Recombinant Peptide Production Softens Escherichia coli Cells and Increases Their Size during C-Limited Fed-Batch Cultivation
Abstract
1. Introduction
2. Results
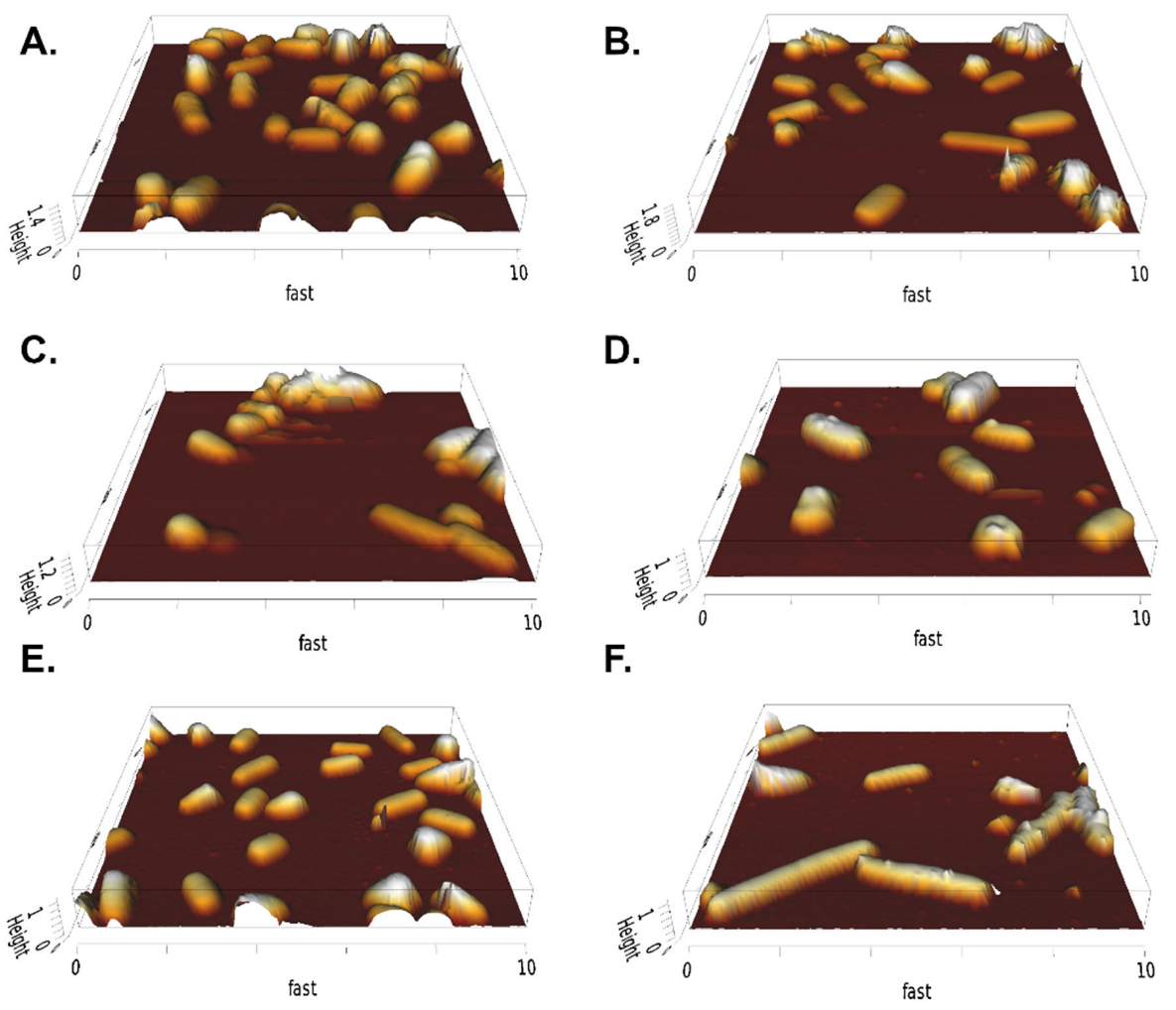
2.1. Establishment and Evaluation of AFM Sample Preparation Protocol
2.2. Bioreactor Cultivations of Producing and Non-Producing Strains
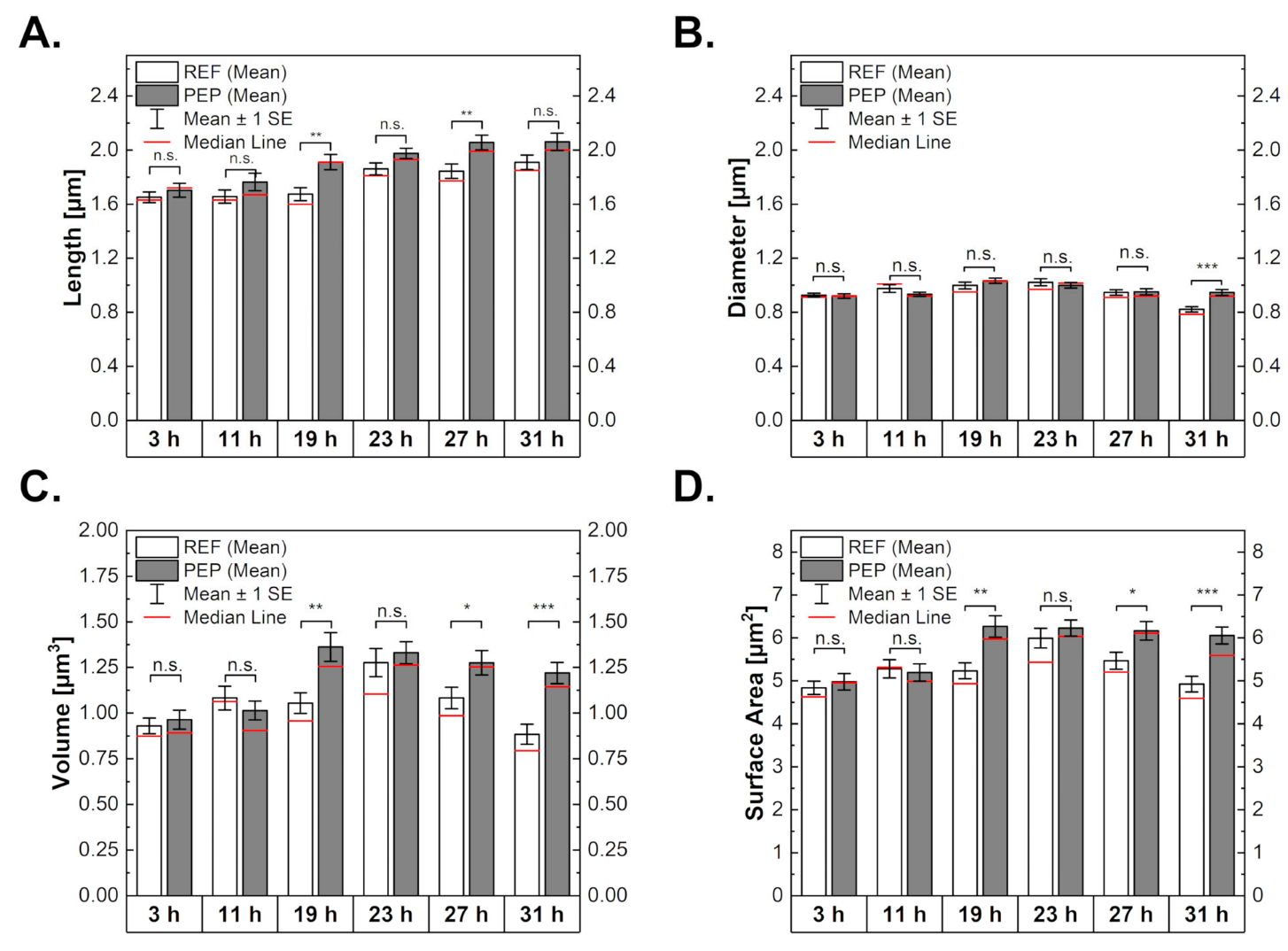
2.3. Influence of Peptide Production on Cell Morphology
2.4. Influence of Peptide Production on Mechanical Properties of Bacterial Cells
2.5. Peptide-Producing Cells Appear Less Solid-Like
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Media
4.3. Cultivations
4.4. Analytics
4.5. AFM Sample Preparation
4.6. AFM Setup
4.7. Bacterial Imaging and Morphological Property Determination
4.8. Force Spectroscopy and Creep Measurements
4.9. Data Analysis
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Persat, A.; Nadell, C.D.; Kim, M.K.; Ingremeau, F.; Siryaporn, A.; Drescher, K.; Wingreen, N.S.; Bassler, B.L.; Gitai, Z.; Stone, H.A. The Mechanical World of Bacteria. Cell 2015, 161, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Cabeen, M.T.; Jacobs-Wagner, C. Bacterial cell shape. Nat. Rev. Microbiol. 2005, 3, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Auer, G.K.; Weibel, D.B. Bacterial Cell Mechanics. Biochemistry 2017, 56, 3710–3724. [Google Scholar] [CrossRef]
- Belgrave, A.M.T.; Wolgemuth, C.W. Elasticity and Biochemistry of Growth Relate Replication Rate to Cell Length and Cross-link Density in Rod-Shaped Bacteria. Biophys. J. 2013, 104, 2607–2611. [Google Scholar] [CrossRef]
- Auer, G.K.; Lee, T.K.; Rajendram, M.; Cesar, S.; Miguel, A.; Huang, K.C.; Weibel, D.B. Mechanical Genomics Identifies Diverse Modulators of Bacterial Cell Stiffness. Cell Syst. 2016, 2, 402–411. [Google Scholar] [CrossRef]
- Ojkic, N.; Serbanescu, D.; Banerjee, S. Surface-to-volume scaling and aspect ratio preservation in rod-shaped bacteria. Elife 2019, 8, e47033. [Google Scholar] [CrossRef]
- Beveridge, T.J. The bacterial surface: General considerations towards design and function. Can. J. Microbiol. 1988, 34, 363–372. [Google Scholar] [CrossRef]
- Beveridge, T.J. Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef]
- Huang, K.C.; Mukhopadhyay, R.; Wen, B.; Gitai, Z.; Wingreen, N.S. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 19282–19287. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.R.; Billings, G.; Odermatt, P.D.; Auer, G.K.; Zhu, L.; Miguel, A.; Chang, F.; Weibel, D.B.; Theriot, J.A.; Huang, K.C. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 2018, 559, 617–621. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical properties of the bacterial outer membrane. Nat. Rev. Microbiol. 2021, 20, 236–248. [Google Scholar] [CrossRef]
- Arnoldi, M.; Fritz, M.; Bäuerlein, E.; Radmacher, M.; Sackmann, E.; Boulbitch, A. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 2000, 62, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.; Theriot, J.A.; Huang, K.C. Response of Escherichia coli growth rate to osmotic shock. Proc. Natl. Acad. Sci. USA 2014, 111, 7807–7812. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Metabolic load and heterologous gene expression. Biotechnol. Adv. 1995, 13, 247–261. [Google Scholar] [CrossRef]
- Fink, M.; Vazulka, S.; Egger, E.; Jarmer, J.; Grabherr, R.; Cserjan-Puschmann, M.; Striedner, G. Microbioreactor Cultivations of Fab-Producing Escherichia coli Reveal Genome-Integrated Systems as Suit-able for Prospective Studies on Direct Fab Expression Effects. Biotechnol. J. 2019, 14, e1800637. [Google Scholar] [CrossRef] [PubMed]
- Wegmuller, S.; Schmid, S. Recombinant Peptide Production in Microbial Cells. Curr. Org. Chem. 2014, 18, 1005–1019. [Google Scholar] [CrossRef]
- Vadillo-Rodríguez, V.; Dutcher, J.R. Viscoelasticity of the bacterial cell envelope. Soft Matter 2011, 7, 4101–4110. [Google Scholar] [CrossRef]
- Arnoldi, M.; Kacher, C.; Bäuerlein, E.; Radmacher, M.; Fritz, M. Elastic properties of the cell wall of Magnetospirillum gryphiswaldense investigated by atomic force microscopy. Appl. Phys. A 1998, 66, S613–S617. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Persat, A. Mechanomicrobiology: How bacteria sense and respond to forces. Nat. Rev. Microbiol. 2020, 18, 227–240. [Google Scholar] [CrossRef]
- Pilizota, T.; Shaevitz, J.W. Origins of Escherichia coli Growth Rate and Cell Shape Changes at High External Osmolality. Biophys. J. 2014, 107, 1962–1969. [Google Scholar] [CrossRef] [PubMed]
- Krieg, M.; Fläschner, G.; Alsteens, D.; Gaub, B.M.; Roos, W.H.; Wuite, G.J.L.; Gaub, H.E.; Gerber, C.; Dufrêne, Y.F.; Müller, D.J. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 2019, 1, 41–57. [Google Scholar] [CrossRef]
- Smolyakov, G.; Formosa-Dague, C.; Severac, C.; Duval, R.E.; Dague, E. High speed indentation measures by FV, QI and QNM introduce a new understanding of bionanomechanical experiments. Micron 2016, 85, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Vadillo-Rodriguez, V.; Beveridge, T.J.; Dutcher, J.R. Surface Viscoelasticity of Individual Gram-Negative Bacterial Cells Measured Using Atomic Force Microscopy. J. Bacteriol. 2008, 190, 4225–4232. [Google Scholar] [CrossRef]
- Vadillo-Rodriguez, V.; Schooling, S.R.; Dutcher, J.R. In Situ Characterization of Differences in the Viscoelastic Response of Individual Gram-Negative and Gram-Positive Bacterial Cells. J. Bacteriol. 2009, 191, 5518–5525. [Google Scholar] [CrossRef]
- Mathelié-Guinlet, M.; Asmar, A.T.; Collet, J.-F.; Dufrêne, Y.F. Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat. Commun. 2020, 11, 1789–1800. [Google Scholar] [CrossRef]
- Rozkov, A.; Enfors, S.-O. Analysis and Control of Proteolysis of Recombinant Proteins in Escherichia coli. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2004; Volume 89, pp. 163–195. [Google Scholar] [CrossRef]
- Li, Z.; Geffers, R.; Jain, G.; Klawonn, F.; Kökpinar, Ö.; Nimtz, M.; Schmidt-Heck, W.; Rinas, U. Transcriptional network analysis identifies key elements governing the recombinant protein production provoked reprogramming of carbon and energy metabolism in Escherichia coli BL21 (DE3). Eng. Rep. 2021, 3, e12393. [Google Scholar] [CrossRef]
- Marisch, K.; Bayer, K.; Cserjan-Puschmann, M.; Luchner, M.; Striedner, G. Evaluation of three industrial Escherichia coli strains in fed-batch cultivations during high-level SOD protein production. Microb. Cell Factories 2013, 12, 58. [Google Scholar] [CrossRef]
- Young, K.D. The Selective Value of Bacterial Shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef] [PubMed]
- Schaechter, M. A brief history of bacterial growth physiology. Front. Microbiol. 2015, 6, 289. [Google Scholar] [CrossRef] [PubMed]
- Vadia, S.; Levin, P.A. Bacterial Size: Can’t Escape the Long Arm of the Law. Curr. Biol. 2017, 27, R339–R341. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.K.; Theriot, J.A. Surface Area to Volume Ratio: A Natural Variable for Bacterial Morphogenesis. Trends Microbiol. 2018, 26, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Dewachter, L.; Verstraeten, N.; Fauvart, M.; Michiels, J. An integrative view of cell cycle control in Escherichia coli. FEMS Microbiol. Rev. 2018, 42, 116–136. [Google Scholar] [CrossRef] [PubMed]
- Whatmore, A.M.; Reed, R.H. Determination of turgor pressure in Bacillus subtilis: A possible role for K+ in turgor regulation. J. Gen. Microbiol. 1990, 136, 2521–2526. [Google Scholar] [CrossRef]
- Deng, Y.; Sun, M.; Shaevitz, J.W. Direct Measurement of Cell Wall Stress Stiffening and Turgor Pressure in Live Bacterial Cells. Phys. Rev. Lett. 2011, 107, 158101. [Google Scholar] [CrossRef]
- Li, S.H.-J.; Li, Z.; Park, J.O.; King, C.G.; Rabinowitz, J.D.; Wingreen, N.S.; Gitai, Z. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 2018, 3, 939–947. [Google Scholar] [CrossRef]
- Basan, M.; Zhu, M.; Dai, X.; Warren, M.; Sévin, D.; Wang, Y.; Hwa, T. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 2015, 11, 836. [Google Scholar] [CrossRef]
- Köppl, C.; Lingg, N.; Fischer, A.; Kröß, C.; Loibl, J.; Buchinger, W.; Schneider, R.; Jungbauer, A.; Striedner, G.; Cserjan-Puschmann, M. Fusion Tag Design Influences Soluble Recombinant Protein Production in Escherichia coli. Int. J. Mol. Sci. 2022, 23, 7678. [Google Scholar] [CrossRef]
- Mairhofer, J.; Wittwer, A.; Cserjan-Puschmann, M.; Striedner, G. Preventing T7 RNA Polymerase Read-through Transcription—A Synthetic Termination Signal Capable of Improving Bioprocess Stability. ACS Synth. Biol. 2014, 4, 265–273. [Google Scholar] [CrossRef]
- Egger, E.; Tauer, C.; Cserjan-Puschmann, M.; Grabherr, R.; Striedner, G. Fast and antibiotic free genome integration into Escherichia coli chromosome. Sci. Rep. 2020, 10, 16510. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Schimek, C.; Egger, E.; Tauer, C.; Striedner, G.; Brocard, C.; Cserjan-Puschmann, M.; Hahn, R. Extraction of recombinant periplasmic proteins under industrially relevant process conditions: Selectivity and yield strongly depend on protein titer and methodology. Biotechnol. Prog. 2020, 36, e2999. [Google Scholar] [CrossRef]
- Fink, M.; Schimek, C.; Cserjan-Puschmann, M.; Reinisch, D.; Brocard, C.; Hahn, R.; Striedner, G. Integrated process development: The key to improve Fab production in E. coli. Biotechnol. J. 2021, 16, 2000562. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.M.; Schofield, D.; Vlahopoulou, J.; Zhou, Y. Detecting cell lysis using viscosity monitoring in E. coli fermentation to prevent product loss. Biotechnol. Prog. 2016, 32, 1069–1076. [Google Scholar] [CrossRef]
- Benítez, R.; Vicente, J.B.; Toca-Herrera, J.-L. afmToolkit: An R Package for Automated AFM Force-Distance Curves Analysis. R J. 2017, 9, 291–308. [Google Scholar] [CrossRef]
- Benítez, R.; Moreno-Flores, S.; Bolós, V.J.; Toca-Herrera, J.L. A new automatic contact point detection algorithm for AFM force curves. Microsc. Res. Tech. 2013, 76, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Iturri, J.; Benitez, R.; Toca-Herrera, J.L. Measuring biomaterials mechanics with atomic force microscopy. Influence of the loading rate and applied force (pyramidal tips). Microsc. Res. Tech. 2019, 82, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Radok, J.R.M. The Contact Problem for Viscoelastic Bodies. J. Appl. Mech. 1960, 27, 438–444. [Google Scholar] [CrossRef]
- Ting, T.C.T. The Contact Stresses between a Rigid Indenter and a Viscoelastic Half-Space. J. Appl. Mech. 1966, 33, 845–854. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Okajima, T.; Raman, A. Measuring viscoelasticity of soft biological samples using atomic force microscopy. Soft Matter 2019, 16, 64–81. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Kotova, S.L.; Khlebnikova, T.M.; Timashev, P.S. A time-shift correction for extraction of viscoelastic parameters from ramp-hold AFM experiments. Jpn. J. Appl. Phys. 2021, 60, SE1002. [Google Scholar] [CrossRef]
- Mainardi, F.; Spada, G. Creep, relaxation and viscosity properties for basic fractional models in rheology. Eur. Phys. J. Spec. Top. 2011, 193, 133–160. [Google Scholar] [CrossRef]
- Bonfanti, A.; Kaplan, J.L.; Charras, G.; Kabla, A.J. Fractional viscoelastic models for power-law materials. Soft Matter 2020, 16, 6002–6020. [Google Scholar] [CrossRef]
- Vadillo-Rodriguez, V.; Dutcher, J.R. Dynamic viscoelastic behavior of individual Gram-negative bacterial cells. Soft Matter 2009, 5, 5012–5019. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, A.; Gibisch, M.; Tyrakowski, D.; Cserjan-Puschmann, M.; Toca-Herrera, J.L.; Striedner, G. Recombinant Peptide Production Softens Escherichia coli Cells and Increases Their Size during C-Limited Fed-Batch Cultivation. Int. J. Mol. Sci. 2023, 24, 2641. https://doi.org/10.3390/ijms24032641
Weber A, Gibisch M, Tyrakowski D, Cserjan-Puschmann M, Toca-Herrera JL, Striedner G. Recombinant Peptide Production Softens Escherichia coli Cells and Increases Their Size during C-Limited Fed-Batch Cultivation. International Journal of Molecular Sciences. 2023; 24(3):2641. https://doi.org/10.3390/ijms24032641
Chicago/Turabian StyleWeber, Andreas, Martin Gibisch, Daniel Tyrakowski, Monika Cserjan-Puschmann, José L. Toca-Herrera, and Gerald Striedner. 2023. "Recombinant Peptide Production Softens Escherichia coli Cells and Increases Their Size during C-Limited Fed-Batch Cultivation" International Journal of Molecular Sciences 24, no. 3: 2641. https://doi.org/10.3390/ijms24032641
APA StyleWeber, A., Gibisch, M., Tyrakowski, D., Cserjan-Puschmann, M., Toca-Herrera, J. L., & Striedner, G. (2023). Recombinant Peptide Production Softens Escherichia coli Cells and Increases Their Size during C-Limited Fed-Batch Cultivation. International Journal of Molecular Sciences, 24(3), 2641. https://doi.org/10.3390/ijms24032641









