Machine Learning Approximations to Predict Epigenetic Age Acceleration in Stroke Patients
Abstract
1. Introduction
2. Results
2.1. Descriptive Analysis
2.2. Model Training
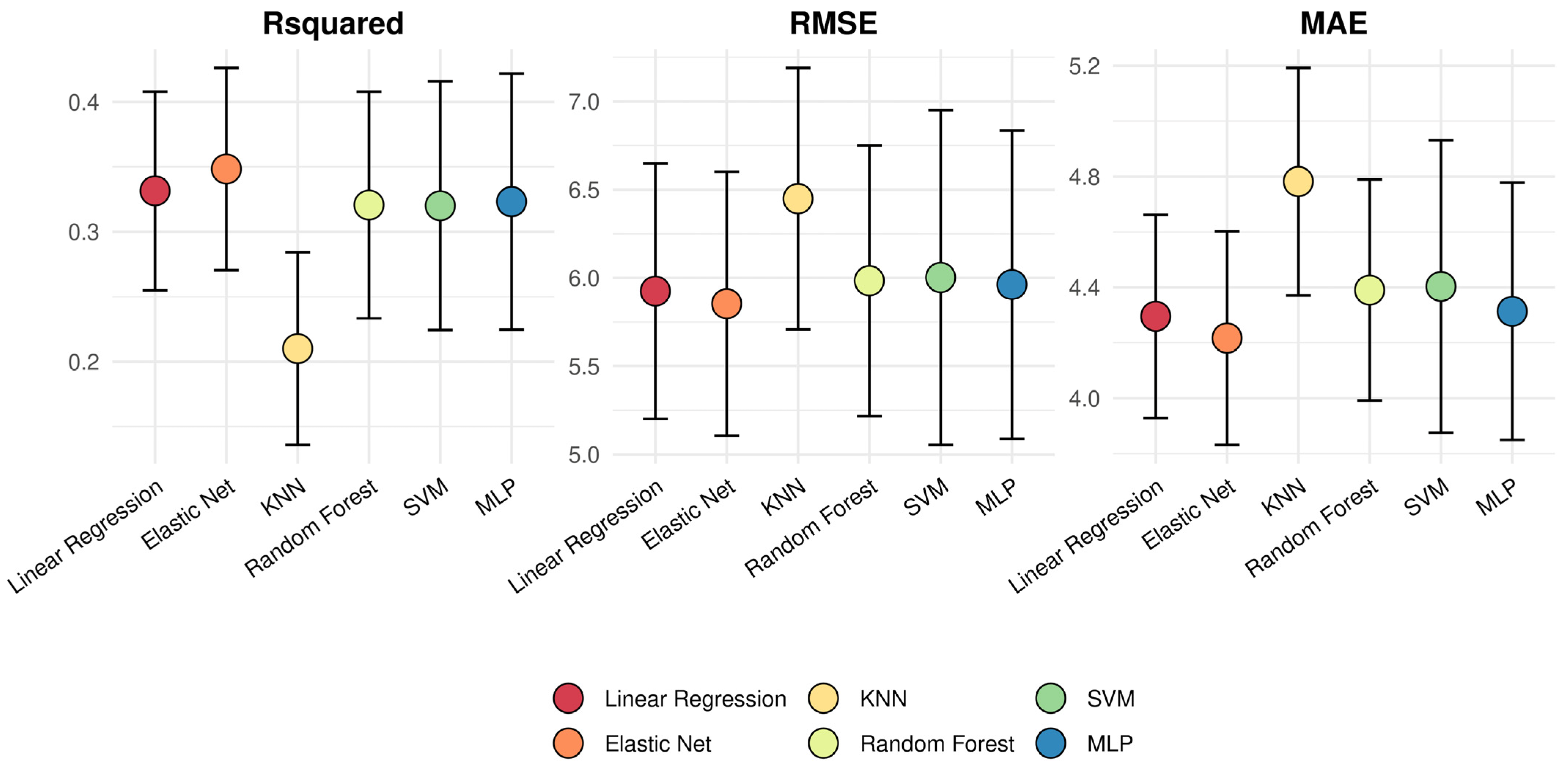
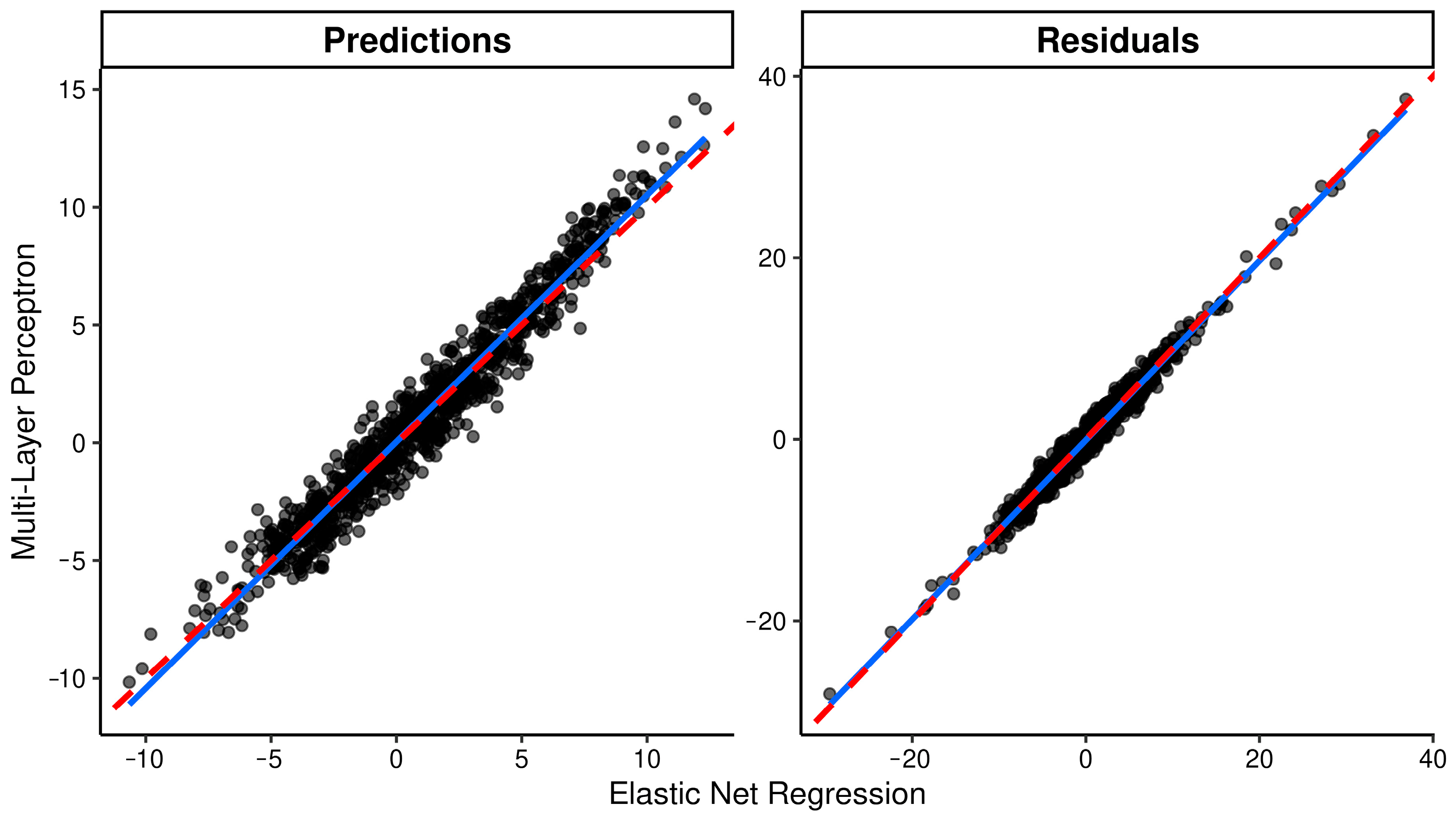
2.3. Model Evaluation
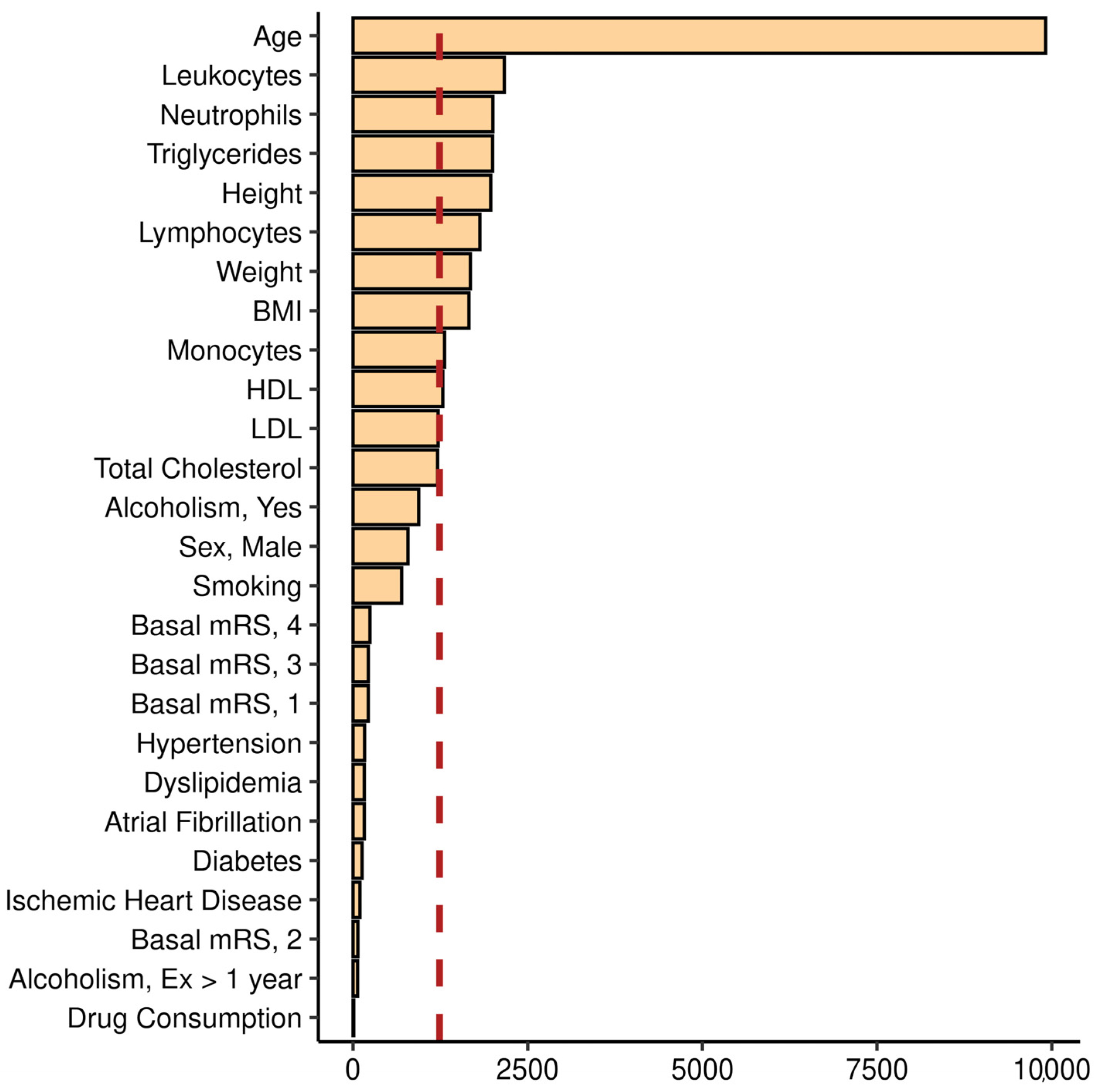
2.4. Model Interpretation
3. Discussion
4. Materials and Methods
4.1. Setting
4.2. Ethics
4.3. Clinical Variables
4.3.1. Vascular Risk Factors
4.3.2. Lifestyle and Diet
4.3.3. Target Organ Damage
4.4. Age Acceleration Estimation
4.4.1. Array-Based DNA Methylation Quantification
4.4.2. Biological Age
4.5. Statistical Analyses
4.5.1. Descriptive Statistics
4.5.2. Missing Data, Imputation, and Data Pre-Processing
4.5.3. Model Training
4.5.4. Model Evaluation
4.5.5. Model Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niccoli, T.; Partridge, L. Ageing as a Risk Factor for Disease. Curr. Biol. 2012, 22, R741–R752. [Google Scholar] [CrossRef] [PubMed]
- Christensen, K.; Doblhammer, G.; Rau, R.; Vaupel, J.W. Ageing Populations: The Challenges Ahead. Lancet 2009, 374, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Diebel, L.W.M.; Rockwood, K. Determination of Biological Age: Geriatric Assessment vs Biological Biomarkers. Curr. Oncol. Rep. 2021, 23, 104. [Google Scholar] [CrossRef]
- Baker, G.T., 3rd; Sprott, R.L. Biomarkers of Aging. Exp. Gerontol. 1988, 23, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic Modifications and Human Disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Menon, U.; Gentry-Maharaj, A.; Ramus, S.J.; Weisenberger, D.J.; Shen, H.; Campan, M.; Noushmehr, H.; Bell, C.G.; Maxwell, A.P.; et al. Age-Dependent DNA Methylation of Genes That Are Suppressed in Stem Cells Is a Hallmark of Cancer. Genome Res. 2010, 20, 440–446. [Google Scholar] [CrossRef]
- Ahuja, N.; Li, Q.; Mohan, A.L.; Baylin, S.B.; Issa, J.P. Aging and DNA Methylation in Colorectal Mucosa and Cancer. Cancer Res. 1998, 58, 5489–5494. [Google Scholar] [PubMed]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA Methylation and Healthy Human Aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Laird, P.W. Principles and Challenges of Genome-Wide DNA Methylation Analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Noroozi, R.; Ghafouri-Fard, S.; Pisarek, A.; Rudnicka, J.; Spólnicka, M.; Branicki, W.; Taheri, M.; Pośpiech, E. DNA Methylation-Based Age Clocks: From Age Prediction to Age Reversion. Ageing Res. Rev. 2021, 68, 101314. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA Methylation-Based Biomarkers and the Epigenetic Clock Theory of Ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-Wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Pelegi-Siso, D.; de Prado, P.; Ronkainen, J.; Bustamante, M.; Gonzalez, J.R. Methylclock: A Bioconductor Package to Estimate DNA Methylation Age Methylclock: A Bioconductor Package to Estimate DNA Methylation Age. Bioinformatics 2021, 37, 1759–1760. [Google Scholar] [CrossRef]
- Ryan, C.P. “Epigenetic Clocks”: Theory and Applications in Human Biology. Am. J. Hum. Biol. 2021, 33, e23488. [Google Scholar] [CrossRef]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA Methylation-Based Measures of Biological Age: Meta-Analysis Predicting Time to Death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef]
- Soriano-Tárraga, C.; Giralt-Steinhauer, E.; Mola-Caminal, M.; Ois, A.; Rodríguez-Campello, A.; Cuadrado-Godia, E.; Fernández-Cadenas, I.; Cullell, N.; Roquer, J.; Jiménez-Conde, J. Biological Age Is a Predictor of Mortality in Ischemic Stroke. Sci. Rep. 2018, 8, 4148. [Google Scholar] [CrossRef]
- Soriano-Tárraga, C.; Lazcano, U.; Jiménez-Conde, J.; Ois, A.; Cuadrado-Godia, E.; Giralt-Steinhauer, E.; Rodríguez-Campello, A.; Gomez-Gonzalez, A.; Avellaneda-Gómez, C.; Vivanco-Hidalgo, R.M.; et al. Biological Age Is a Novel Biomarker to Predict Stroke Recurrence. J. Neurol. 2021, 268, 285–292. [Google Scholar] [CrossRef]
- Soriano-Tárraga, C.; Mola-Caminal, M.; Giralt-Steinhauer, E.; Ois, A.; Rodríguez-Campello, A.; Cuadrado-Godia, E.; Gómez-González, A.; Vivanco-Hidalgo, R.M.; Fernández-Cadenas, I.; Cullell, N.; et al. Biological Age Is Better than Chronological as Predictor of 3-Month Outcome in Ischemic Stroke. Neurology 2017, 89, 830–836. [Google Scholar] [CrossRef]
- Jiménez-Balado, J.; Giralt-Steinhauer, E.; Fernández-Pérez, I.; Rey, L.; Cuadrado-Godia, E.; Ois, Á.; Rodríguez-Campello, A.; Soriano-Tárraga, C.; Lazcano, U.; Macias-Gómez, A.; et al. Epigenetic Clock Explains White Matter Hyperintensity Burden Irrespective of Chronological Age. Biology 2022, 12, 33. [Google Scholar] [CrossRef]
- Roetker, N.S.; Pankow, J.S.; Bressler, J.; Morrison, A.C.; Boerwinkle, E. Prospective Study of Epigenetic Age Acceleration and Incidence of Cardiovascular Disease Outcomes in the ARIC Study (Atherosclerosis Risk in Communities). Circ. Genom. Precis. Med. 2018, 11, e001937. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Chen, B.H.; Hernandez, D.G.; Singleton, A.B.; Ferrucci, L.; Bandinelli, S.; Salfati, E.; Manson, J.A.E.; Quach, A.; et al. Menopause Accelerates Biological Aging. Proc. Natl. Acad. Sci. USA 2016, 113, 9327–9332. [Google Scholar] [CrossRef] [PubMed]
- Breitling, L.P.; Saum, K.U.; Perna, L.; Schöttker, B.; Holleczek, B.; Brenner, H. Frailty Is Associated with the Epigenetic Clock but Not with Telomere Length in a German Cohort. Clin. Epigenet. 2016, 8, 21. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M.F. Epigenetics and the Environment: Emerging Patterns and Implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Horvath, S.; Erhart, W.; Brosch, M.; Ammerpohl, O.; von Schönfels, W.; Ahrens, M.; Heits, N.; Bell, J.T.; Tsai, P.C.; Spector, T.D.; et al. Obesity Accelerates Epigenetic Aging of Human Liver. Proc. Natl. Acad. Sci. USA 2014, 111, 15538–15543. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, G.; Gonzalez, J.R. Bayesian Neural Networks for the Optimisation of Biological Clocks in Humans. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ashiqur Rahman, S.; Giacobbi, P.; Pyles, L.; Mullett, C.; Doretto, G.; Adjeroh, D.A. Deep Learning for Biological Age Estimation. Brief. Bioinform. 2021, 22, 1767–1781. [Google Scholar] [CrossRef]
- Carter, A.; Bares, C.; Lin, L.; Reed, B.G.; Bowden, M.; Zucker, R.A.; Zhao, W.; Smith, J.A.; Becker, J.B. Sex-Specific and Generational Effects of Alcohol and Tobacco Use on Epigenetic Age Acceleration in the Michigan Longitudinal Study. Drug Alcohol Depend. Rep. 2022, 4, 100077. [Google Scholar] [CrossRef]
- Quach, A.; Levine, M.E.; Tanaka, T.; Lu, A.T.; Chen, B.H.; Ferrucci, L.; Ritz, B.; Bandinelli, S.; Neuhouser, M.L.; Beasley, J.M.; et al. Epigenetic Clock Analysis of Diet, Exercise, Education, and Lifestyle Factors. Aging 2017, 9, 419–446. [Google Scholar] [CrossRef]
- Sampathkumar, N.K.; Bravo, J.I.; Chen, Y.; Danthi, P.S.; Donahue, E.K.; Lai, R.W.; Lu, R.; Randall, L.T.; Vinson, N.; Benayoun, B.A. Widespread Sex Dimorphism in Aging and Age-Related Diseases. Hum. Genet. 2020, 139, 333–356. [Google Scholar] [CrossRef] [PubMed]
- Samaras, T.T. How Height Is Related to Our Health and Longevity: A Review. Nutr. Health 2012, 21, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Liang, M. Epigenetic Mechanisms and Hypertension. Hypertension 2018, 72, 1244–1254. [Google Scholar] [CrossRef]
- Soriano-Tárraga, C.; Jiménez-Conde, J.; Giralt-Steinhauer, E.; Mola-Caminal, M.; Vivanco-Hidalgo, R.M.; Ois, A.; Rodríguez-Campello, A.; Cuadrado-Godia, E.; Sayols-Baixeras, S.; Elosua, R.; et al. Epigenome-Wide Association Study Identifies TXNIP Gene Associated with Type 2 Diabetes Mellitus and Sustained Hyperglycemia. Hum. Mol. Genet. 2016, 25, 609–619. [Google Scholar] [CrossRef]
- Roberts, J.D.; Vittinghoff, E.; Lu, A.T.; Alonso, A.; Wang, B.; Sitlani, C.M.; Mohammadi-Shemirani, P.; Fornage, M.; Kornej, J.; Brody, J.A.; et al. Epigenetic Age and the Risk of Incident Atrial Fibrillation. Circulation 2021, 144, 1899–1911. [Google Scholar] [CrossRef]
- Sadighi Akha, A.A. Aging and the Immune System: An Overview. J. Immunol. Methods 2018, 463, 21–26. [Google Scholar] [CrossRef]
- Tsaprouni, L.G.; Yang, T.P.; Bell, J.; Dick, K.J.; Kanoni, S.; Nisbet, J.; Viñuela, A.; Grundberg, E.; Nelson, C.P.; Meduri, E.; et al. Cigarette Smoking Reduces DNA Methylation Levels at Multiple Genomic Loci but the Effect Is Partially Reversible upon Cessation. Epigenetics 2014, 9, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, Q.; Javed, R.; Zhong, J.; Gao, H.; Liang, H. Effect of Tobacco Smoking on the Epigenetic Age of Human Respiratory Organs. Clin. Epigenet. 2019, 11, 183. [Google Scholar] [CrossRef]
- Drew, L. Turning Back Time with Epigenetic Clocks. Nature 2022, 601, S20–S22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Guo, Y. Epigenetic Clock: DNA Methylation in Aging. Stem Cells Int. 2020, 2020, 1047896. [Google Scholar] [CrossRef]
- Oblak, L.; van der Zaag, J.; Higgins-Chen, A.T.; Levine, M.E.; Boks, M.P. A Systematic Review of Biological, Social and Environmental Factors Associated with Epigenetic Clock Acceleration. Ageing Res. Rev. 2021, 69, 101348. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA Methylation GrimAge Strongly Predicts Lifespan and Healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.L. Computational risk management. In Predictive Data Mining Models, 2nd ed.; Wu, D.D., Ed.; Springer: Singapore, 2020; ISBN 9789811396649. [Google Scholar]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The Process and the Challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Redman, T.C. If Your Data Is Bad, Your Machine Learning Tools Are Useless. Harvard Business Review, 2 April 2018. Available online: https://hbr.org/2018/04/if-your-data-is-bad-your-machine-learning-tools-are-useless (accessed on 26 January 2023).
- Soriano-Tárraga, C.; Giralt-Steinhauer, E.; Mola-Caminal, M.; Vivanco-Hidalgo, R.M.; Ois, A.; Rodríguez-Campello, A.; Cuadrado-Godia, E.; Sayols-Baixeras, S.; Elosua, R.; Roquer, J.; et al. Ischemic Stroke Patients Are Biologically Older than Their Chronological Age. Aging 2016, 8, 2655–2666. [Google Scholar] [CrossRef]
- Lowe, R.; Slodkowicz, G.; Goldman, N.; Rakyan, V.K. The Human Blood DNA Methylome Displays a Highly Distinctive Profile Compared with Other Somatic Tissues. Epigenetics 2015, 10, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Roquer, J.; Rodríguez-Campello, A.; Gomis, M.; Jiménez-Conde, J.; Cuadrado-Godia, E.; Vivanco, R.; Giralt, E.; Sepúlveda, M.; Pont-Sunyer, C.; Cucurella, G.; et al. Acute Stroke Unit Care and Early Neurological Deterioration in Ischemic Stroke. J. Neurol. 2008, 255, 1012–1017. [Google Scholar] [CrossRef]
- Rodríguez-Campello, A.; Jiménez-Conde, J.; Ois, Á.; Cuadrado-Godia, E.; Giralt-Steinhauer, E.; Schroeder, H.; Romeral, G.; Llop, M.; Soriano-Tárraga, C.; Garralda-Anaya, M.; et al. Dietary Habits in Patients with Ischemic Stroke: A Case-Control Study. PLoS ONE 2014, 9, e114716. [Google Scholar] [CrossRef]
- Rovira, M.-A.; Grau, M.; Castañer, O.; Covas, M.-I.; Schröder, H. Dietary Supplement Use and Health-Related Behaviors in a Mediterranean Population. J. Nutr. Educ. Behav. 2013, 45, 386–391. [Google Scholar] [CrossRef]
- Schröder, H.; Covas, M.I.; Marrugat, J.; Vila, J.; Pena, A.; Alcántara, M.; Masiá, R. Use of a Three-Day Estimated Food Record, a 72-Hour Recall and a Food-Frequency Questionnaire for Dietary Assessment in a Mediterranean Spanish Population. Clin. Nutr. 2001, 20, 429–437. [Google Scholar] [CrossRef]
- Stevens, P.E.; Levin, A. Evaluation and Management of Chronic Kidney Disease: Synopsis of the Kidney Disease: Improving Global Outcomes 2012 Clinical Practice Guideline. Ann. Intern. Med. 2013, 158, 825–830. [Google Scholar] [CrossRef]
- Ledig, C.; Heckemann, R.A.; Hammers, A.; Lopez, J.C.; Newcombe, V.F.J.; Makropoulos, A.; Lötjönen, J.; Menon, D.K.; Rueckert, D. Robust Whole-Brain Segmentation: Application to Traumatic Brain Injury. Med. Image Anal. 2015, 21, 40–58. [Google Scholar] [CrossRef] [PubMed]
- Pidsley, R.; Y Wong, C.C.; Volta, M.; Lunnon, K.; Mill, J.; Schalkwyk, L.C. A Data-Driven Approach to Preprocessing Illumina 450K Methylation Array Data. BMC Genom. 2013, 14, 293. [Google Scholar] [CrossRef] [PubMed]
- Touleimat, N.; Tost, J. Complete Pipeline for Infinium® Human Methylation 450K BeadChip Data Processing Using Subset Quantile Normalization for Accurate DNA Methylation Estimation. Epigenomics 2012, 4, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Sayols-Baixeras, S.; Lluís-Ganella, C.; Subirana, I.; Salas, L.A.; Vilahur, N.; Corella, D.; Muñoz, D.; Segura, A.; Jimenez-Conde, J.; Moran, S.; et al. Identification of a New Locus and Validation of Previously Reported Loci Showing Differential Methylation Associated with Smoking. The REGICOR Study. Epigenetics 2015, 10, 1156–1165. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Sayols-Baixeras, S.; Subirana, I.; Sentí, M.; Pérez-Fernández, S.; de Castro Moura, M.; Esteller, M.; Marrugat, J.; Elosua, R. DNA Methylation Biomarkers of Myocardial Infarction and Cardiovascular Disease. Clin. Epigenet. 2021, 13, 86. [Google Scholar] [CrossRef]
- McEwen, L.M.; Jones, M.J.; Lin, D.T.S.; Edgar, R.D.; Husquin, L.T.; MacIsaac, J.L.; Ramadori, K.E.; Morin, A.M.; Rider, C.F.; Carlsten, C.; et al. Systematic Evaluation of DNA Methylation Age Estimation with Common Preprocessing Methods and the Infinium MethylationEPIC BeadChip Array. Clin. Epigenet. 2018, 10, 123. [Google Scholar] [CrossRef]
- McCrory, C.; Fiorito, G.; McLoughlin, S.; Polidoro, S.; Cheallaigh, C.N.; Bourke, N.; Karisola, P.; Alenius, H.; Vineis, P.; Layte, R.; et al. Epigenetic Clocks and Allostatic Load Reveal Potential Sex-Specific Drivers of Biological Aging. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 495–503. [Google Scholar] [CrossRef]
- Troyanskaya, O.; Cantor, M.; Sherlock, G.; Brown, P.; Hastie, T.; Tibshirani, R.; Botstein, D.; Altman, R.B. Missing Value Estimation Methods for DNA Microarrays. Bioinformatics 2001, 17, 520–525. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Liljequist, D.; Elfving, B.; Roaldsen, K.S. Intraclass Correlation—A Discussion and Demonstration of Basic Features. PLoS ONE 2019, 14, e0219854. [Google Scholar] [CrossRef]
| Demographic Variables | |
|---|---|
| Age, years | 72.68 (±12.4) |
| Biological Age, years | 73.76 (±10.4) |
| Age Acceleration, years | 1.081 (±7.3) |
| Sex, Female | 404 (42.4%) |
| Morphometric Measurements | |
| Weight, kg | 73.3 (±13.4) |
| Height, cm | 163.7 (±9.2) |
| BMI, kg/m2 | 26.7 (±4.6) |
| Vascular Risk Factors | |
| Hypertension | 701 (73.6%) |
| Diabetes | 352 (37.0%) |
| Hyperlipidemia | 448 (47.1%) |
| Ischemic heart disease | 128 (13.4%) |
| Atrial fibrillation | 293 (30.8%) |
| Laboratory Determinations | |
| Leukocytes, u/mcL | 8917 (±2937) |
| Neutrophils, u/mcL | 6285 (±2732) |
| Lymphocytes, % | 21.8 (±9.3) |
| Monocytes, % | 6 (±2.2) |
| Total cholesterol, mg/dL | 174.2 (±41) |
| Triglycerides, mg/dL | 124.5 (±63.2) |
| HDL, mg/dL | 47.1 (±12.5) |
| LDL, mg/dL | 103.1 (±34.2) |
| Lifestyle | |
| Active smoker | 284 (29.8%) |
| Alcoholism | |
| No | 673 (70.7%) |
| Previous alcoholism > 1 year | 42 (4.41%) |
| Yes | 237 (24.9%) |
| Drug consumption | 22 (2.31%) |
| Previous Functional Status | |
| Baseline mRS | |
| 0 | 611 (64.2%) |
| 1 | 113 (11.9%) |
| 2 | 100 (10.5%) |
| 3 | 93 (9.8%) |
| 4 | 33 (3.5%) |
| 5 | 2 (0.2%) |
| Variable | Correlation® | Average Age-A (±SD) | p-Value |
|---|---|---|---|
| Demographic Variables | |||
| Age, years | −0.55 | <0.001 * | |
| Biological Age, years | 0.05 | 0.1273 | |
| Sex: | <0.001 * | ||
| Female | −1.22 (±6.6) | ||
| Male | 2.78 (±7.27) | ||
| Morphometric Measurements | |||
| Weight (kg) | 0.23 | <0.001 * | |
| Height (cm) | 0.3 | <0.001 * | |
| BMI (kg/m2) | 0.07 | 0.02 * | |
| Vascular Risk Factors | |||
| Hypertension | 0.78 | ||
| No | 1.19 (±7.23) | ||
| Yes | 1.04 (±7.28) | ||
| Diabetes | 0.952 | ||
| No | 1.07 (±7.39) | ||
| Yes | 1.1 (±7.06) | ||
| Hyperlipidemia | 0.229 | ||
| No | 0.81 (±7.60) | ||
| Yes | 1.38 (±6.86) | ||
| Ischemic heart disease | 0.01 * | ||
| No | 1.3 (±7.34) | ||
| Yes | −0.36 (±6.59) | ||
| Atrial fibrillation | <0.001 * | ||
| No | 1.98 (±7.26) | ||
| Yes | −0.93 (±6.87) | ||
| Laboratory Determinations | |||
| Leukocytes, u/mcL | 0.14 | <0.001 * | |
| Neutrophils, u/mcL | 0.13 | <0.001 * | |
| Lymphocytes, % | −0.02 | 0.517 | |
| Monocytes, % | −0.004 | 0.898 | |
| Total cholesterol, mg/dL | −0.008 | 0.803 | |
| Triglycerides, mg/dL | 0.12 | <0.001 * | |
| HDL, mg/dL | −0.15 | <0.001 * | |
| LDL, mg/dL | 0.05 | 0.0868 | |
| Lifestyle | |||
| Smoking | <0.001 * | ||
| No | −0.29 (±6.93) | ||
| Yes | 4.31 (±7.00) | ||
| Alcoholism | <0.001 * | ||
| No | −0.19 (±7.22) | ||
| Previous alcoholism > 1 year | 1.57 (±5.35) | ||
| Yes | 4.6 (±6.49) | ||
| Drug consumption | <0.001 * | ||
| No | 0.97 (±7.27) | ||
| Yes | 5.88 (±4.92) | ||
| Previous Functional Status | |||
| Baseline mRS | −0.22 | <0.001 * | |
| r | R² | RMSE | MAE | ICCC | |
|---|---|---|---|---|---|
| Linear Regression | 0.583 | 0.34 | 5.969 | 4.287 | 0.51 |
| Elastic net | 0.598 | 0.358 | 5.917 | 4.248 | 0.49 |
| K nearest-neighbors | 0.516 | 0.266 | 6.401 | 4.747 | 0.37 |
| Random Forest | 0.579 | 0.335 | 5.992 | 4.371 | 0.49 |
| Support Vector Machine | 0.584 | 0.341 | 6.083 | 4.467 | 0.44 |
| Multi-Layer Perceptron | 0.615 | 0.378 | 5.852 | 4.263 | 0.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Pérez, I.; Jiménez-Balado, J.; Lazcano, U.; Giralt-Steinhauer, E.; Rey Álvarez, L.; Cuadrado-Godia, E.; Rodríguez-Campello, A.; Macias-Gómez, A.; Suárez-Pérez, A.; Revert-Barberá, A.; et al. Machine Learning Approximations to Predict Epigenetic Age Acceleration in Stroke Patients. Int. J. Mol. Sci. 2023, 24, 2759. https://doi.org/10.3390/ijms24032759
Fernández-Pérez I, Jiménez-Balado J, Lazcano U, Giralt-Steinhauer E, Rey Álvarez L, Cuadrado-Godia E, Rodríguez-Campello A, Macias-Gómez A, Suárez-Pérez A, Revert-Barberá A, et al. Machine Learning Approximations to Predict Epigenetic Age Acceleration in Stroke Patients. International Journal of Molecular Sciences. 2023; 24(3):2759. https://doi.org/10.3390/ijms24032759
Chicago/Turabian StyleFernández-Pérez, Isabel, Joan Jiménez-Balado, Uxue Lazcano, Eva Giralt-Steinhauer, Lucía Rey Álvarez, Elisa Cuadrado-Godia, Ana Rodríguez-Campello, Adrià Macias-Gómez, Antoni Suárez-Pérez, Anna Revert-Barberá, and et al. 2023. "Machine Learning Approximations to Predict Epigenetic Age Acceleration in Stroke Patients" International Journal of Molecular Sciences 24, no. 3: 2759. https://doi.org/10.3390/ijms24032759
APA StyleFernández-Pérez, I., Jiménez-Balado, J., Lazcano, U., Giralt-Steinhauer, E., Rey Álvarez, L., Cuadrado-Godia, E., Rodríguez-Campello, A., Macias-Gómez, A., Suárez-Pérez, A., Revert-Barberá, A., Estragués-Gázquez, I., Soriano-Tarraga, C., Roquer, J., Ois, A., & Jiménez-Conde, J. (2023). Machine Learning Approximations to Predict Epigenetic Age Acceleration in Stroke Patients. International Journal of Molecular Sciences, 24(3), 2759. https://doi.org/10.3390/ijms24032759






