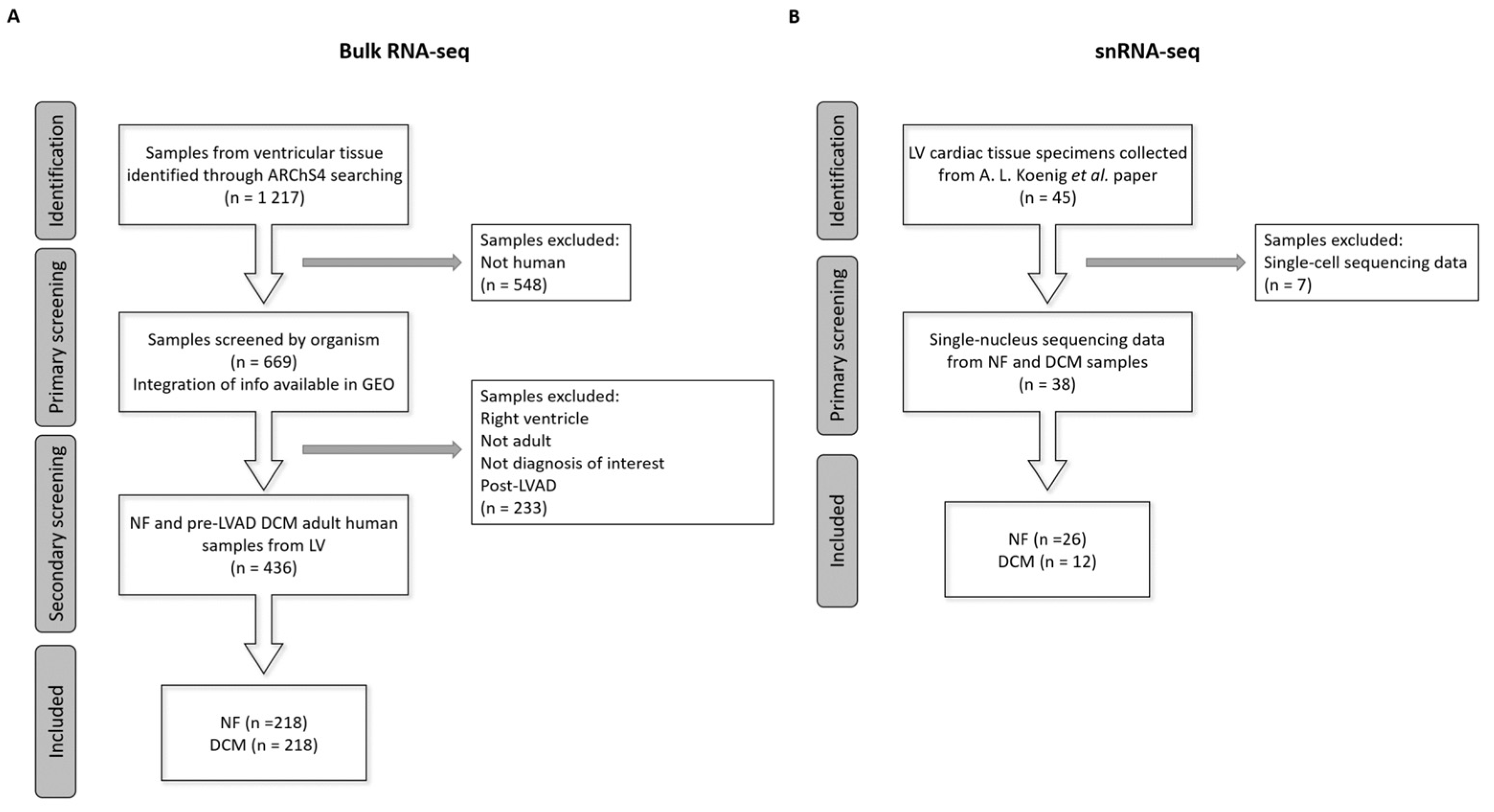
4.1. Data Collection
4.2. Gene Expression Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seferović, P.M.; Polovina, M.; Bauersachs, J.; Arad, M.; Ben Gal, T.; Lund, L.H.; Felix, S.B.; Arbustini, E.; Caforio, A.L.P.; Farmakis, D.; et al. Heart failure in cardiomyopathies: A position paper from the Heart Failure Association of the European Society of Cardiology. Eur. J. Heart Fail. 2019, 21, 553–576. [Google Scholar] [CrossRef] [PubMed]
- Visseren, F.L.J.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Back, M.; Benetos, A.; Biffi, A.; Boavida, J.M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Domae, K.; Kainuma, S.; Matsuura, R.; Yoshioka, D.; Hata, H.; Yoshikawa, Y.; Toda, K.; Sawa, Y. Long-term outcome of a dilated cardiomyopathy patient after mitral valve surgery combined with tissue-engineered myoblast sheets—Report of a case. Surg. Case Rep. 2018, 4, 142. [Google Scholar] [CrossRef]
- Tenge, T.; Roth, S.; M‘Pembele, R.; Buse, G.L.; Boenner, F.; Ballázs, C.; Tudorache, I.; Boeken, U.; Lichtenberg, A.; Neukirchen, M.; et al. Impact of Left Ventricular Assist Devices on Days Alive and Out of Hospital in Hemodynamically Stable Patients with End-Stage Heart Failure: A Propensity Score Matched Study. Life 2022, 12, 1966. [Google Scholar] [CrossRef] [PubMed]
- Japp, A.G.; Gulati, A.; Cook, S.A.; Cowie, M.R.; Prasad, S.K. The Diagnosis and Evaluation of Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2016, 67, 2996–3010. [Google Scholar] [CrossRef]
- Jankowska, E.A.; Rozentryt, P.; Witkowska, A.; Nowak, J.; Hartmann, O.; Ponikowska, B.; Borodulin-Nadzieja, L.; Banasiak, W.; Polonski, L.; Filippatos, G.; et al. Iron deficiency: An ominous sign in patients with systolic chronic heart failure. Eur. Heart J. 2010, 31, 1872–1880. [Google Scholar] [CrossRef]
- Murphy, C.J.; Oudit, G.Y. Iron-Overload Cardiomyopathy: Pathophysiology, Diagnosis, and Treatment. J. Card. Fail. 2010, 16, 888–900. [Google Scholar] [CrossRef]
- Loncar, G.; Obradovic, D.; Thiele, H.; von Haehling, S.; Lainscak, M. Iron deficiency in heart failure. ESC Heart Fail. 2021, 8, 2368–2379. [Google Scholar] [CrossRef]
- Zhang, H.; Zhabyeyev, P.; Wang, S.; Oudit, G.Y. Role of iron metabolism in heart failure: From iron deficiency to iron overload. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1925–1937. [Google Scholar] [CrossRef]
- Dev, S.; Babitt, J.L. Overview of iron metabolism in health and disease. Hemodial. Int. 2017, 21 (Suppl. 1), S6–S20. [Google Scholar] [CrossRef]
- Vogt, A.-C.; Arsiwala, T.; Mohsen, M.; Vogel, M.; Manolova, V.; Bachmann, M. On Iron Metabolism and Its Regulation. Int. J. Mol. Sci. 2021, 22, 4591. [Google Scholar] [CrossRef]
- Mackenzie, E.L.; Iwasaki, K.; Tsuji, Y. Intracellular Iron Transport and Storage: From Molecular Mechanisms to Health Implications. Antioxid. Redox Signal. 2008, 10, 997–1030. [Google Scholar] [CrossRef]
- Muckenthaler, M.U.; Rivella, S.; Hentze, M.W.; Galy, B. A Red Carpet for Iron Metabolism. Cell 2017, 168, 344–361. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef]
- Sukhbaatar, N.; Weichhart, T. Iron Regulation: Macrophages in Control. Pharmaceuticals 2018, 11, 137. [Google Scholar] [CrossRef]
- Kozłowska, B.; Sochanowicz, B.; Kraj, L.; Palusińska, M.; Kołsut, P.; Szymański, Ł.; Lewicki, S.; Kruszewski, M.; Załęska-Kocięcka, M.; Leszek, P. Clinical and Molecular Aspects of Iron Metabolism in Failing Myocytes. Life 2022, 12, 1203. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, K.; Shapiro, J.S.; Goodman, L.; Ardehali, H. Iron and Heart Failure: Diagnosis, Therapies, and Future Directions. JACC Basic Transl. Sci. 2020, 5, 300–313. [Google Scholar] [CrossRef]
- Ravingerová, T.; Kindernay, L.; Barteková, M.; Ferko, M.; Adameová, A.; Zohdi, V.; Bernátová, I.; Ferenczyová, K.; Lazou, A. The Molecular Mechanisms of Iron Metabolism and Its Role in Cardiac Dysfunction and Cardioprotection. Int. J. Mol. Sci. 2020, 21, 7889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis—An update. Front. Pharmacol. 2014, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Anand, I.S.; Gupta, P. Anemia and Iron Deficiency in Heart Failure: Current Concepts and Emerging Therapies. Circulation 2018, 138, 80–98. [Google Scholar] [CrossRef]
- McDonagh, T.; Damy, T.; Doehner, W.; Lam, C.S.; Sindone, A.; Van Der Meer, P.; Cohen-Solal, A.; Kindermann, I.; Manito, N.; Pfister, O.; et al. Screening, diagnosis and treatment of iron deficiency in chronic heart failure: Putting the 2016 European Society of Cardiology heart failure guidelines into clinical practice. Eur. J. Heart Fail. 2018, 20, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Campodonico, J.; Nicoli, F.; Motta, I.; De Amicis, M.M.; Bonomi, A.; Cappellini, M.; Agostoni, P. Prognostic role of transferrin saturation in heart failure patients. Eur. J. Prev. Cardiol. 2021, 28, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Anker, S.D.; Comin Colet, J.; Filippatos, G.; Willenheimer, R.; Dickstein, K.; Drexler, H.; Lüscher, T.F.; Bart, B.; Banasiak, W.; Niegowska, J.; et al. Ferric Carboxymaltose in Patients with Heart Failure and Iron Deficiency. N. Engl. J. Med. 2009, 361, 2436–2448. [Google Scholar] [CrossRef] [PubMed]
- Caravita, S.; Faini, A.; Vignati, C.; Pelucchi, S.; Salvioni, E.; Cattadori, G.; Baratto, C.; Torlasco, C.; Contini, M.; Villani, A.; et al. Intravenous iron therapy improves the hypercapnic ventilatory response and sleep disordered breathing in chronic heart failure. Eur. J. Heart Fail. 2022, 24, 1940–1949. [Google Scholar] [CrossRef]
- Jankowska, E.A.; Kirwan, B.-A.; Kosiborod, M.; Butler, J.; Anker, S.D.; McDonagh, T.; Dorobantu, M.; Drozdz, J.; Filippatos, G.; Keren, A.; et al. The effect of intravenous ferric carboxymaltose on health-related quality of life in iron-deficient patients with acute heart failure: The results of the AFFIRM-AHF study. Eur. Heart J. 2011, 42, 3011–3020. [Google Scholar] [CrossRef]
- Zhang, H.; Jamieson, K.L.; Grenier, J.; Nikhanj, A.; Tang, Z.; Wang, F.; Wang, S.; Seidman, J.G.; Seidman, C.E.; Thompson, R.; et al. Myocardial Iron Deficiency and Mitochondrial Dysfunction in Advanced Heart Failure in Humans. J. Am. Heart Assoc. 2022, 11, e022853. [Google Scholar] [CrossRef] [PubMed]
- Alnuwaysir, R.I.S.; Hoes, M.F.; van Veldhuisen, D.J.; van der Meer, P.; Beverborg, N.G. Iron Deficiency in Heart Failure: Mechanisms and Pathophysiology. J. Clin. Med. 2021, 11, 125. [Google Scholar] [CrossRef]
- Berdoukas, V.; Coates, T.D.; Cabantchik, Z.I. Iron and oxidative stress in cardiomyopathy in thalassemia. Free. Radic. Biol. Med. 2015, 88 Pt A, 3–9. [Google Scholar] [CrossRef]
- Fang, X.; Ardehali, H.; Min, J.; Wang, F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat. Rev. Cardiol. 2022, 20, 7–23. [Google Scholar] [CrossRef]
- Ayer, A.; Zarjou, A.; Agarwal, A.; Stocker, R. Heme Oxygenases in Cardiovascular Health and Disease. Physiol. Rev. 2016, 96, 1449–1508. [Google Scholar] [CrossRef]
- Zhang, H.; Huo, J.; Jiang, W.; Shan, Q. Integrated microarray analysis to identify potential biomarkers and therapeutic targets in dilated cardiomyopathy. Mol. Med. Rep. 2020, 22, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Koenig, A.L.; Shchukina, I.; Amrute, J.; Andhey, P.S.; Zaitsev, K.; Lai, L.; Bajpai, G.; Bredemeyer, A.; Smith, G.; Jones, C.; et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res. 2022, 1, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, Z.; Yang, X.; Weng, S.; Xu, H.; Guo, C.; Xing, Z.; Liu, L.; Wang, L.; Dang, Q.; et al. Exploring Key Genes to Construct a Diagnosis Model of Dilated Cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 865096. [Google Scholar] [CrossRef] [PubMed]
- Ohgami, R.S.; Campagna, D.R.; Greer, E.L.; Antiochos, B.; McDonald, A.; Chen, J.; Sharp, J.J.; Fujiwara, Y.; Barker, J.E.; Fleming, M.D. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet. 2005, 37, 1264–1269. [Google Scholar] [CrossRef]
- Maeder, M.T.; Khammy, O.; dos Remedios, C.; Kaye, D.M. Myocardial and Systemic Iron Depletion in Heart Failure: Implications for Anemia Accompanying Heart Failure. J. Am. Coll. Cardiol. 2011, 58, 474–480. [Google Scholar] [CrossRef]
- Li, P.-L.; Liu, H.; Chen, G.-P.; Li, L.; Shi, H.-J.; Nie, H.-Y.; Liu, Z.; Hu, Y.-F.; Yang, J.; Zhang, P.; et al. STEAP3 (Six-Transmembrane Epithelial Antigen of Prostate 3) Inhibits Pathological Cardiac Hypertrophy. Hypertension 2020, 76, 1219–1230. [Google Scholar] [CrossRef]
- Kumar, S.; Bandyopadhyay, U. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 2005, 157, 175–188. [Google Scholar] [CrossRef]
- Son, G.H.; Park, S.H.; Kim, Y.; Kim, J.Y.; Kim, J.W.; Chung, S.; Kim, Y.-H.; Kim, H.; Hwang, J.-J.; Seo, J.-S. Postmortem mRNA expression patterns in left ventricular myocardial tissues and their implications for forensic diagnosis of sudden cardiac death. Mol. Cells 2014, 37, 241–247. [Google Scholar] [CrossRef]
- Khechaduri, A.; Bayeva, M.; Chang, H.-C.; Ardehali, H. Heme Levels Are Increased in Human Failing Hearts. J. Am. Coll. Cardiol. 2013, 61, 1884–1893. [Google Scholar] [CrossRef]
- Bellner, L.; Martinelli, L.; Halilovic, A.; Patil, K.; Puri, N.; Dunn, M.W.; Regan, R.F.; Schwartzman, M.L. Heme Oxygenase-2 Deletion Causes Endothelial Cell Activation Marked by Oxidative Stress, Inflammation, and Angiogenesis. J. Pharmacol. Exp. Ther. 2009, 331, 925–932. [Google Scholar] [CrossRef] [PubMed]
- He, J.Z.; Ho, J.J.D.; Gingerich, S.; Courtman, D.W.; Marsden, P.A.; Ward, M.E. Enhanced Translation of Heme Oxygenase-2 Preserves Human Endothelial Cell Viability during Hypoxia. J. Biol. Chem. 2010, 285, 9452–9461. [Google Scholar] [CrossRef] [PubMed]
- Lundvig, D.M.; Scharstuhl, A.; Cremers, N.A.; Pennings, S.W.; te Paske, J.; van Rheden, R.; van Run-van Breda, C.; Regan, R.F.; Russel, F.G.; Carels, C.E.; et al. Delayed cutaneous wound closure in HO-2 deficient mice despite normal HO-1 expression. J. Cell. Mol. Med. 2014, 18, 2488–2498. [Google Scholar] [CrossRef] [PubMed]
- Andriopoulos, B., Jr.; Corradini, E.; Xia, Y.; Faasse, S.A.; Chen, S.; Grgurevic, L.; Knutson, M.D.; Pietrangelo, A.; Vukicevic, S.; Lin, H.Y.; et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 2009, 41, 482–487. [Google Scholar] [CrossRef]
- Vela, D. Balance of cardiac and systemic hepcidin and its role in heart physiology and pathology. Lab. Investig. 2018, 98, 315–326. [Google Scholar] [CrossRef]
- Fang, X.; Cai, Z.; Wang, H.; Han, D.; Cheng, Q.; Zhang, P.; Gao, F.; Yu, Y.; Song, Z.; Wu, Q.; et al. Loss of Cardiac Ferritin H Facilitates Cardiomyopathy via Slc7a11-Mediated Ferroptosis. Circ. Res. 2020, 127, 486–501. [Google Scholar] [CrossRef]
- Yuan, H.; Li, X.; Zhang, X.; Kang, R.; Tang, D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 2016, 478, 838–844. [Google Scholar] [CrossRef]
- Kim, E.H.; Shin, D.; Lee, J.; Jung, A.R.; Roh, J.-L. CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer. Cancer Lett. 2018, 432, 180–190. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, M.; Shen, M.; Kong, D.; Zhang, F.; Shao, J.; Tan, S.; Wang, S.; Chen, A.; Cao, P.; et al. The BRD7-P53-SLC25A28 axis regulates ferroptosis in hepatic stellate cells. Redox Biol. 2020, 36, 101619. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’Ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Sciomer, S.; Rellini, C.; Agostoni, P.; Moscucci, F. A new pathophysiology in heart failure patients. Artif. Organs 2020, 44, 1303–1305. [Google Scholar] [CrossRef]
- Chiesa, M.; Colombo, G.I.; Piacentini, L. DaMiRseq-an R/Bioconductor package for data mining of RNA-Seq data: Normalization, feature selection and classification. Bioinformatics 2018, 34, 1416–1418. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, P.; Zhou, B.; Song, J.; Li, Z.; Zhang, M.; Guo, G.; Wang, Y.; Chen, X.; Han, L.; et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020, 22, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Ringner, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, Y.; Yan, Y.; Niu, F.; Wang, Y.; Chen, X.; Su, G.; Liu, Y.; Zhao, X.; Qian, L.; Liu, P.; et al. Ferroptosis: A cell death connecting oxidative stress, inflammation and cardiovascular diseases. Cell Death Discov. 2021, 7, 193. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | Gene Name | logFC | Adj p Value | |
|---|---|---|---|---|
| UP-REGULATED GENES | HBA2 | Hemoglobin Subunit Alpha 2 | 3.7 | 1.9 × 10−42 |
| HBA1 | Hemoglobin Subunit Alpha 1 | 3.5 | 6.0 × 10−42 | |
| P3H2 | Prolyl 3–Hydroxylase 2 | 1.6 | 7.5 × 10−42 | |
| CYP11A1 | Cytochrome P450 Family 11 Subfamily A Member 1 | 1.6 | 4.0 × 10−60 | |
| ABCG2 | ATP Binding Cassette Subfamily G Member 2 | 1.6 | 4.2 × 10−53 | |
| HAAO | 3–Hydroxyanthranilate 3,4–Dioxygenase | 1.3 | 2.9 × 10−55 | |
| ALOX15 | Arachidonate 15–Lipoxygenase | 1.1 | 2.9 × 10−15 | |
| SNCA | Synuclein Alpha | 1.0 | 5.8 × 10−33 | |
| CYP4F3 | Cytochrome P450 Family 4 Subfamily F Member 3 | 1.0 | 1.3 × 10−25 | |
| P3H3 | Prolyl 3–Hydroxylase 3 | 1.0 | 1.1 × 10−32 | |
| CYP2J2 | Cytochrome P450 Family 2 Subfamily J Member 2 | 0.9 | 2.0 × 10−43 | |
| ATP6V1G2 | ATPase H+ Transporting V1 Subunit G2 | 0.9 | 5.2 × 10−25 | |
| CH25H | Cholesterol 25–Hydroxylase | 0.8 | 3.2 × 10−6 | |
| HPX | Hemopexin | 0.8 | 2.9 × 10−25 | |
| ARHGAP1 | Rho GTPase Activating Protein 1 | 0.7 | 2.7 × 10−57 | |
| DOWN–REGULATED GENES | EGLN1 | Egl–9 Family Hypoxia Inducible Factor 1 | −0.6 | 1.6 × 10−23 |
| CYP1A1 | Cytochrome P450 Family 1 Subfamily A Member 1 | −0.6 | 7.7 × 10−4 | |
| HMOX1 | Heme Oxygenase 1 | −0.6 | 5.7 × 10−7 | |
| TFRC | Transferrin Receptor | −0.8 | 6.7 × 10−19 | |
| STEAP4 | STEAP4 Metalloreductase | −0.9 | 1.2 × 10−26 | |
| STEAP3 | STEAP3 Metalloreductase | −0.9 | 6.4 × 10−48 | |
| HMOX2 | Heme Oxygenase 2 | −0.9 | 4.8 × 10−67 | |
| ALOX5 | Arachidonate 5–Lipoxygenase | −0.9 | 2.7 × 10−29 | |
| NECTIN1 | Nectin Cell Adhesion Molecule 1 | −1.0 | 1.1 × 10−38 | |
| SLC11A1 | Solute Carrier Family 11 Member 1 | −1.4 | 1.6 × 10−35 | |
| PPEF1 | Protein Phosphatase With EF-Hand Domain 1 | −1.8 | 1.1 × 10−36 | |
| CYP4Z1 | Cytochrome P450 Family 4 Subfamily Z Memb 1 | −2.2 | 6.4 × 10−54 | |
| CYP4B1 | Cytochrome P450 Family 4 Subfamily B Memb 1 | −2.3 | 2.0 × 10−48 | |
| AOX1 | Aldehyde Oxidase 1 | −2.3 | 1.7 × 10−59 | |
| ALOX15B | Arachidonate 15–Lipoxygenase Type B | −2.6 | 4.9 × 10−36 |
| Gene | Function | Cardiomyocytes | Fibroblasts | Myeloid Cells | Endocardial Cells | Endothelial Cells |
|---|---|---|---|---|---|---|
| ABCG2 | Exports of heme from cells | ↑ | ↑ | ↑ | ↑ | − |
| ALAS2 | The rate-limiting enzyme in heme synthesis | ↑ | − | − | − | − |
| BMP6 | Major mediator in the negative feedback loop between iron and hepcidin | ↑ | ↑ | ↑ | ↑ | − |
| CYBRD1 | Enzymatically reduces Fe3+ to Fe2+ using ascorbate (vitamin C) as an electron donor | − | − | − | ↑ | − |
| FLVCR2 | Imports heme into the cell | − | ↓ | − | − | − |
| HBA1/2 | Constitutes hemoglobin | ↑ | ↑ | ↑ | ↑ | ↑ |
| HMOX1/2 | Degrades heme into iron, carbon monoxide, and biliverdin | ↓ | ↓ | − | − | − |
| NEO1 | Associates with the BMP receptor complex via BMP receptor type Ia | − | ↑ | − | − | − |
| SCARA5 | Binds and internalizes L-ferritin | ↓ | ↓ | ↓ | − | ↓ |
| SLC25A28 | Inner membrane transporter of Fe2+ into mitochondria | ↑ | − | − | − | ↑ |
| SLC39A8 | Takes up unbound Fe2+ into the cell | − | − | ↓ | − | − |
| SLC48A1 | Transports heme across the endosomal membrane | − | ↓ | − | ↓ | − |
| STEAP3/4 | Transmembrane proteins with 4 homologous members | ↓ | − | ↓ | ↓ | ↓ |
| TFRC | Upon internalization of the Tf-TfR complex by endocytosis | − | ↓ | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massaiu, I.; Campodonico, J.; Mapelli, M.; Salvioni, E.; Valerio, V.; Moschetta, D.; Myasoedova, V.A.; Cappellini, M.D.; Pompilio, G.; Poggio, P.; et al. Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy. Int. J. Mol. Sci. 2023, 24, 2887. https://doi.org/10.3390/ijms24032887
Massaiu I, Campodonico J, Mapelli M, Salvioni E, Valerio V, Moschetta D, Myasoedova VA, Cappellini MD, Pompilio G, Poggio P, et al. Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy. International Journal of Molecular Sciences. 2023; 24(3):2887. https://doi.org/10.3390/ijms24032887
Chicago/Turabian StyleMassaiu, Ilaria, Jeness Campodonico, Massimo Mapelli, Elisabetta Salvioni, Vincenza Valerio, Donato Moschetta, Veronika A. Myasoedova, Maria Domenica Cappellini, Giulio Pompilio, Paolo Poggio, and et al. 2023. "Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy" International Journal of Molecular Sciences 24, no. 3: 2887. https://doi.org/10.3390/ijms24032887
APA StyleMassaiu, I., Campodonico, J., Mapelli, M., Salvioni, E., Valerio, V., Moschetta, D., Myasoedova, V. A., Cappellini, M. D., Pompilio, G., Poggio, P., & Agostoni, P. (2023). Dysregulation of Iron Metabolism-Linked Genes at Myocardial Tissue and Cell Levels in Dilated Cardiomyopathy. International Journal of Molecular Sciences, 24(3), 2887. https://doi.org/10.3390/ijms24032887







