Characterization of a Neutral Sphingomyelinase Activity in Human Serum and Plasma
Abstract
1. Introduction
2. Results
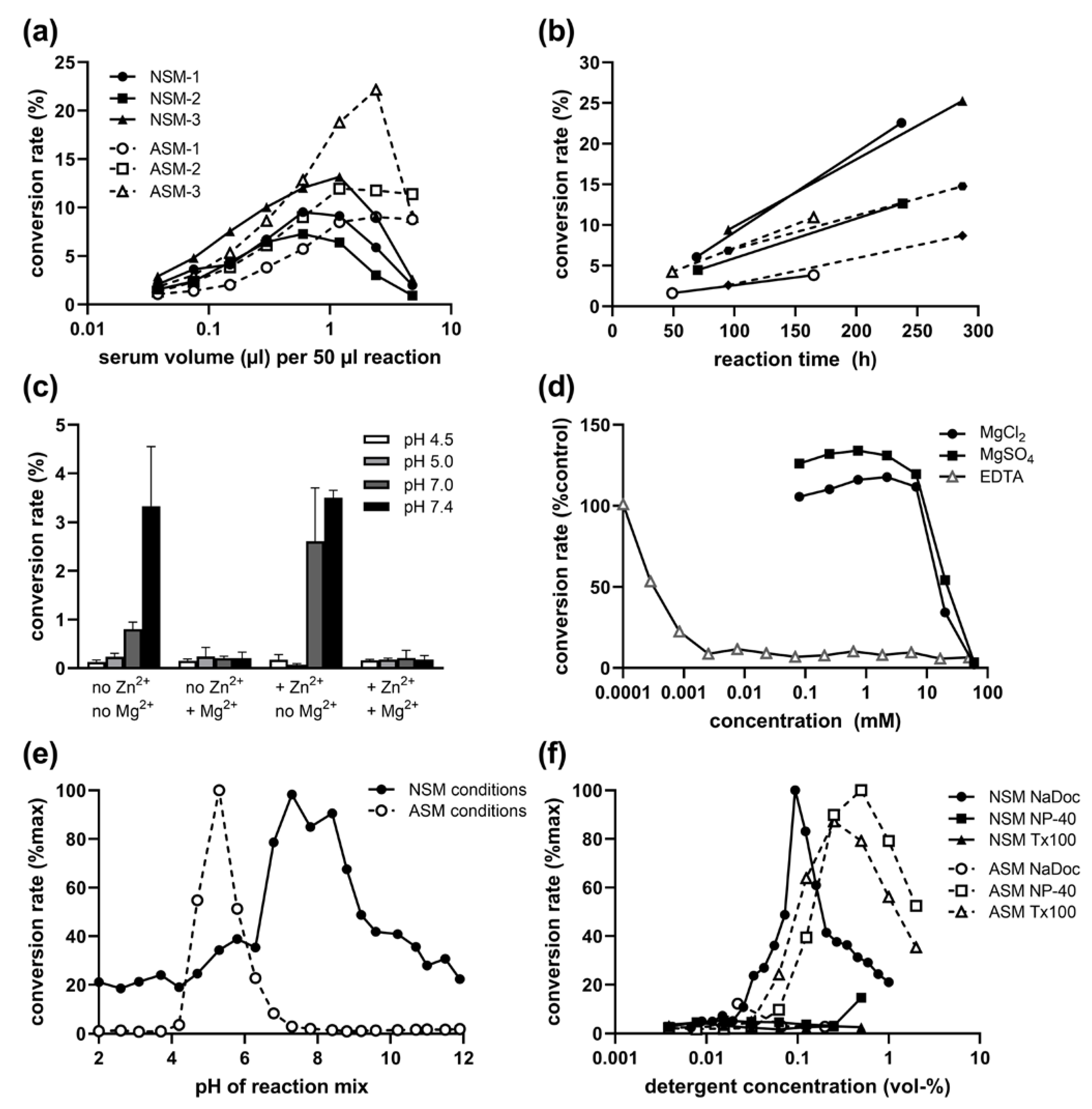
2.1. Presence of NSM Activity in Human Serum: Volume and Time Dependence
2.2. Inhibitory Effect of Mg2+ on Serum NSM Activity
2.3. pH and Detergent Profile of Serum NSM Activity
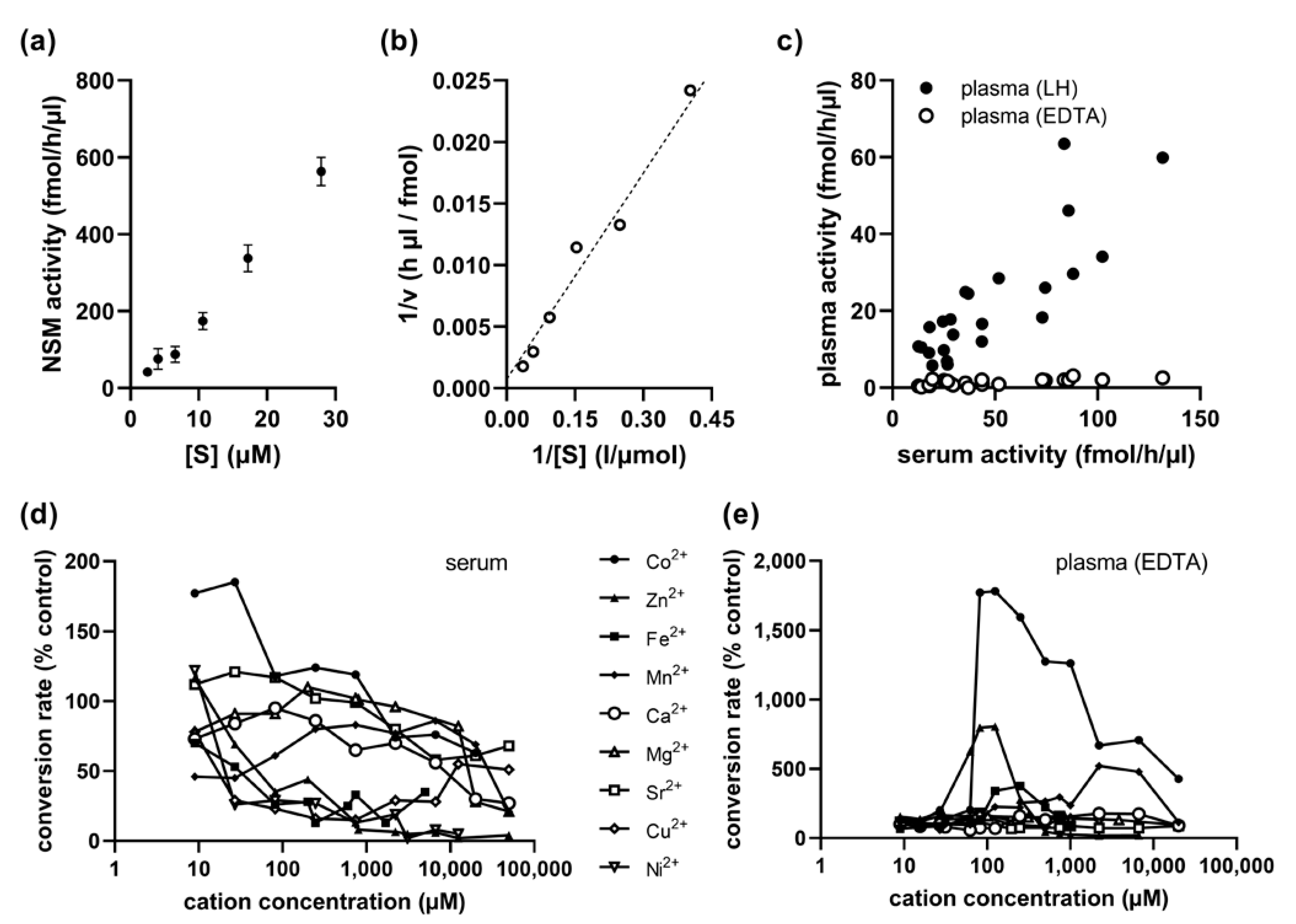
2.4. Kinetics of Serum NSM Activity
2.5. Cobalt Cation Dependence of NSM in EDTA-Anticoagulated Plasma
2.6. No Association of Serum NSM with Sex, Age, or ASM Activity
2.7. Very Low to Undetectable Serum NSM Activities in Rodents
3. Discussion
4. Materials and Methods
4.1. Blood Samples
4.2. ASM and NSM Activity Assay
4.3. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muhle, C.; Bilbao Canalejas, R.D.; Kornhuber, J. Sphingomyelin Synthases in Neuropsychiatric Health and Disease. Neurosignals 2019, 27, 54–76. [Google Scholar] [CrossRef] [PubMed]
- van Blitterswijk, W.J.; van der Luit, A.H.; Veldman, R.J.; Verheij, M.; Borst, J. Ceramide: Second messenger or modulator of membrane structure and dynamics? Biochem. J. 2003, 369, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. The Ceramide-centric universe of lipid-mediated cell regulation: Stress encounters of the lipid kind. J. Biol. Chem. 2002, 277, 25847–25850. [Google Scholar] [CrossRef] [PubMed]
- Kornhuber, J.; Rhein, C.; Muller, C.P.; Muhle, C. Secretory sphingomyelinase in health and disease. Biol. Chem. 2015, 396, 707–736. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.D. Alkaline sphingomyelinase: An old enzyme with novel implications. Biochim. Biophys Acta 2006, 1761, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Snook, C.F.; Tani, M.; Matmati, N.; Marchesini, N.; Hannun, Y.A. The extended family of neutral sphingomyelinases. Biochemistry 2006, 45, 11247–11256. [Google Scholar] [CrossRef]
- Wu, B.X.; Rajagopalan, V.; Roddy, P.L.; Clarke, C.J.; Hannun, Y.A. Identification and characterization of murine mitochondria-associated neutral sphingomyelinase (MA-nSMase), the mammalian sphingomyelin phosphodiesterase 5. J. Biol. Chem. 2010, 285, 17993–18002. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Jin, S.; Tan, F.; Xu, Y.; Lu, Y.; Wu, T. Physiological functions and therapeutic applications of neutral sphingomyelinase and acid sphingomyelinase. Biomed Pharm. 2021, 139, 111610. [Google Scholar] [CrossRef] [PubMed]
- Sawai, H.; Domae, N.; Nagan, N.; Hannun, Y.A. Function of the cloned putative neutral sphingomyelinase as lyso-platelet activating factor-phospholipase C. J. Biol. Chem. 1999, 274, 38131–38139. [Google Scholar] [CrossRef]
- Zumbansen, M.; Stoffel, W. Neutral sphingomyelinase 1 deficiency in the mouse causes no lipid storage disease. Mol. Cell Biol. 2002, 22, 3633–3638. [Google Scholar] [CrossRef]
- Marchesini, N.; Luberto, C.; Hannun, Y.A. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J. Biol. Chem. 2003, 278, 13775–13783. [Google Scholar] [CrossRef]
- Stoffel, W.; Jenke, B.; Block, B.; Zumbansen, M.; Koebke, J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc. Natl. Acad Sci. USA 2005, 102, 4554–4559. [Google Scholar] [CrossRef] [PubMed]
- Krut, O.; Wiegmann, K.; Kashkar, H.; Yazdanpanah, B.; Kronke, M. Novel tumor necrosis factor-responsive mammalian neutral sphingomyelinase-3 is a C-tail-anchored protein. J. Biol. Chem. 2006, 281, 13784–13793. [Google Scholar] [CrossRef] [PubMed]
- Muhle, C.; Amova, V.; Biermann, T.; Bayerlein, K.; Richter-Schmidinger, T.; Kraus, T.; Reichel, M.; Gulbins, E.; Kornhuber, J. Sex-dependent decrease of sphingomyelinase activity during alcohol withdrawal treatment. Cell Physiol. Biochem. 2014, 34, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Muhle, C.; Weinland, C.; Gulbins, E.; Lenz, B.; Kornhuber, J. Peripheral Acid Sphingomyelinase Activity Is Associated with Biomarkers and Phenotypes of Alcohol Use and Dependence in Patients and Healthy Controls. Int. J. Mol. Sci. 2018, 19, 4028. [Google Scholar] [CrossRef] [PubMed]
- Muhle, C.; Wagner, C.J.; Farber, K.; Richter-Schmidinger, T.; Gulbins, E.; Lenz, B.; Kornhuber, J. Secretory Acid Sphingomyelinase in the Serum of Medicated Patients Predicts the Prospective Course of Depression. J. Clin. Med. 2019, 8, 846. [Google Scholar] [CrossRef]
- Muhle, C.; Huttner, H.B.; Walter, S.; Reichel, M.; Canneva, F.; Lewczuk, P.; Gulbins, E.; Kornhuber, J. Characterization of acid sphingomyelinase activity in human cerebrospinal fluid. PLoS ONE 2013, 8, e62912. [Google Scholar] [CrossRef] [PubMed]
- Muhle, C.; Reichel, M.; Gulbins, E.; Kornhuber, J. Sphingolipids in psychiatric disorders and pain syndromes. Handb. Exp. Pharmacol. 2013, 216, 431–456. [Google Scholar] [CrossRef]
- Pavlic, A.; Bahram Sangani, N.; Kerins, J.; Nicolaes, G.; Schurgers, L.; Reutelingsperger, C. Vascular Smooth Muscle Cell Neutral Sphingomyelinase 2 in the Release of Exosomes and Vascular Calcification. Int. J. Mol. Sci. 2022, 23, 9178. [Google Scholar] [CrossRef]
- Sindhu, S.; Leung, Y.H.; Arefanian, H.; Madiraju, S.R.M.; Al-Mulla, F.; Ahmad, R.; Prentki, M. Neutral sphingomyelinase-2 and cardiometabolic diseases. Obes. Rev. 2021, 22, e13248. [Google Scholar] [CrossRef]
- Zoicas, I.; Muhle, C.; Schmidtner, A.K.; Gulbins, E.; Neumann, I.D.; Kornhuber, J. Anxiety and Depression Are Related to Higher Activity of Sphingolipid Metabolizing Enzymes in the Rat Brain. Cells 2020, 9, 1239. [Google Scholar] [CrossRef]
- Schumacher, F.; Carpinteiro, A.; Edwards, M.J.; Wilson, G.C.; Keitsch, S.; Soddemann, M.; Wilker, B.; Kleuser, B.; Becker, K.A.; Muller, C.P.; et al. Stress induces major depressive disorder by a neutral sphingomyelinase 2-mediated accumulation of ceramide-enriched exosomes in the blood plasma. J. Mol. Med. (Berl) 2022, 100, 1493–1508. [Google Scholar] [CrossRef] [PubMed]
- Kalinichenko, L.S.; Muhle, C.; Jia, T.; Anderheiden, F.; Datz, M.; Eberle, A.L.; Eulenburg, V.; Granzow, J.; Hofer, M.; Hohenschild, J.; et al. Adult alcohol drinking and emotional tone are mediated by neutral sphingomyelinase during development in males. Cereb. Cortex. 2022, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kalinichenko, L.S.; Muhle, C.; Jia, T.; Anderheiden, F.; Datz, M.; Eberle, A.L.; Eulenburg, V.; Granzow, J.; Hofer, M.; Hohenschild, J.; et al. Neutral sphingomyelinase mediates the co-morbidity trias of alcohol abuse, major depression and bone defects. Mol. Psychiatry 2021, 26, 7403–7416. [Google Scholar] [CrossRef] [PubMed]
- Mah, M.; Febbraio, M.; Turpin-Nolan, S. Circulating Ceramides- Are Origins Important for Sphingolipid Biomarkers and Treatments? Front. Endocrinol. (Lausanne) 2021, 12, 684448. [Google Scholar] [CrossRef] [PubMed]
- Airola, M.V.; Hannun, Y. Sphingolipid metabolism and neutral sphingomyelinases. Handb. Exp. Pharmacol. 2013, 215, 57–76. [Google Scholar] [CrossRef]
- Hofmann, K.; Tomiuk, S.; Wolff, G.; Stoffel, W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc. Natl. Acad. Sci. USA 2000, 97, 5895–5900. [Google Scholar] [CrossRef]
- Cheng, Y.; Nilsson, A.; Tomquist, E.; Duan, R.D. Purification, characterization, and expression of rat intestinal alkaline sphingomyelinase. J. Lipid. Res. 2002, 43, 316–324. [Google Scholar] [CrossRef]
- Duan, R.D.; Nilsson, A. Purification of a newly identified alkaline sphingomyelinase in human bile and effects of bile salts and phosphatidylcholine on enzyme activity. Hepatology 1997, 26, 823–830. [Google Scholar] [CrossRef]
- Duan, R.D.; Bergman, T.; Xu, N.; Wu, J.; Cheng, Y.; Duan, J.; Nelander, S.; Palmberg, C.; Nilsson, A. Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J. Biol. Chem. 2003, 278, 38528–38536. [Google Scholar] [CrossRef]
- Duan, R.D.; Hertervig, E.; Nyberg, L.; Hauge, T.; Sternby, B.; Lillienau, J.; Farooqi, A.; Nilsson, A. Distribution of alkaline sphingomyelinase activity in human beings and animals. Tissue and species differences. Dig. Dis. Sci. 1996, 41, 1801–1806. [Google Scholar] [CrossRef]
- Duan, R.D.; Nyberg, L.; Nilsson, A. Alkaline sphingomyelinase activity in rat gastrointestinal tract: Distribution and characteristics. Biochim. Biophys Acta. 1995, 1259, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, F.; Nilsson, A.; Duan, R.D. Pancreatic trypsin cleaves intestinal alkaline sphingomyelinase from mucosa and enhances the sphingomyelinase activity. Am. J. Physiol. Gastrointest Liver Physiol. 2004, 287, G967–G973. [Google Scholar] [CrossRef] [PubMed]
- Di Marzio, L.; Di Leo, A.; Cinque, B.; Fanini, D.; Agnifili, A.; Berloco, P.; Linsalata, M.; Lorusso, D.; Barone, M.; De Simone, C.; et al. Detection of alkaline sphingomyelinase activity in human stool: Proposed role as a new diagnostic and prognostic marker of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, A.; Liu, F.; Illes, K.; Nagar, B. Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition. J. Biol. Chem. 2017, 292, 7087–7094. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Ghosh, N. Neutral sphingomyelinase from human urine. Purification and preparation of monospecific antibodies. J. Biol. Chem. 1989, 264, 12554–12561. [Google Scholar] [CrossRef]
- Hirshfeld, D.; Loyter, A. Sphingomyelinase of chicken erythrocyte membranes. Arch. Biochem. Biophys 1975, 167, 186–192. [Google Scholar] [CrossRef]
- Sawai, H.; Okamoto, Y.; Luberto, C.; Mao, C.; Bielawska, A.; Domae, N.; Hannun, Y.A. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 39793–39798. [Google Scholar] [CrossRef]
- Okazaki, T.; Bielawska, A.; Domae, N.; Bell, R.M.; Hannun, Y.A. Characteristics and partial purification of a novel cytosolic, magnesium-independent, neutral sphingomyelinase activated in the early signal transduction of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J. Biol. Chem. 1994, 269, 4070–4077. [Google Scholar] [CrossRef]
- Ghosh, N.; Sabbadini, R.; Chatterjee, S. Identification, partial purification, and localization of a neutral sphingomyelinase in rabbit skeletal muscle: Neutral sphingomyelinase in skeletal muscle. Mol. Cell Biochem. 1998, 189, 161–168. [Google Scholar] [CrossRef]
- Ago, H.; Oda, M.; Takahashi, M.; Tsuge, H.; Ochi, S.; Katunuma, N.; Miyano, M.; Sakurai, J. Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus. J. Biol. Chem. 2006, 281, 16157–16167. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Itoh, H.; Yoshida, A.; Higashi, S.; Ikezawa, H.; Ikeda, K. Activation of sphingomyelinase from Bacillus cereus by Zn2+ hitherto accepted as a strong inhibitor. Arch. Biochem. Biophys 2005, 436, 227–236. [Google Scholar] [CrossRef]
- Carre, J.B.; Morand, O.; Homayoun, P.; Roux, F.; Bourre, J.M.; Baumann, N. Purified rat brain microvessels exhibit both acid and neutral sphingomyelinase activities. J. Neurochem. 1989, 52, 1294–1299. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Hannun, Y.A. Inhibition of the neutral magnesium-dependent sphingomyelinase by glutathione. J. Biol. Chem. 1997, 272, 16281–16287. [Google Scholar] [CrossRef] [PubMed]
- Luberto, C.; Hassler, D.F.; Signorelli, P.; Okamoto, Y.; Sawai, H.; Boros, E.; Hazen-Martin, D.J.; Obeid, L.M.; Hannun, Y.A.; Smith, G.K. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem. 2002, 277, 41128–41139. [Google Scholar] [CrossRef]
- Rojas, C.; Barnaeva, E.; Thomas, A.G.; Hu, X.; Southall, N.; Marugan, J.; Chaudhuri, A.D.; Yoo, S.W.; Hin, N.; Stepanek, O.; et al. DPTIP, a newly identified potent brain penetrant neutral sphingomyelinase 2 inhibitor, regulates astrocyte-peripheral immune communication following brain inflammation. Sci. Rep. 2018, 8, 17715. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, S.H.; Ahn, K.H.; Kim, S.K.; Choi, J.M.; Ji, J.E.; Won, J.H.; Park, Y.H.; Lim, C.; Kim, S.; et al. Identification and evaluation of neutral sphingomyelinase 2 inhibitors. Arch. Pharm. Res. 2011, 34, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Hannun, Y.A. Neutral sphingomyelinases and nSMase2: Bridging the gaps. Biochim. Biophys Acta. 2006, 1758, 1893–1901. [Google Scholar] [CrossRef]
- Rhein, C.; Tripal, P.; Seebahn, A.; Konrad, A.; Kramer, M.; Nagel, C.; Kemper, J.; Bode, J.; Muhle, C.; Gulbins, E.; et al. Functional implications of novel human acid sphingomyelinase splice variants. PLoS ONE 2012, 7, e35467. [Google Scholar] [CrossRef]
- Rhein, C.; Reichel, M.; Kramer, M.; Rotter, A.; Lenz, B.; Muhle, C.; Gulbins, E.; Kornhuber, J. Alternative splicing of SMPD1 coding for acid sphingomyelinase in major depression. J. Affect Disord 2017, 209, 10–15. [Google Scholar] [CrossRef]
- Rhein, C.; Zoicas, I.; Marx, L.M.; Zeitler, S.; Hepp, T.; von Zimmermann, C.; Muhle, C.; Richter-Schmidinger, T.; Lenz, B.; Erim, Y.; et al. mRNA Expression of SMPD1 Encoding Acid Sphingomyelinase Decreases upon Antidepressant Treatment. Int. J. Mol. Sci. 2021, 22, 5700. [Google Scholar] [CrossRef] [PubMed]
- Lenz, B.; Muhle, C.; Braun, B.; Weinland, C.; Bouna-Pyrrou, P.; Behrens, J.; Kubis, S.; Mikolaiczik, K.; Muschler, M.R.; Saigali, S.; et al. Prenatal and adult androgen activities in alcohol dependence. Acta Psychiatr. Scand. 2017, 136, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Muhle, C.; Kornhuber, J. Assay to measure sphingomyelinase and ceramidase activities efficiently and safely. J. Chromatogr. A 2017, 1481, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Carmody, W.R. An easily prepared wide range buffer series. J Chem Educ 1961, 38, 559–560. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mühle, C.; Kornhuber, J. Characterization of a Neutral Sphingomyelinase Activity in Human Serum and Plasma. Int. J. Mol. Sci. 2023, 24, 2467. https://doi.org/10.3390/ijms24032467
Mühle C, Kornhuber J. Characterization of a Neutral Sphingomyelinase Activity in Human Serum and Plasma. International Journal of Molecular Sciences. 2023; 24(3):2467. https://doi.org/10.3390/ijms24032467
Chicago/Turabian StyleMühle, Christiane, and Johannes Kornhuber. 2023. "Characterization of a Neutral Sphingomyelinase Activity in Human Serum and Plasma" International Journal of Molecular Sciences 24, no. 3: 2467. https://doi.org/10.3390/ijms24032467
APA StyleMühle, C., & Kornhuber, J. (2023). Characterization of a Neutral Sphingomyelinase Activity in Human Serum and Plasma. International Journal of Molecular Sciences, 24(3), 2467. https://doi.org/10.3390/ijms24032467






