Alzheimer’s Precision Neurology: Epigenetics of Cytochrome P450 Genes in Circulating Cell-Free DNA for Disease Prediction and Mechanism
Abstract
1. Introduction
2. Results
2.1. Study Groups
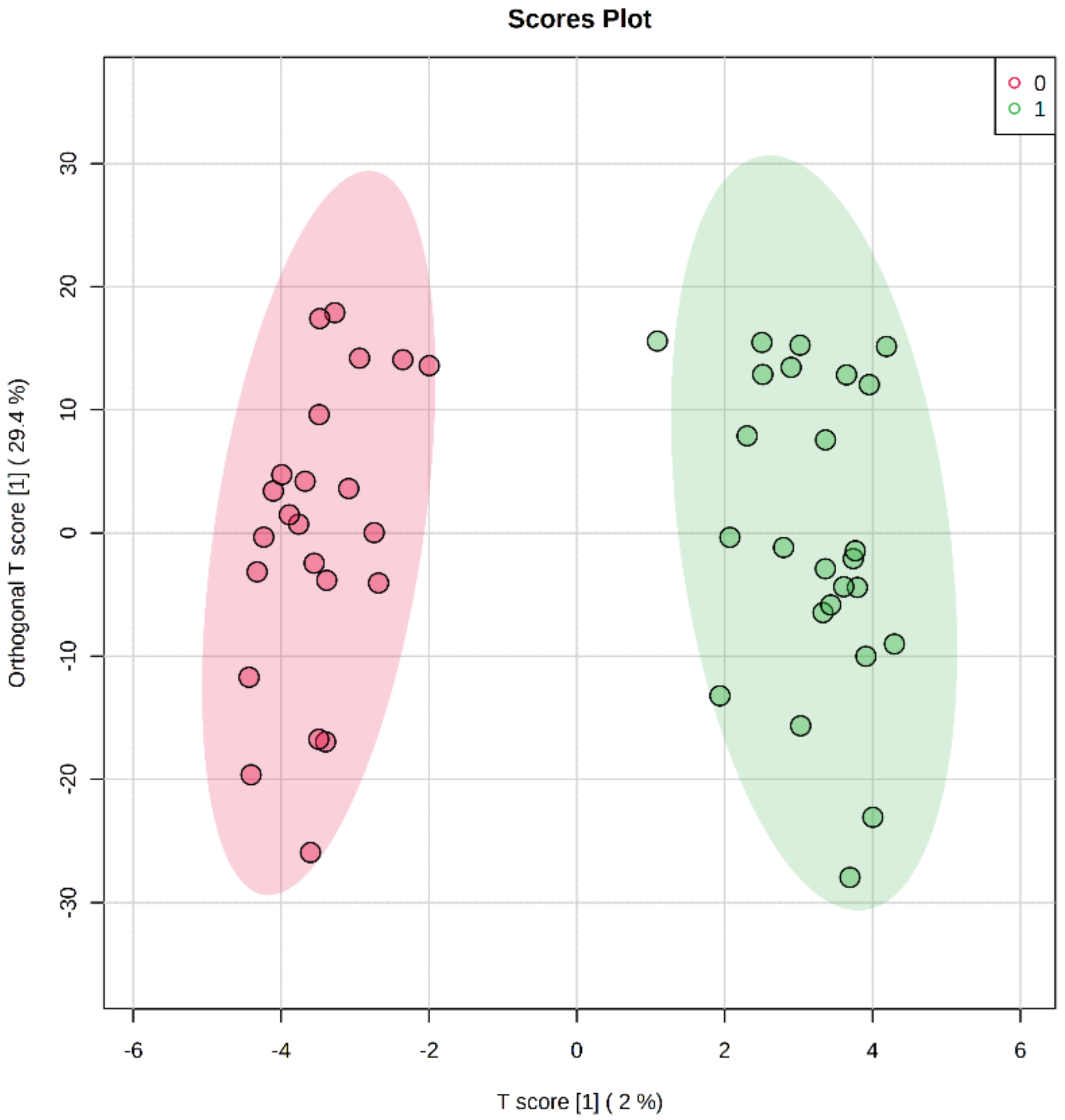
2.2. Differentially Methylated Cytosines in CYP Genes
2.3. Regression Analysis
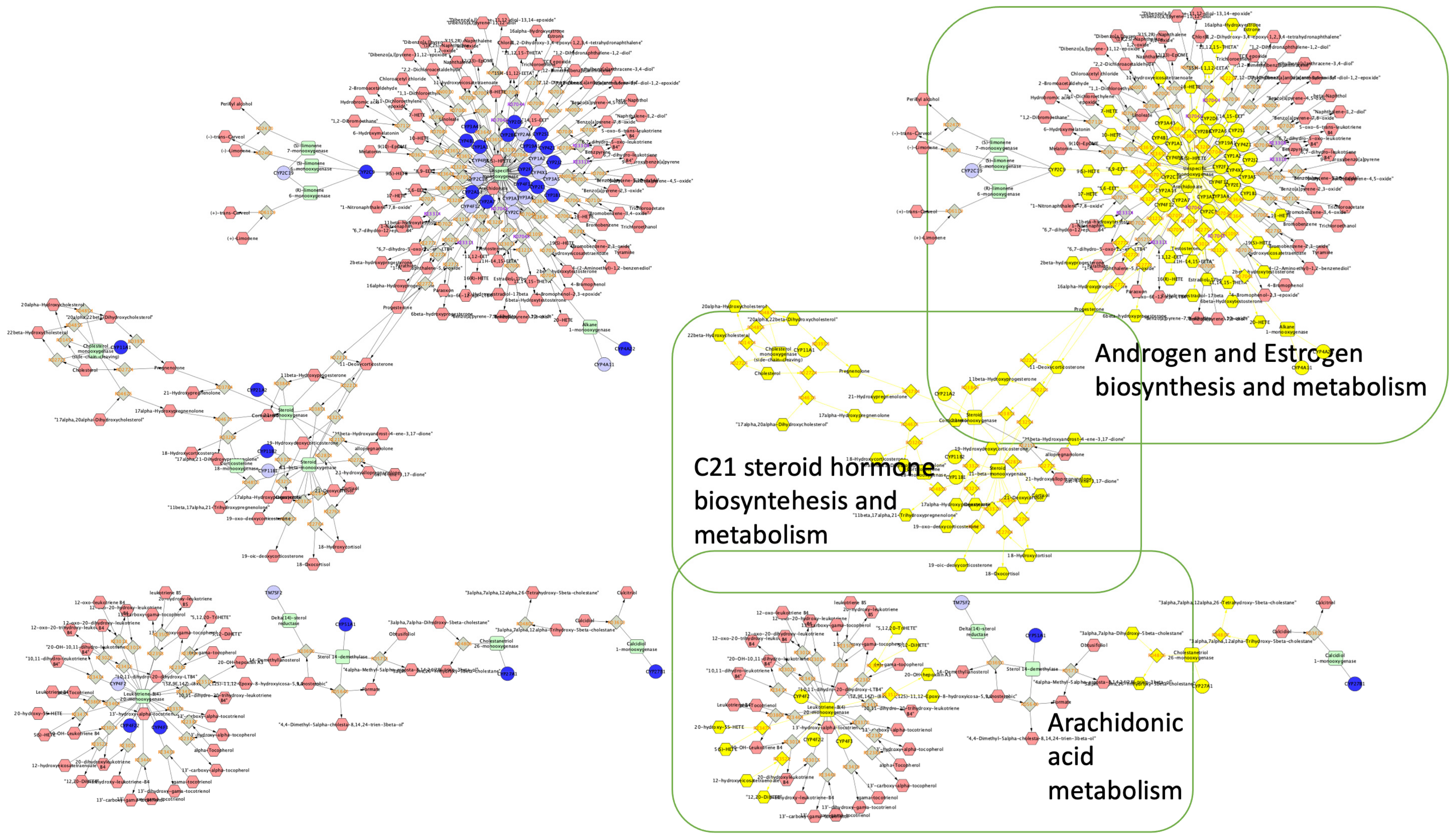
2.4. Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Data Analysis
4.2. Regression Analysis
4.3. Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Shireby, G.; Dempster, E.L.; Policicchio, S.; Smith, R.G.; Pishva, E.; Chioza, B.; Davies, J.P.; Burrage, J.; Lunnon, K.; Seiler Vellame, D.; et al. DNA methylation signatures of Alzheimer’s disease neuropathology in the cortex are primarily driven by variation in non-neuronal cell-types. Nat. Commun. 2022, 13, 5620. [Google Scholar] [CrossRef] [PubMed]
- Konki, M.; Lindgren, N.; Kyläniemi, M.; Venho, R.; Laajala, E.; Ghimire, B.; Lahesmaa, R.; Kaprio, J.; Rinne, J.O.; Lund, R.J. Plasma cell-free DNA methylation marks for episodic memory impairment: A pilot twin study. Sci. Rep. 2020, 10, 14192. [Google Scholar] [CrossRef]
- Nidadavolu, L.S.; Feger, D.; Wu, Y.; Grodstein, F.; Gross, A.L.; Bennett, D.A.; Walston, J.D.; Oh, E.S.; Abadir, P.M. Circulating Cell-Free Genomic DNA Is Associated with an Increased Risk of Dementia and with Change in Cognitive and Physical Function. J. Alzheimers Dis. 2022, 89, 1233–1240. [Google Scholar] [CrossRef]
- Hampel, H.; Toschi, N.; Babiloni, C.; Baldacci, F.; Black, K.L.; Bokde, A.L.W.; Bun, R.S.; Cacciola, F.; Cavedo, E.; Chiesa, P.A.; et al. Revolution of Alzheimer Precision Neurology. Passageway of Systems Biology and Neurophysiology. J. Alzheimer’s Dis. JAD 2018, 64, S47–S105. [Google Scholar] [CrossRef]
- Altuna, M.; Urdánoz-Casado, A.; de Gordoa, J.S.-R.; Zelaya, M.V.; Labarga, A.; Lepesant, J.M.J.; Roldán, M.; Blanco-Luquin, I.; Perdones, Á.; Larumbe, R.; et al. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin. Epigenet. 2019, 11, 91. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Radhakrishna, U.; Gordevičius, J.; Aydas, B.; Yilmaz, A.; Jafar, F.; Imam, K.; Maddens, M.; Challapalli, K.; Metpally, R.P.; et al. Artificial Intelligence and Circulating Cell-Free DNA Methylation Profiling: Mechanism and Detection of Alzheimer’s Disease. Cells 2022, 11, 1744. [Google Scholar] [CrossRef]
- Kuban, W.; Daniel, W.A. Cytochrome P450 expression and regulation in the brain. Drug Metab. Rev. 2021, 53, 1–29. [Google Scholar] [CrossRef]
- Stocco, M.R.; Tyndale, R.F. Cytochrome P450 enzymes and metabolism of drugs and neurotoxins within the mammalian brain. Adv. Pharmacol. 2022, 95, 73–106. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Mabarak, C.; Camacho-Carranza, R.; Espinosa-Aguirre, J.J. Cytochrome P450 in the central nervous system as a therapeutic target in neurodegenerative diseases. Drug Metab. Rev. 2018, 50, 95–108. [Google Scholar] [CrossRef]
- Nebert, D.W.; Wikvall, K.; Miller, W.L. Human cytochromes P450 in health and disease. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2013, 368, 20120431. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Chew, H.; Solomon, V.A.; Fonteh, A.N. Involvement of Lipids in Alzheimer’s Disease Pathology and Potential Therapies. Front. Physiol. 2020, 11, 598. [Google Scholar] [CrossRef]
- Feringa, F.M.; van der Kant, R. Cholesterol and Alzheimer’s Disease; From Risk Genes to Pathological Effects. Front. Aging Neurosci. 2021, 13, 690372. [Google Scholar] [CrossRef]
- Clarke, J.A.; Cutler, M.; Gong, I.; Schwarz, U.I.; Freeman, D.; Dasgupta, M. Cytochrome P450 2D6 phenotyping in an elderly population with dementia and response to galantamine in dementia: A pilot study. Am. J. Geriatr. Pharmacother. 2011, 9, 224–233. [Google Scholar] [CrossRef]
- Borkowski, K.; Pedersen, T.L.; Seyfried, N.T.; Lah, J.J.; Levey, A.I.; Hales, C.M.; Dammer, E.B.; Blach, C.; Louie, G.; Kaddurah-Daouk, R.; et al. Association of plasma and CSF cytochrome P450, soluble epoxide hydrolase, and ethanolamide metabolism with Alzheimer’s disease. Alzheimers Res. Ther. 2021, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Benedet, A.L.; Yu, L.; Labbe, A.; Mathotaarachchi, S.; Pascoal, T.A.; Shin, M.; Kang, M.S.; Gauthier, S.; Rouleau, G.A.; Poirier, J.; et al. CYP2C19 variant mitigates Alzheimer disease pathophysiology in vivo and postmortem. Neurol. Genet. 2018, 4, e216. [Google Scholar] [CrossRef]
- Ma, S.L.; Tang, N.L.S.; Wat, K.H.Y.; Tang, J.H.Y.; Lau, K.H.; Law, C.B.; Chiu, J.; Tam, C.C.W.; Poon, T.K.; Lin, K.L.; et al. Effect of CYP2D6 and CYP3A4 Genotypes on the Efficacy of Cholinesterase Inhibitors in Southern Chinese Patients with Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Dement. 2019, 34, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Benedet, A.L.; Yu, L.; Labbe, A.; Mathotaarachchi, S.S.; Shin, M.; Pascoal, T.A.; Kang, M.-S.; Beaudry, T.; Gauthier, S.; Bennett, D.A.; et al. P2-093: Polymorphism in Cytochrome P450 Gene is Associated with Alzheimer’s Pathology. Alzheimer’s Dement. 2016, 12, P646. [Google Scholar] [CrossRef]
- Adwan, L.; Zawia, N.H. Epigenetics: A novel therapeutic approach for the treatment of Alzheimer’s disease. Pharmacol. Ther. 2013, 139, 41–50. [Google Scholar] [CrossRef]
- Rasmi, Y.; Shokati, A.; Hassan, A.; Aziz, S.G.; Bastani, S.; Jalali, L.; Moradi, F.; Alipour, S. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci. Rep. 2023, 14, 28–37. [Google Scholar] [CrossRef]
- Hedlund, E.; Gustafsson, J.A.; Warner, M. Cytochrome P450 in the brain; a review. Curr. Drug Metab. 2001, 2, 245–263. [Google Scholar] [CrossRef]
- Bufill, E.; Ribosa-Nogue, R.; Blesa, R. The Therapeutic Potential of Epigenetic Modifications in Alzheimer’s Disease. In Alzheimer’s Disease: Drug Discovery; Huang, X., Ed.; Exon Publications: Brisbane, QL, Australia, 2020. [Google Scholar]
- Majidazar, R.; Rezazadeh-Gavgani, E.; Sadigh-Eteghad, S.; Naseri, A. Pharmacotherapy of Alzheimer’s disease: An overview of systematic reviews. Eur. J. Clin. Pharmacol. 2022, 78, 1567–1587. [Google Scholar] [CrossRef]
- Dhar, G.A.; Saha, S.; Mitra, P.; Nag Chaudhuri, R. DNA methylation and regulation of gene expression: Guardian of our health. Nucl. Int. J. Cytol. Allied Top. 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Varma, V.R.; Büşra Lüleci, H.; Oommen, A.M.; Varma, S.; Blackshear, C.T.; Griswold, M.E.; An, Y.; Roberts, J.A.; O’Brien, R.; Pletnikova, O.; et al. Abnormal brain cholesterol homeostasis in Alzheimer’s disease-a targeted metabolomic and transcriptomic study. NPJ Aging Mech. Dis. 2021, 7, 11. [Google Scholar] [CrossRef]
- Spector, A.A.; Kim, H.Y. Cytochrome P450 epoxygenase pathway of polyunsaturated fatty acid metabolism. Biochim. Biophys. Acta 2015, 1851, 356–365. [Google Scholar] [CrossRef]
- Akwa, Y. Steroids and Alzheimer’s Disease: Changes Associated with Pathology and Therapeutic Potential. Int. J. Mol. Sci. 2020, 21, 4812. [Google Scholar] [CrossRef]
- Mielke, M.M. Sex and Gender Differences in Alzheimer’s Disease Dementia. Psychiatr. Times 2018, 35, 14–17. [Google Scholar]
- Vest, R.S.; Pike, C.J. Gender, sex steroid hormones, and Alzheimer’s disease. Horm. Behav. 2013, 63, 301–307. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomics of Cognitive Dysfunction and Neuropsychiatric Disorders in Dementia. Int. J. Mol. Sci. 2020, 21, 3059. [Google Scholar] [CrossRef]
- Pinchaud, K.; Maguin-Gaté, K.; Olivier, J.-L. Dietary arachidonic acid: A Janus face actor in brain and Alzheimer’s disease? OCL 2018, 25, D406. [Google Scholar] [CrossRef]
- Thomas, M.H.; Olivier, J.L. Arachidonic acid in Alzheimer’s disease. J. Neurol. Neuromed. 2016, 1, 1–6. [Google Scholar]
- Cacabelos, R.; Cacabelos, P.; Torrellas, C. Personalized Medicine of Alzheimer’s Disease. In Handbook of Pharmacogenomics and Stratified Medicine; Elsevier: Amsterdam, The Netherlands, 2014; pp. 563–615. [Google Scholar] [CrossRef]
- Martins-Ferreira, R.; Leal, B.; Chaves, J.; Ciudad, L.; Samoes, R.; Martins da Silva, A.; Pinho Costa, P.; Ballestar, E. Circulating cell-free DNA methylation mirrors alterations in cerebral patterns in epilepsy. Clin. Epigenet. 2022, 14, 188. [Google Scholar] [CrossRef]
- Loyfer, N.; Magenheim, J.; Peretz, A.; Cann, G.; Bredno, J.; Klochendler, A.; Fox-Fisher, I.; Shabi-Porat, S.; Hecht, M.; Pelet, T.; et al. A DNA methylation atlas of normal human cell types. Nature 2023, 613, 355–364. [Google Scholar] [CrossRef]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgard, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Vishweswaraiah, S.; Aydas, B.; Yilmaz, A.; Metpally, R.P.; Carey, D.J.; Crist, R.C.; Berrettini, W.H.; Wilson, G.D.; Imam, K.; et al. Artificial intelligence and leukocyte epigenomics: Evaluation and prediction of late-onset Alzheimer’s disease. PLoS ONE 2021, 16, e0248375. [Google Scholar] [CrossRef]
- Yan, Y.Y.; Guo, Q.R.; Wang, F.H.; Adhikari, R.; Zhu, Z.Y.; Zhang, H.Y.; Zhou, W.M.; Yu, H.; Li, J.Q.; Zhang, J.Y. Cell-Free DNA: Hope and Potential Application in Cancer. Front. Cell Dev. Biol. 2021, 9, 639233. [Google Scholar] [CrossRef]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Barták, B.K.; Kalmár, A.; Galamb, O.; Wichmann, B.; Nagy, Z.B.; Tulassay, Z.; Dank, M.; Igaz, P.; Molnár, B. Blood Collection and Cell-Free DNA Isolation Methods Influence the Sensitivity of Liquid Biopsy Analysis for Colorectal Cancer Detection. Pathol. Oncol. Res. POR 2019, 25, 915–923. [Google Scholar] [CrossRef]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Hardy, T.; Zeybel, M.; Day, C.P.; Dipper, C.; Masson, S.; McPherson, S.; Henderson, E.; Tiniakos, D.; White, S.; French, J.; et al. Plasma DNA methylation: A potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2017, 66, 1321–1328. [Google Scholar] [CrossRef]
- Ramirez, K.; Fernández, R.; Collet, S.; Kiyar, M.; Delgado-Zayas, E.; Gómez-Gil, E.; Van Den Eynde, T.; T’Sjoen, G.; Guillamon, A.; Mueller, S.C.; et al. Epigenetics Is Implicated in the Basis of Gender Incongruence: An Epigenome-Wide Association Analysis. Front. Neurosci. 2021, 15, 701017. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metab. Off. J. Metab. Soc. 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed]
| Clinical Factor | Discovery Set | Validation Set | ||||
|---|---|---|---|---|---|---|
| Controls (n = 13) | Cases (n = 15) | p-Value | Controls (n = 10) | Cases (n = 10) | p-Value | |
| Age, mean (SD) | 78.0 (9.6) | 82.4 (5.4) | 0.09 * | 79.5 (8.6) | 80.9 (9.4) | 0.78 * |
| Sex (F/M) | 9/4 | 9/6 | 0.61 ^ | 6/4 | 5/5 | 0.65 ^ |
| BMI, mean (SD) | 25.6 (4.7) | 26.0 (4.4) | 0.7 * | 26.5 (5.7) | 27.5 (4.2) | 0.25 * |
| Diabetes, n | 4 | 4 | 0.65 ^ | 2 | 2 | 1.0 ^ |
| Hypertension, n | 7 | 11 | 0.28 ^ | 8 | 7 | 0.60 ^ |
| History of stroke/TIA | 2 | 3 | 0.8 ^ | 1 | 1 | 0.65 ^ |
| Family history of Alzheimer’s disease, mean (SD) | 3 | 7 | 0.07 ^ | 3 | 3 | 0.87 ^ |
| Geriatric depression score, mean (SD) | 2.07 (1.7) | 2.35 (2.23) | 0.60 * | 1.3 | 1.7 | 0.29 * |
| SLUMS total score, mean (SD) | 25 (3.39) | 13.16 (6.64) | 0.12 * | 24.4 | 12.1 | 0.02 * |
| CLOX 1, mean (SD) | 12.07 (1.55) | 9.0 (3.39) | 0.21 * | 12 | 7.7 | 0.60 * |
| CLOX 2, mean (SD) | 12.58 (2.39) | 10.07 (3.68) | 0.29 * | 13.25 | 10.2 | 0.09 * |
| Clinical Dementia Rating, mean (SD) | 0 | 1.03 | 0.013 * | 0.05 | 1.0 | 0.075 * |
| MMSE score, mean (SD) | 29 (0.81) | 20.3 (4.79) | <0.001 * | 29 | 20.4 | 0.002 * |
| CpG Methylation Predictive Algorithms | Study Group | AUC (95% CI) | Sensitivity | Specificity |
|---|---|---|---|---|
| cg17852385/cg23101118 + cg14355428/cg22536554 | Training/Discovery | 0.974 (0.957~0.992) | 100 % | 92.3% |
| 10-fold Cross-Validation | 0.928 (0.787~1.000) | 100 % | 92.3% | |
| cg17852385/cg23101118 + cg22195884/cg22536554 | Training/Discovery | 0.972 (0.954~0.991) | 100% | 86.3% |
| 10-fold Cross-Validation | 0.928 (0.787~1.000) | 100 % | 92.3% | |
| cg17852385/cg23101118 + cg07014416/cg22536554 | Training/Discovery | 0.977 (0.963~0.991) | 88.1% | 92.3% |
| 10-fold Cross-Validation | 0.913 (0.771~1.000) | 86.7% | 92.3% | |
| cg07014416/cg22536554 + cg17011709/cg17852385 | Training/Discovery | 0.977 (0.964~0.990) | 93.3% | 92.3% |
| 10-fold Cross-Validation | 0.918 (0.796~1.000) | 93.3% | 92.3% | |
| cg07014416/cg22536554 + cg02604290/cg17852385 | Training/Discovery | 0.988 (0.978~0.997) | 94.1% | 100% |
| 10-fold Cross-Validation | 0.810 (0.610~1.000) | 93.3% | 84.6% | |
| cg07014416/cg22536554 + cg02604290/cg17852385 + cg01689657/cg13608716 | Training/Discovery | 0.991 (0.984~0.998) | 94.1% | 92.3% |
| 10-fold Cross-Validation | 0.908 (0.763~1.000) | 93.3% | 92.3% | |
| cg17852385/cg23101118 + cg07014416/cg22536554 + cg01689657/cg13608716 | Training/Discovery | 0.990 (0.982~0.997) | 88.1% | 92.3% |
| 10-fold Cross-Validation | 0.908 (0.765~1.000) | 86.7% | 92.3% |
| CpG Predictive Models | AUC (95% CI) | Sensitivity | Specificity |
|---|---|---|---|
| cg17852385/cg23101118 + cg14355428/cg22536554 | 0.942 (0.905~0.979) | 90.0 % | 90.0% |
| cg17852385/cg23101118 + cg22195884/cg22536554 | 0.940 (0.908~0.972) | 87.8% | 90.0% |
| cg17852385/cg23101118 + cg07014416/cg22536554 | 0.932 (0.898~0.966) | 81.1% | 90% |
| cg07014416/cg22536554 + cg17011709/cg17852385 | 0.915 (0.874~0.955) | 88.1% | 90.0% |
| cg07014416/cg22536554 + cg02604290/cg17852385 | 0.919 (0.876~0.961) | 80.0% | 90.0% |
| cg07014416/cg22536554 + cg02604290/cg17852385 + cg01689657/cg13608716 | 0.925 (0.886~0.964) | 80.0% | 88.0% |
| cg17852385/cg23101118 + cg07014416/cg22536554 + cg01689657/cg13608716 | 0.941 (0.911~0.972) | 80.1% | 89.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahado-Singh, R.O.; Vishweswaraiah, S.; Turkoglu, O.; Graham, S.F.; Radhakrishna, U. Alzheimer’s Precision Neurology: Epigenetics of Cytochrome P450 Genes in Circulating Cell-Free DNA for Disease Prediction and Mechanism. Int. J. Mol. Sci. 2023, 24, 2876. https://doi.org/10.3390/ijms24032876
Bahado-Singh RO, Vishweswaraiah S, Turkoglu O, Graham SF, Radhakrishna U. Alzheimer’s Precision Neurology: Epigenetics of Cytochrome P450 Genes in Circulating Cell-Free DNA for Disease Prediction and Mechanism. International Journal of Molecular Sciences. 2023; 24(3):2876. https://doi.org/10.3390/ijms24032876
Chicago/Turabian StyleBahado-Singh, Ray O., Sangeetha Vishweswaraiah, Onur Turkoglu, Stewart F. Graham, and Uppala Radhakrishna. 2023. "Alzheimer’s Precision Neurology: Epigenetics of Cytochrome P450 Genes in Circulating Cell-Free DNA for Disease Prediction and Mechanism" International Journal of Molecular Sciences 24, no. 3: 2876. https://doi.org/10.3390/ijms24032876
APA StyleBahado-Singh, R. O., Vishweswaraiah, S., Turkoglu, O., Graham, S. F., & Radhakrishna, U. (2023). Alzheimer’s Precision Neurology: Epigenetics of Cytochrome P450 Genes in Circulating Cell-Free DNA for Disease Prediction and Mechanism. International Journal of Molecular Sciences, 24(3), 2876. https://doi.org/10.3390/ijms24032876









