Uterine Flushing Fluid-Derived Let-7b Targets CXCL10 to Regulate Uterine Receptivity in Goats during Embryo Implantation
Abstract
1. Introduction
2. Results
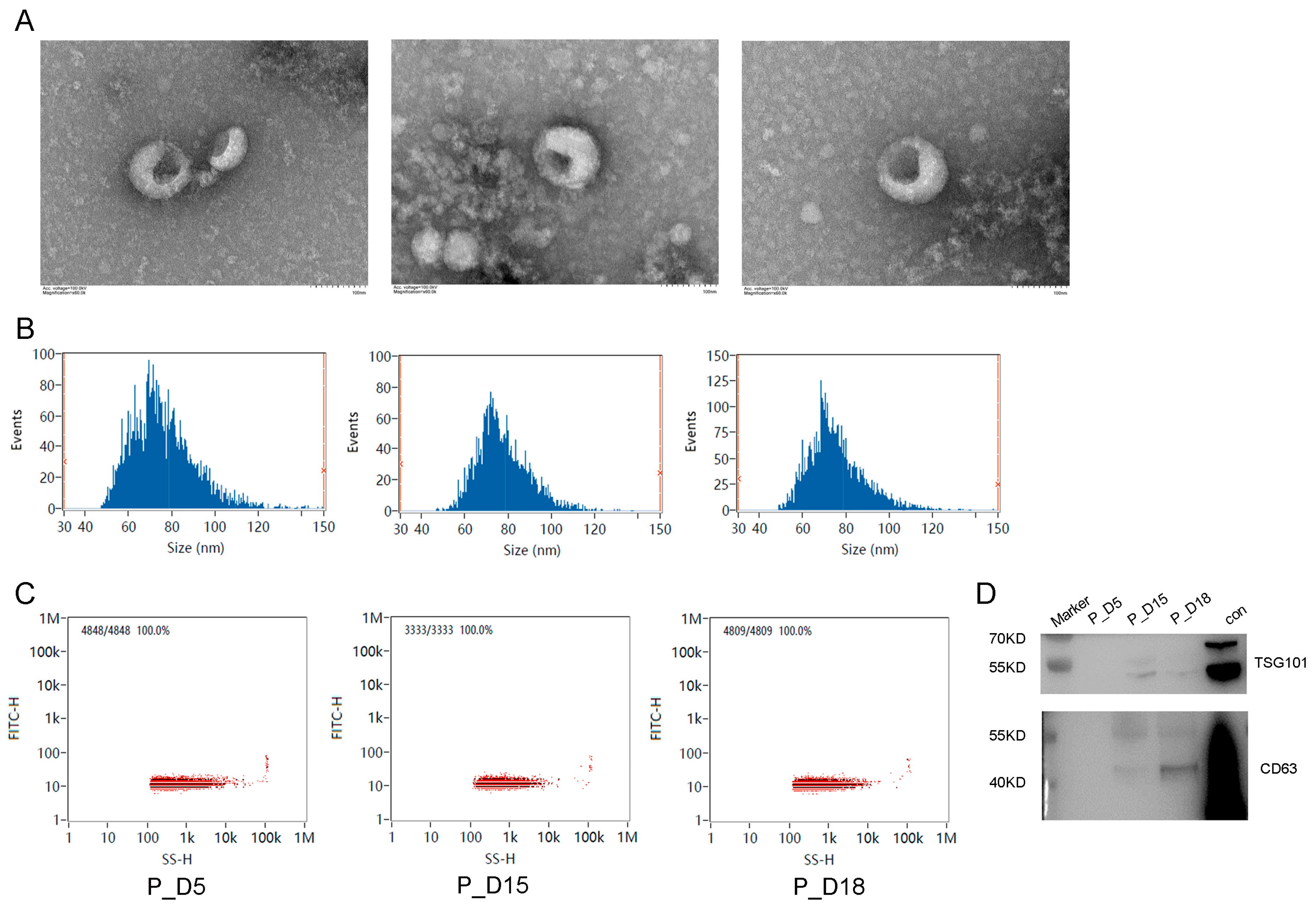
2.1. Exosome Identification
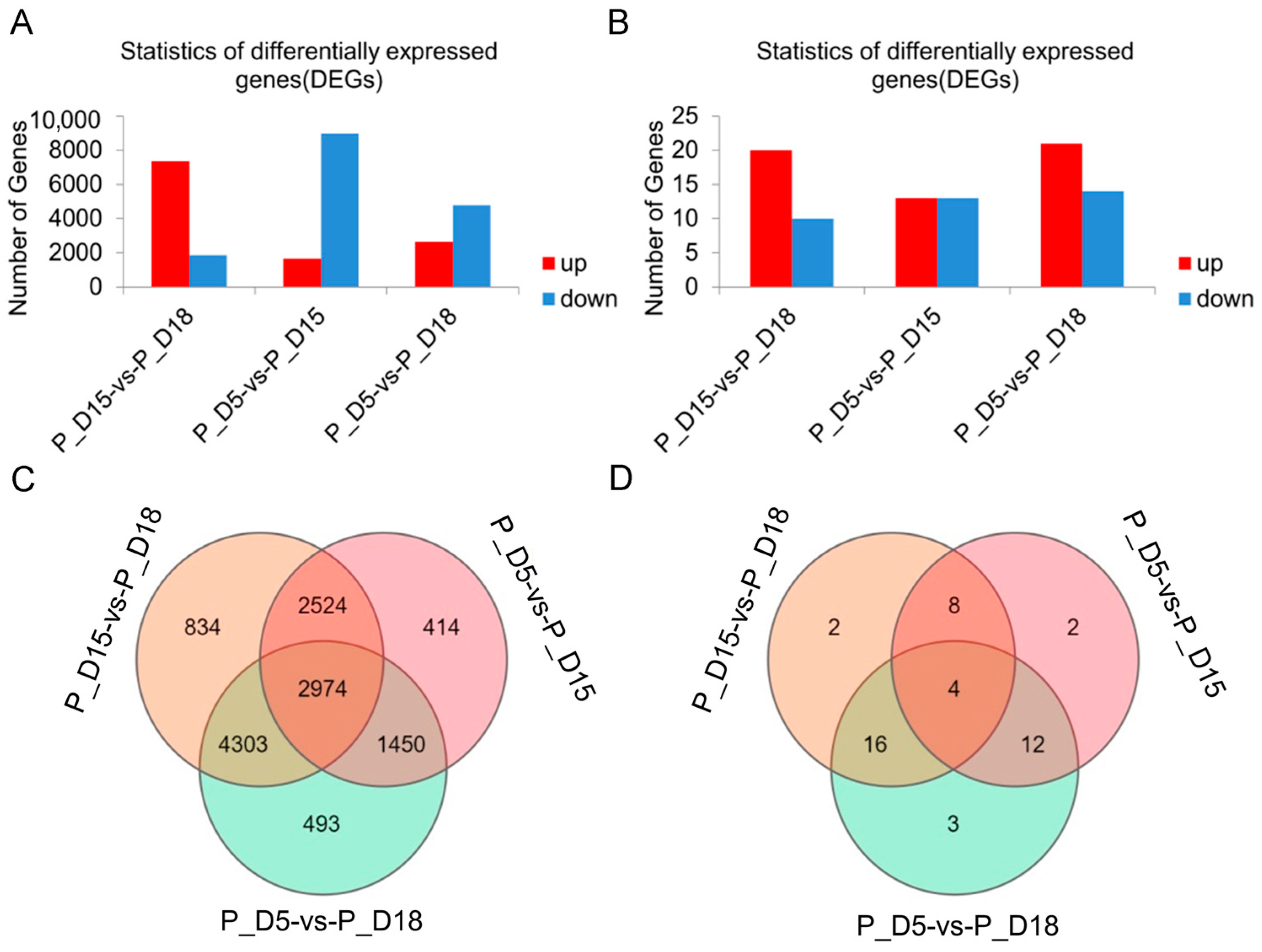
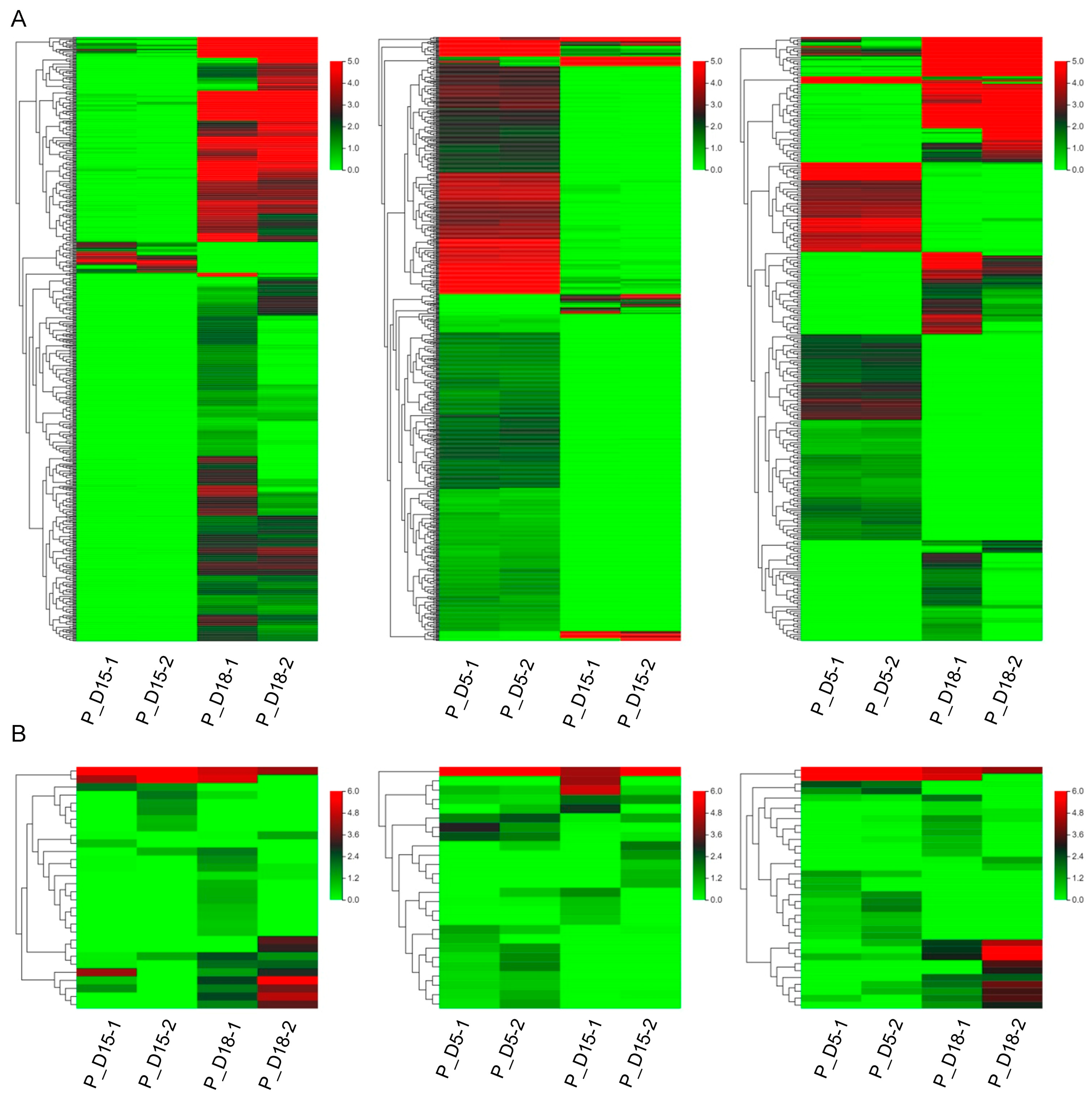
2.2. Differential mRNA and miRNA Expression Analysis
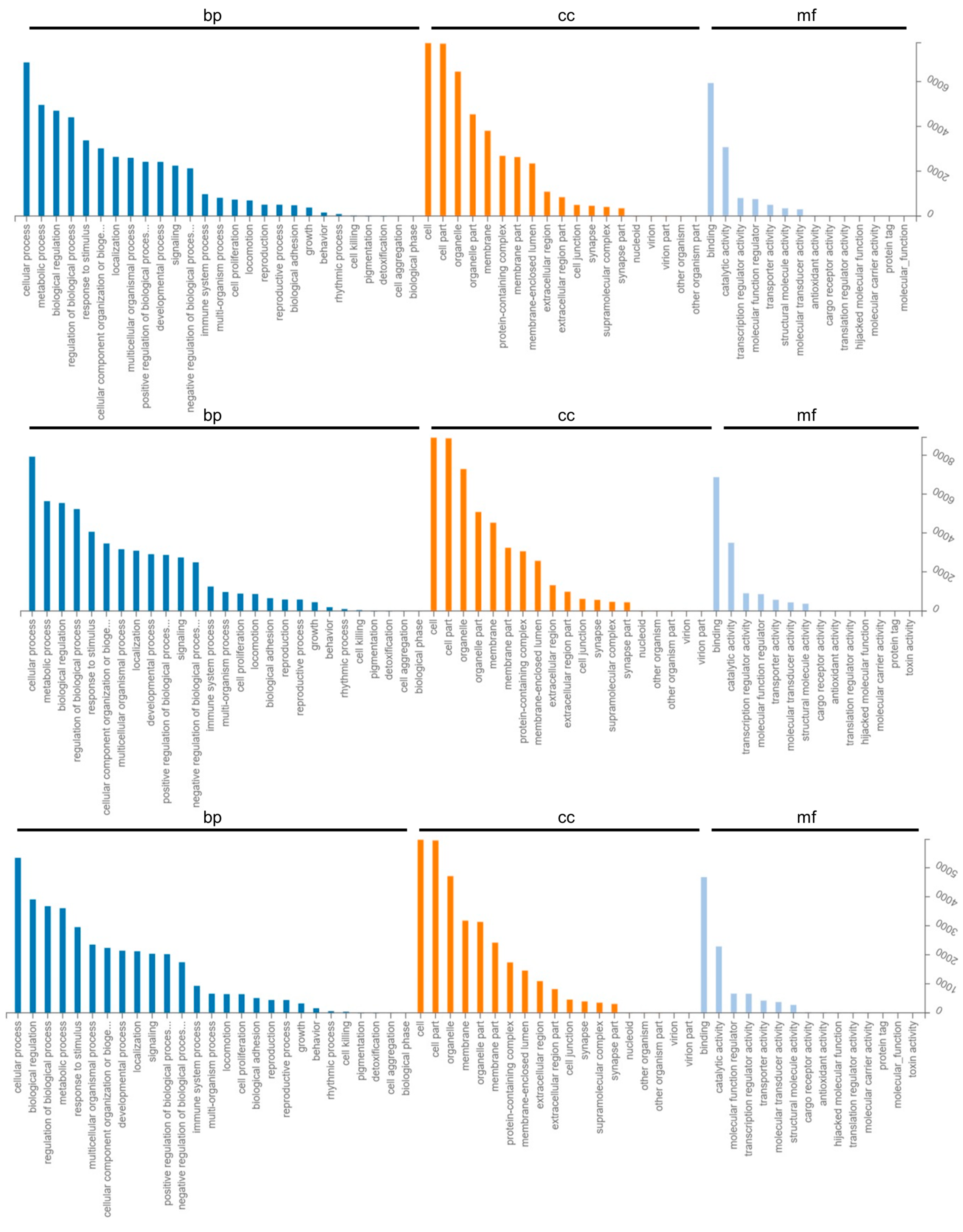
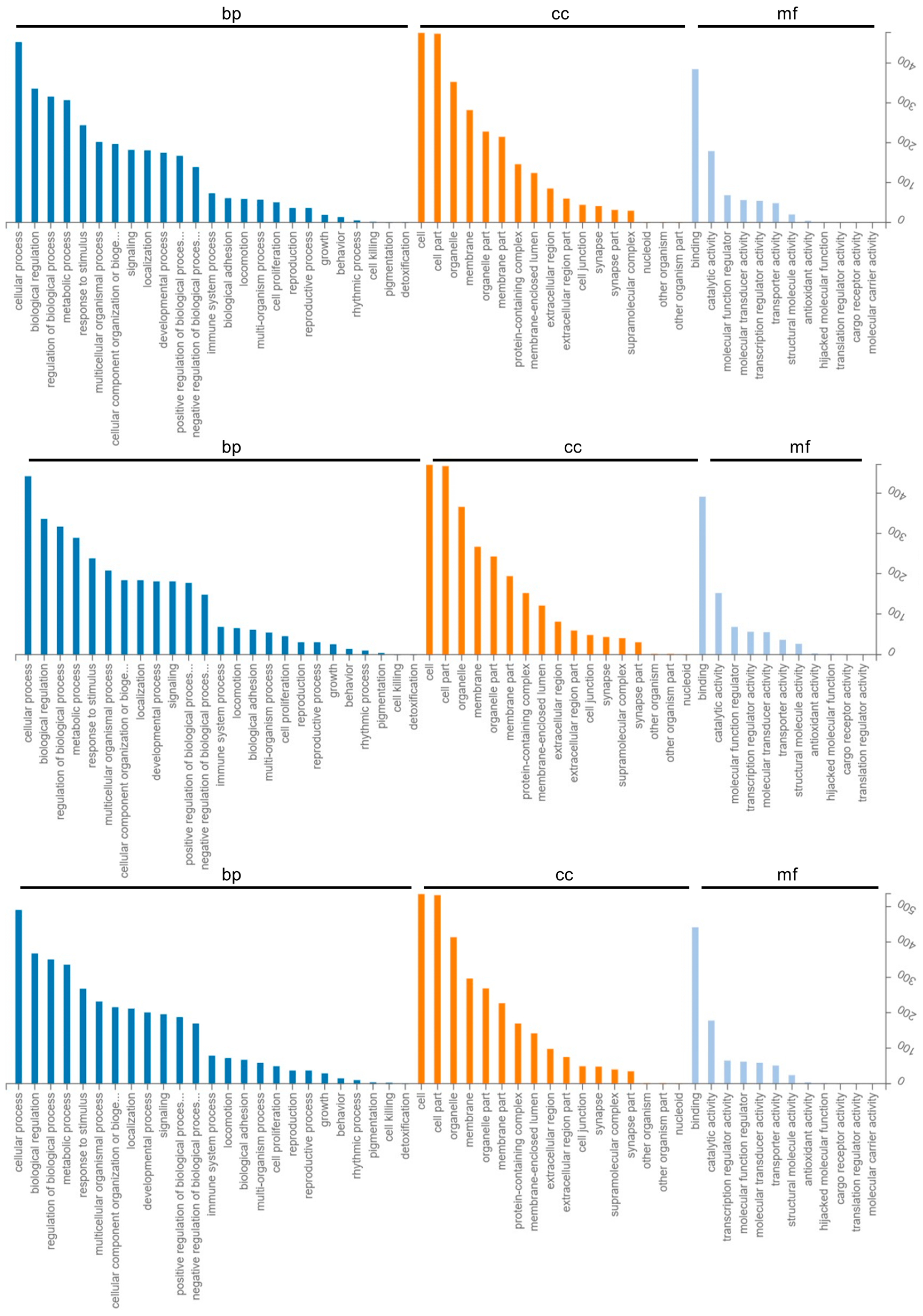
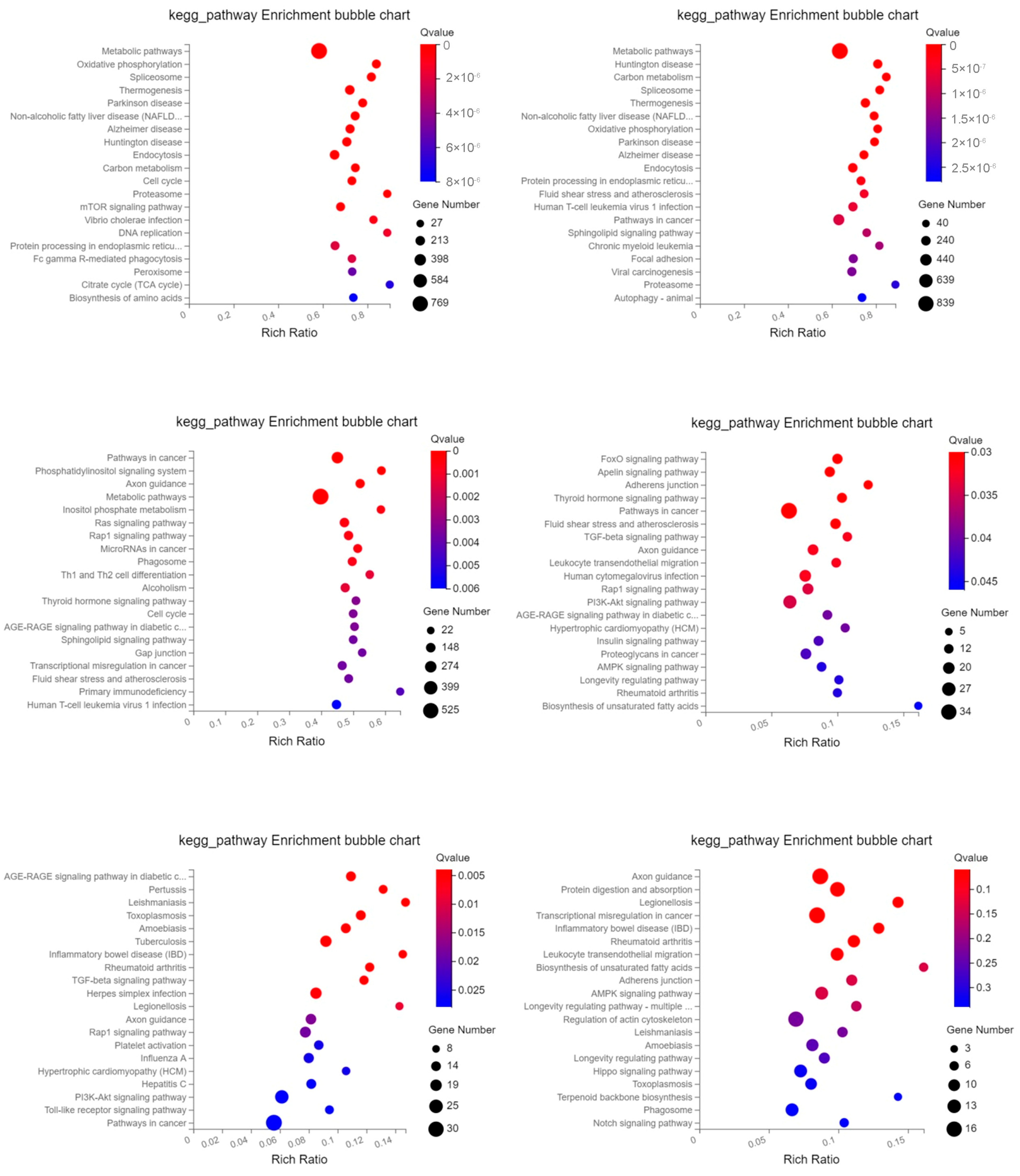
2.3. GO Enrichment and KEGG Pathway Analysis of Differentially Expressed Genes
2.4. Validation of Deep Sequencing Results by Quantitative RT-PCR
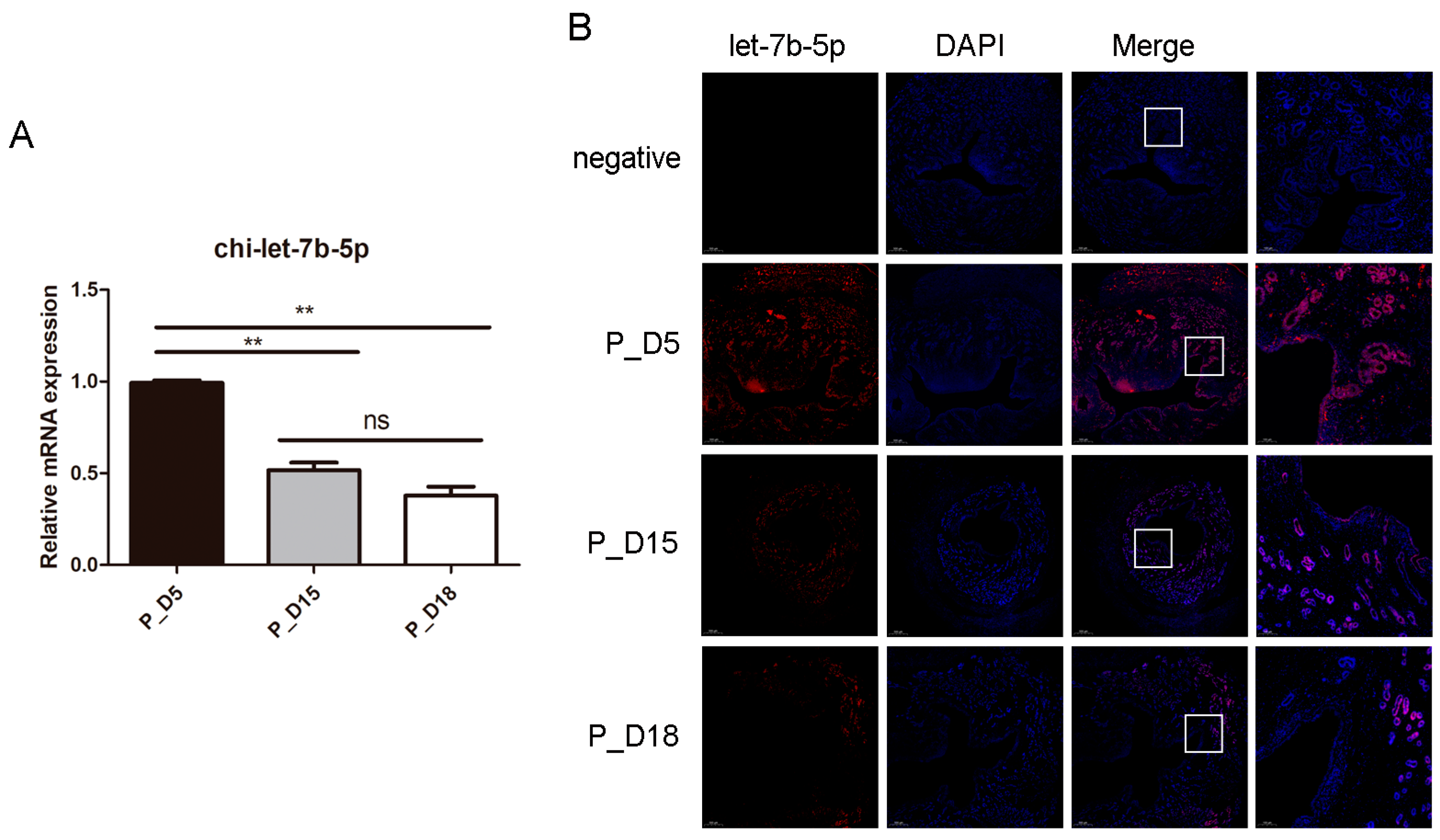
2.5. Expression and Localization of chi-let-7b-5p
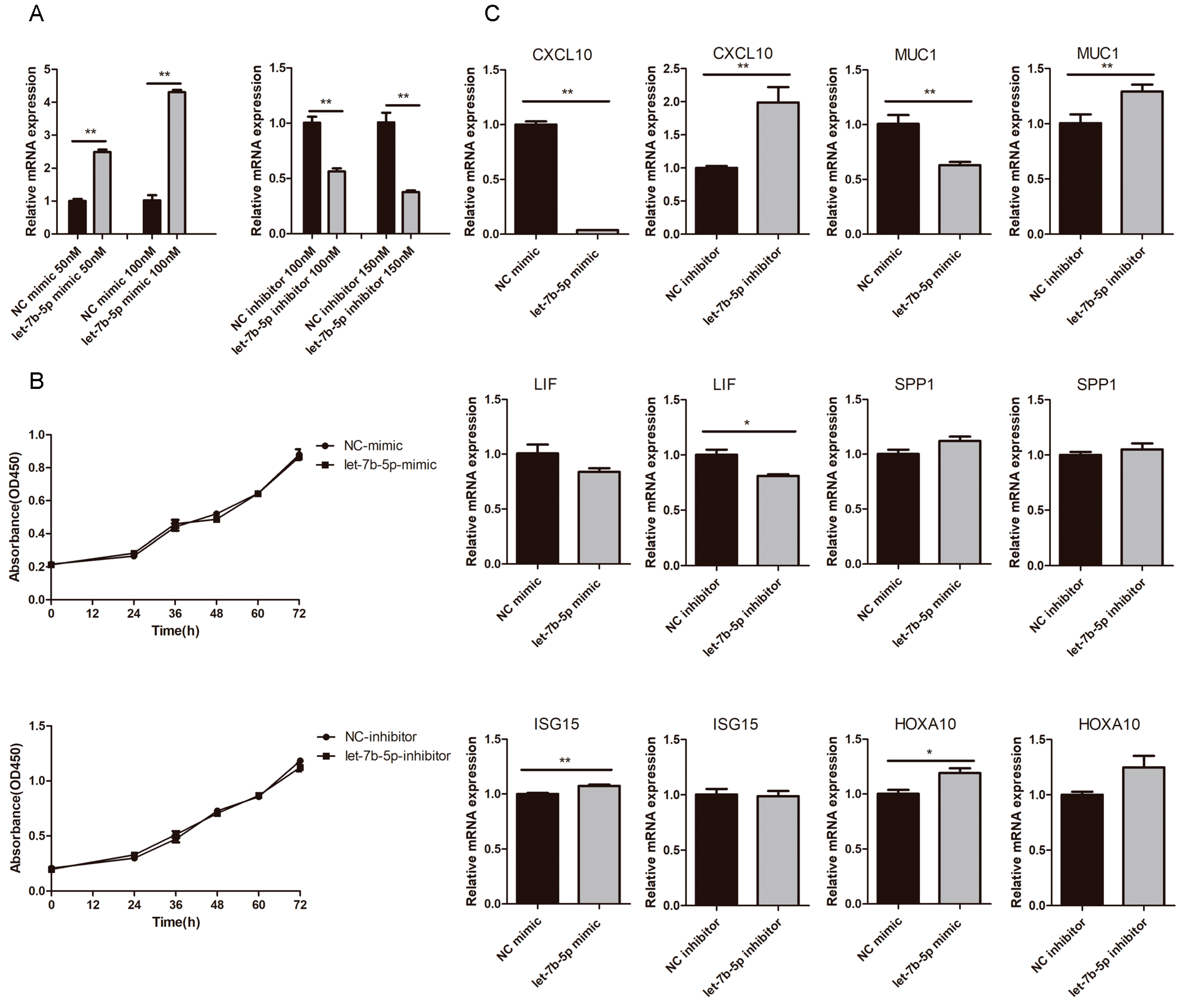
2.6. Effects of chi-let-7b-5p on Proliferation of gEEC and Expression of Genes Related to Embryo Implantation
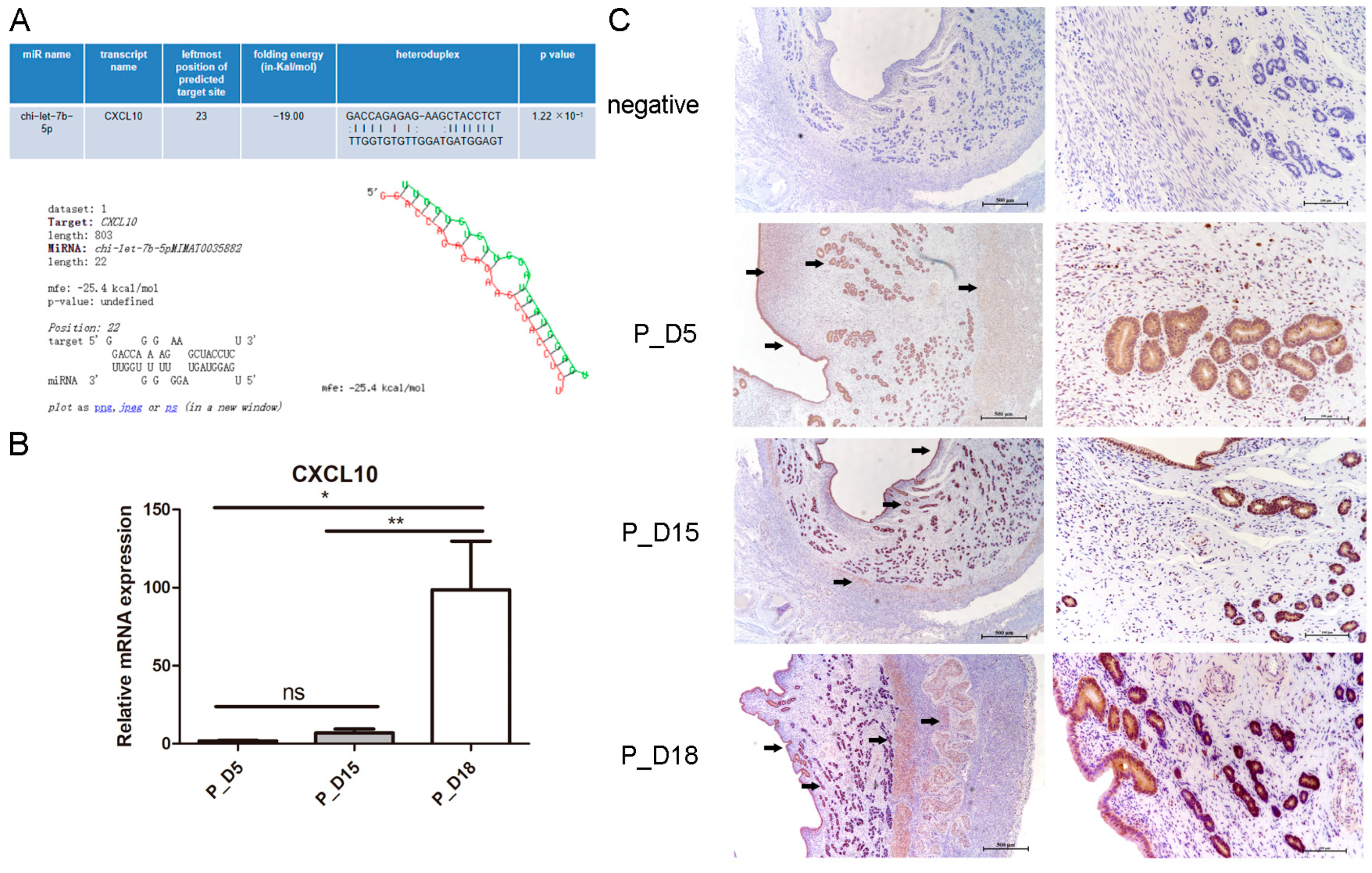
2.7. Expression and Localization of CXCL10
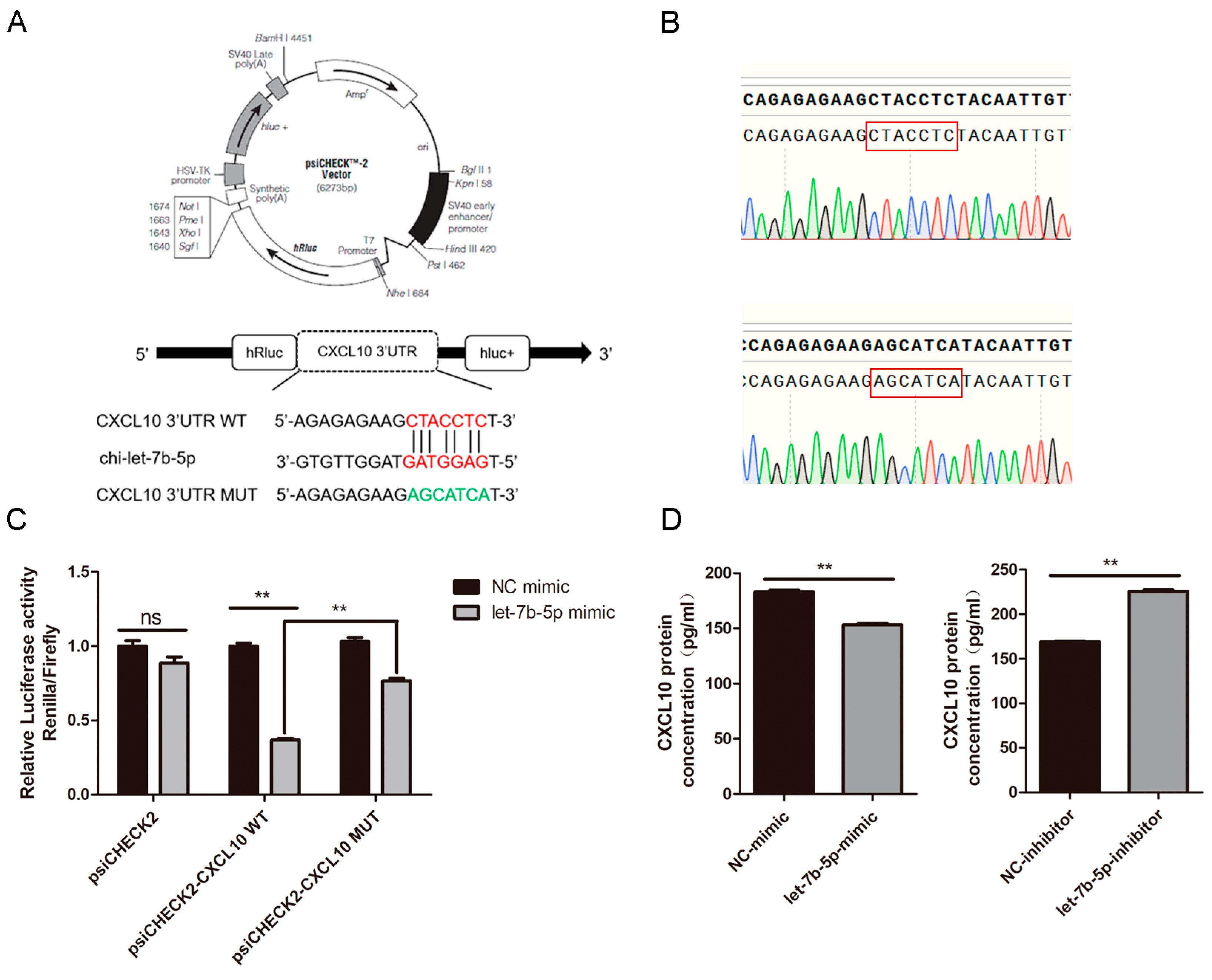
2.8. CXCL10 Is the Direct Target Gene of chi-let-7b-5p
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Exosome Isolation
4.2. Transmission Electron Microscopy
4.3. Nanoparticle Tracking Analysis
4.4. Western Blot Analysis
4.5. RNA Extraction and Library Construction
4.6. Sequencing Data Analysis
4.7. Differentially Expressed miRNAs and mRNAs
4.8. GO and KEGG Enrichment Analysis
4.9. RT-PCR
4.10. Fluorescence In Situ Hybridization
4.11. Cell Transfection and CCK8
4.12. Immunohistochemistry
4.13. Dual-Luciferase Reporter Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Urato, A.C.; Norwitz, E.R. A guide towards pre-pregnancy management of defective implantation and placentation. Best Pract. Res. Clin. Obstet. Gynaecol. 2011, 25, 367–387. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.W.; Williams, E.J.; Evans, A.C. A review of the causes of poor fertility in high milk producing dairy cows. Anim. Reprod. Sci. 2011, 123, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Salomon, C.; Guanzon, D.; Scholz-Romero, K.; Longo, S.; Correa, P.; Illanes, S.E.; Rice, G.E. Placental Exosomes as Early Biomarker of Preeclampsia: Potential Role of Exosomal MicroRNAs Across Gestation. J. Clin. Endocrinol. Metab. 2017, 102, 3182–3194. [Google Scholar] [CrossRef]
- Shao, J.; Zaro, J.; Shen, Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomed. 2020, 15, 9355–9371. [Google Scholar] [CrossRef]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef]
- Burns, G.; Brooks, K.; Wildung, M.; Navakanitworakul, R.; Christenson, L.K.; Spencer, T.E. Extracellular vesicles in luminal fluid of the ovine uterus. PLoS ONE 2014, 9, e90913. [Google Scholar] [CrossRef]
- da Silveira, J.C.; Winger, Q.A.; Bouma, G.J.; Carnevale, E.M. Effects of age on follicular fluid exosomal microRNAs and granulosa cell transforming growth factor-β signalling during follicle development in the mare. Reprod. Fertil. Dev. 2015, 27, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Nguyen, H.P.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Liu, W.M.; Cheng, R.R.; Niu, Z.R.; Chen, A.C.; Ma, M.Y.; Li, T.; Chiu, P.C.; Pang, R.T.; Lee, Y.L.; Ou, J.P.; et al. Let-7 derived from endometrial extracellular vesicles is an important inducer of embryonic diapause in mice. Sci. Adv. 2020, 6, eaaz7070. [Google Scholar] [CrossRef]
- Xia, H.F.; Jin, X.H.; Song, P.P.; Cui, Y.; Liu, C.M.; Ma, X. Temporal and spatial regulation of let-7a in the uterus during embryo implantation in the rat. J. Reprod. Dev. 2010, 56, 73–78. [Google Scholar] [CrossRef]
- Yu, D.; Liu, X.; Han, G.; Liu, Y.; Zhao, X.; Wang, D.; Bian, X.; Gu, T.; Wen, L. The let-7 family of microRNAs suppresses immune evasion in head and neck squamous cell carcinoma by promoting PD-L1 degradation. Cell Commun. Signal. 2019, 17, 173. [Google Scholar] [CrossRef]
- Boubaker, N.S.; Gurtner, A.; Trabelsi, N.; Manni, I.; Said, R.; Ayed, H.; Ksentini, M.; Karray, O.; Saadi, A.; Essid, M.A.; et al. Evaluating prognostic utility of preoperative Neutrophil to Lymphocyte Ratio and hsa-let-7g/c up-regulation in patients with urinary bladder cancer. Cancer Biomark 2020, 27, 63–73. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, B.C.; Dos Anjos, L.G.; Uno, M.; Cunha, I.W.D.; Soares, F.A.; Baiocchi, G.; Baracat, E.C.; Carvalho, K.C. Let-7 miRNA’s Expression Profile and Its Potential Prognostic Role in Uterine Leiomyosarcoma. Cells 2019, 8, 1452. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Kim, S.J.; Choi, Y.B.; Lee, B.C. Improvement of cloned embryos development by co-culturing with parthenotes: A possible role of exosomes/microvesicles for embryos paracrine communication. Cell Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Stenqvist, A.C.; Nagaeva, O.; Baranov, V.; Mincheva-Nilsson, L. Exosomes secreted by human placenta carry functional Fas ligand and TRAIL molecules and convey apoptosis in activated immune cells, suggesting exosome-mediated immune privilege of the fetus. J. Immunol. 2013, 191, 5515–5523. [Google Scholar] [CrossRef]
- Jia, L.; Zhou, X.; Huang, X.; Xu, X.; Jia, Y.; Wu, Y.; Yao, J.; Wu, Y.; Wang, K. Maternal and umbilical cord serum-derived exosomes enhance endothelial cell proliferation and migration. Faseb J. 2018, 32, 4534–4543. [Google Scholar] [CrossRef]
- Gurung, S.; Greening, D.W.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation. Mol. Hum. Reprod. 2020, 26, 510–520. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Xie, T.; Yuan, M.; Sheng, X.; Qi, X.; Xing, K.; Liu, F.; Guo, Y.; Xiao, L.; et al. Exosomes from bovine endometrial epithelial cells ensure trophoblast cell development by miR-218 targeting secreted frizzled related protein 2. J. Cell Physiol. 2021, 236, 4565–4579. [Google Scholar] [CrossRef]
- Markkandan, K.; Ahn, K.; Lee, D.J.; Kim, T.I.; Dang, C.; Hong, S.E.; Yoon, H.B.; Lim, H.J.; Hong, C.P. Profiling and identification of pregnancy-associated circulating microRNAs in dairy cattle. Genes Genom. 2018, 40, 1111–1117. [Google Scholar] [CrossRef]
- Wu, J.; Huang, H.; Huang, W.; Wang, L.; Xia, X.; Fang, X. Analysis of exosomal lncRNA, miRNA and mRNA expression profiles and ceRNA network construction in endometriosis. Epigenomics 2020, 12, 1193–1213. [Google Scholar] [CrossRef]
- Ren, G.L.; Zhu, J.; Li, J.; Meng, X.M. Noncoding RNAs in acute kidney injury. J. Cell Physiol. 2019, 234, 2266–2276. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J.; Bu, H.; Wang, H.; Zhang, Y.; Shen, Q.; Li, M.; Lu, Z.; Rong, X.; Zheng, D.; et al. Circular RNA expression alteration identifies a novel circulating biomarker in serum exosomal for detection of alcohol dependence. Addict. Biol. 2021, 26, e13031. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, X.Y.; Yang, P.W.; Yang, S.L.; Hu, D.; Zhang, H.W.; Sui, C. Activation of Uterine Smad3 Pathway Is Crucial for Embryo Implantation. Curr. Med. Sci. 2019, 39, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Riley, J.K.; Carayannopoulos, M.O.; Wyman, A.H.; Chi, M.; Ratajczak, C.K.; Moley, K.H. The PI3K/Akt pathway is present and functional in the preimplantation mouse embryo. Dev. Biol. 2005, 284, 377–386. [Google Scholar] [CrossRef]
- Riley, J.K.; Carayannopoulos, M.O.; Wyman, A.H.; Chi, M.; Moley, K.H. Phosphatidylinositol 3-kinase activity is critical for glucose metabolism and embryo survival in murine blastocysts. J. Biol. Chem. 2006, 281, 6010–6019. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Yu, Q. The PI3K/Akt signaling pathway exerts effects on the implantation of mouse embryos by regulating the expression of RhoA. Int. J. Mol. Med. 2014, 33, 1089–1096. [Google Scholar] [CrossRef]
- Ochoa-Bernal, M.A.; Fazleabas, A.T. Physiologic Events of Embryo Implantation and Decidualization in Human and Non-Human Primates. Int. J. Mol. Sci. 2020, 21, 1973. [Google Scholar] [CrossRef]
- Zhao, W.X.; Lin, J.H. Notch signaling pathway and human placenta. Int. J. Med. Sci. 2012, 9, 447–452. [Google Scholar] [CrossRef]
- Inyawilert, W.; Fu, T.Y.; Lin, C.T.; Tang, P.C. Let-7-mediated suppression of mucin 1 expression in the mouse uterus during embryo implantation. J. Reprod. Dev. 2015, 61, 138–144. [Google Scholar] [CrossRef]
- Sakumoto, R.; Iga, K.; Hayashi, K.G.; Fujii, S.; Kanahara, H.; Hosoe, M.; Furusawa, T. Gene expression of CCL8 and CXCL10 in peripheral blood leukocytes during early pregnancy in cows. J. Anim. Sci. Biotechnol. 2018, 9, 46. [Google Scholar] [CrossRef]
- Han, J.; Gu, M.J.; Yoo, I.; Choi, Y.; Jang, H.; Kim, M.; Yun, C.H.; Ka, H. Analysis of cysteine-X-cysteine motif chemokine ligands 9, 10, and 11, their receptor CXCR3, and their possible role on the recruitment of immune cells at the maternal-conceptus interface in pigs. Biol. Reprod. 2017, 97, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, J.; Yang, C.; Lee, K.F.; Lee, Y.L.; Chiu, P.C.; Zhang, Y.; Duan, Y.G.; Liu, K.; Yeung, W.S. Expression of microRNA let-7 in cleavage embryos modulates cell fate determination and formation of mouse blastocysts. Biol. Reprod. 2022, 107, 1452–1463. [Google Scholar] [CrossRef]
- Peter, M.E. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle 2009, 8, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Bronevetsky, Y.; Burt, T.D.; McCune, J.M. Lin28b Regulates Fetal Regulatory T Cell Differentiation through Modulation of TGF-β Signaling. J. Immunol. 2016, 197, 4344–4350. [Google Scholar] [CrossRef]
- McLendon, B.A.; Seo, H.; Kramer, A.C.; Burghardt, R.C.; Bazer, F.W.; Johnson, G.A. Pig conceptuses secrete interferon gamma to recruit T cells to the endometrium during the peri-implantation period. Biol. Reprod. 2020, 103, 1018–1029. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006. Chapter 3, Unit 3.22. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.; Star, R.A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal. Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef]
- Chen, T.S.; Arslan, F.; Yin, Y.; Tan, S.S.; Lai, R.C.; Choo, A.B.; Padmanabhan, J.; Lee, C.N.; de Kleijn, D.P.; Lim, S.K. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. J. Transl. Med. 2011, 9, 47. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Abdi, H. Bonferroni and Sidak corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics; Sage: Thousand Oaks, CA, USA, 2007; pp. 103–107. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wang, A.H.; Wu, Q.X.; Sheng, H.X.; Jin, Y.P. Establishment and characteristics of immortal goat endometrial epithelial cells and stromal cells with hTERT. J. Anim. Vet. Adv. 2010, 9, 2738–2747. [Google Scholar] [CrossRef]

| Name | Forward (5′ → 3′) | Reverse (5′ → 3′) |
|---|---|---|
| CXCL10 | CTGCCCACGTGTCGAGATTA | TGCCTCTTTCCGTGTTCGAG |
| MUC1 | TACTCTGCCTACCACACCCA | CTGGACTCTCAGCAGACGTG |
| LIF | TGCTGCCTTACTTGGGTGAG | GCCTTTCAAGGGCCTCTCTT |
| SPP1 | TGTTAAAGCAGGGTGGGAGAC | AGGGTGTTACCATGAAGCCAC |
| ISG15 | GACACCAGAACCCACGG | GGAAAGCAGGCACATTGA |
| HOXA10 | GTACCTTACTCGAGAGCGGC | TTGCCTGGAGCTTCATCAGG |
| GAPDH | TCTGCTGATGCCCCCATGTT | TGACCTTGCCCACGGCCTTG |
| chi-let-7b-5p | GCGCGTGAGGTAGTAGGTTGT | AGTGCAGGGTCCGAGGTATT |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| chi-let-7b-5p stem loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCAC | |
| FTH1 | CGTGATGACTGGGAGAACGG | TTGTGCAGTTCCAGTAGCGA |
| PTGS2 | TGTATCCCGCCCTTCTGGTA | CCGGCTTCTACCATGGTCTC |
| CTNNB1 | AATCAGCTGGCCTGGTTTGA | GCTTGGTTAGTGTGTCAGGC |
| ACSL4 | AACTTCGGCAGTGGACTCAC | CCGCAATCATCCATTCAGCC |
| CD24 | GCTGCTCTTACCTACGCAGA | CAGAGTACCATCGCTTGCCT |
| GPX4 | TGTGGTTTACGGATCCTGGC | CCCTTGGGCTGGACTTTCAT |
| chi-miR-199b-5p | GCTCGACGCCCAGTGTTTAGACTAT | miRNA qPCR 3′primer |
| chi-miR-136-5p | ACTCCATTTGTTTTGATGATGG | miRNA qPCR 3′primer |
| chi-miR-106b-3p | CCGCACTGTGGGTACTTGCT | miRNA qPCR 3′primer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, X.; Li, J.; Fang, H.; Yu, S.; Zhang, H.; Zhao, Y.; Zhang, L.; Wang, A.; Jin, Y.; Zhou, D. Uterine Flushing Fluid-Derived Let-7b Targets CXCL10 to Regulate Uterine Receptivity in Goats during Embryo Implantation. Int. J. Mol. Sci. 2023, 24, 2799. https://doi.org/10.3390/ijms24032799
Ning X, Li J, Fang H, Yu S, Zhang H, Zhao Y, Zhang L, Wang A, Jin Y, Zhou D. Uterine Flushing Fluid-Derived Let-7b Targets CXCL10 to Regulate Uterine Receptivity in Goats during Embryo Implantation. International Journal of Molecular Sciences. 2023; 24(3):2799. https://doi.org/10.3390/ijms24032799
Chicago/Turabian StyleNing, Xinnuan, Jie Li, Hui Fang, Siyuan Yu, Hongxia Zhang, Yanan Zhao, Lu Zhang, Aihua Wang, Yaping Jin, and Dong Zhou. 2023. "Uterine Flushing Fluid-Derived Let-7b Targets CXCL10 to Regulate Uterine Receptivity in Goats during Embryo Implantation" International Journal of Molecular Sciences 24, no. 3: 2799. https://doi.org/10.3390/ijms24032799
APA StyleNing, X., Li, J., Fang, H., Yu, S., Zhang, H., Zhao, Y., Zhang, L., Wang, A., Jin, Y., & Zhou, D. (2023). Uterine Flushing Fluid-Derived Let-7b Targets CXCL10 to Regulate Uterine Receptivity in Goats during Embryo Implantation. International Journal of Molecular Sciences, 24(3), 2799. https://doi.org/10.3390/ijms24032799





