Disruption of the Expression of the Placental Clock and Melatonin Genes in Preeclampsia
Abstract
1. Introduction
2. Results
2.1. Circadian Expression of Clock Genes in Placental Cells
2.2. Trophoblast-Macrophage Cooperation in the Regulation of Placental Circadian Rhythms
2.3. Circadian Rhythm Impairment in Preeclamptic Macrophages
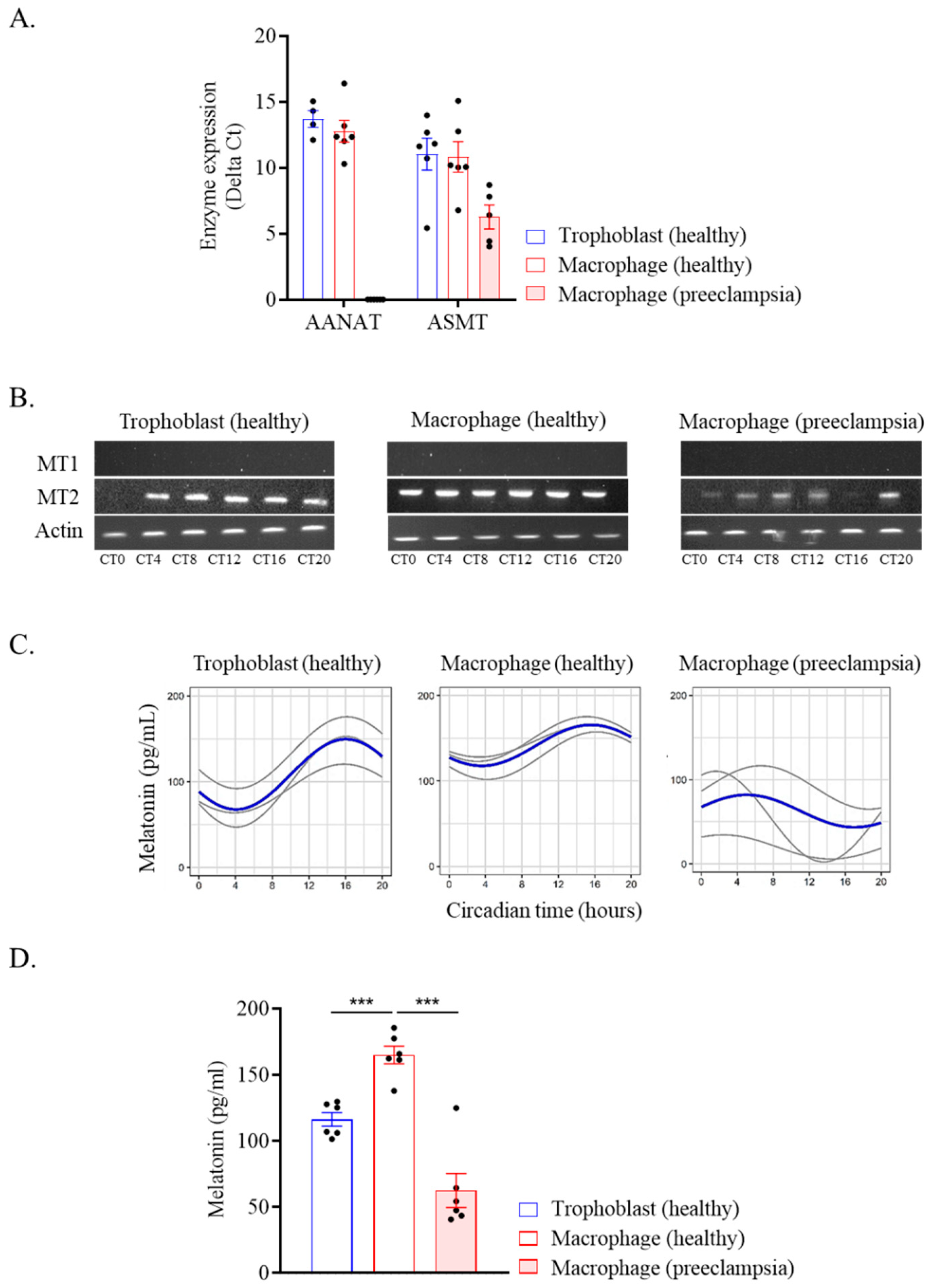
2.4. Melatonin Production by Placental Cells
3. Discussion
4. Materials and Methods
4.1. Placenta Collection
4.2. Cell Isolation and Culture
4.3. Study Design of Circadian Rhythm in Placental Cells
4.4. Study of Circadian Gene Expression
4.5. Study of Melatonin and Its Receptors
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diallo, A.B.; Coiffard, B.; Leone, M.; Mezouar, S.; Mege, J.L. For Whom the Clock Ticks: Clinical Chronobiology for Infectious Diseases. Front. Immunol. 2020, 11, 1457. [Google Scholar] [CrossRef]
- Coiffard, B.; Diallo, A.B.; Culver, A.; Mezouar, S.; Hammad, E.; Vigne, C.; Nicolino-Brunet, C.; Dignat-George, F.; Baumstarck, K.; Boucekine, M.; et al. Circadian Rhythm Disruption and Sepsis in Severe Trauma Patients. Shock 2019, 52, 29–36. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U.; Albrecht, U. The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef]
- Honma, S. The mammalian circadian system: A hierarchical multi-oscillator structure for generating circadian rhythm. J. Physiol. Sci. 2018, 68, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L.A.J. Circadian Clock Proteins and Immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef]
- Serón-Ferré, M.; Forcelledo, M.L.; Torres-Farfan, C.; Valenzuela, F.J.; Rojas, A.; Vergara, M.; Rojas-Garcia, P.P.; Recabarren, M.P.; Valenzuela, G.J. Impact of Chronodisruption during Primate Pregnancy on the Maternal and Newborn Temperature Rhythms. PLoS ONE 2013, 8, e57710. [Google Scholar] [CrossRef] [PubMed]
- Torrealba, F.; Parraguez, V.H.; Reyes, T.; Valenzuela, G.; Serón-Ferré, M. Prenatal development of the retinohypothalamic pathway and the suprachiasmatic nucleus in the sheep. J. Comp. Neurol. 1993, 338, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Olson, S.L.; Turek, F.W.; Levine, J.E.; Horton, T.H.; Takahashi, J.S. Circadian Clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr. Biol. 2004, 14, 1367–1373. [Google Scholar] [CrossRef]
- Ditisheim, A.J.; Dibner, C.; Philippe, J.; Pechére-Bertschi, A. Biological rhythms and preeclampsia. Front. Endocrinol. 2013, 4, 47. [Google Scholar] [CrossRef]
- Rana, S.; Lemoine, E.; Granger, J.; Karumanchi, S.A. Preeclampsia. Circ. Res. 2019, 124, 1094–1112. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Xu, X.H.; Jin, L. Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Jauniaux, E. What is the placenta? Am. J. Obstet. Gynecol. 2015, 213, S6-e1. [Google Scholar] [CrossRef] [PubMed]
- Murphy, V.E.; Smith, R.; Giles, W.B.; Clifton, V.L. Endocrine regulation of human fetal growth: The role of the mother, placenta, and fetus. Endocr. Rev. 2006, 27, 141–169. [Google Scholar] [CrossRef]
- Mezouar, S.; Katsogiannou, M.; Ben Amara, A.; Bretelle, F.; Mege, J.-L. Placental macrophages: Origin, heterogeneity, function and role in pregnancy-associated infections. Placenta 2021, 103, 94–103. [Google Scholar] [CrossRef]
- Moffett-King, A. Natural killer cells and pregnancy. Nat. Rev. Immunol. 2002. [CrossRef]
- Erlebacher, A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 2013, 31, 387–411. [Google Scholar] [CrossRef]
- Brown, S.A. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol. Metab. 2016, 27, 415–426. [Google Scholar] [CrossRef]
- Miller, B.H.; Takahashi, J.S. Central circadian control of female reproductive function. Front. Endocrinol. 2014, 4, 195. [Google Scholar] [CrossRef]
- Pérez, S.; Murias, L.; Fernández-Plaza, C.; Díaz, I.; González, C.; Otero, J.; Díaz, E. Evidence for clock genes circadian rhythms in human full-term placenta. Syst. Biol. Reprod. Med. 2015, 61, 360–366. [Google Scholar] [CrossRef]
- Mark, P.J.; Crew, R.C.; Wharfe, M.D.; Waddell, B.J. Rhythmic Three-Part Harmony: The Complex Interaction of Maternal, Placental and Fetal Circadian Systems. J. Biol. Rhythms 2017, 32, 534–549. [Google Scholar] [CrossRef]
- Clarkson-Townsend, D.A.; Everson, T.M.; Deyssenroth, M.A.; Burt, A.A.; Hermetz, K.E.; Hao, K.; Chen, J.; Marsit, C.J. Maternal circadian disruption is associated with variation in placental DNA methylation. PLoS ONE 2019, 14, e0215745. [Google Scholar] [CrossRef]
- Frigato, E.; Lunghi, L.; Ferretti, M.E.; Biondi, C.; Bertolucci, C. Evidence for circadian rhythms in human trophoblast cell line that persist in hypoxia. Biochem. Biophys. Res. Commun. 2009, 378, 108–111. [Google Scholar] [CrossRef]
- Tamura, H.; Nakamura, Y.; Terron, M.P.; Flores, L.J.; Manchester, L.C.; Tan, D.X.; Sugino, N.; Reiter, R.J. Melatonin and pregnancy in the human. Reprod. Toxicol. 2008, 25, 291–303. [Google Scholar] [CrossRef]
- Ekmekcioglu, C. Melatonin receptors in humans: Biological role and clinical relevance. Biomed. Pharmacother. 2006, 60, 97–108. [Google Scholar] [CrossRef]
- Dubocovich, M.L.; Delagrange, P.; Krause, D.N.; Sugden, D.; Cardinali, D.P.; Olcese, J. Nomenclature, Classification, and Pharmacology of G Protein-Coupled Melatonin Receptors. Pharmocological Rev. 2010, 62, 343–380. [Google Scholar] [CrossRef]
- Lanoix, D.; Beghdadi, H.; Lafond, J.; Vaillancourt, C. Human placental trophoblasts synthesize melatonin and express its receptors. J. Pineal Res. 2008, 45, 50–60. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Korkmaz, A.; Rosales-Corral, S.A. Melatonin and stable circadian rhythms optimize maternal, placental and fetal physiology. Hum. Reprod. Update 2014, 20, 293–307. [Google Scholar] [CrossRef]
- Soliman, A.; Lacasse, A.-A.; Lanoix, D.; Sagrillo-Fagundes, L.; Boulard, V.; Vaillancourt, C. Placental melatonin system is present throughout pregnancy and regulates villous trophoblast differentiation. J. Pineal Res. 2015, 59, 38–46. [Google Scholar] [CrossRef]
- Izumo, M.; Johnson, C.H.; Yamazaki, S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: Temperature compensation and damping. Proc. Natl. Acad. Sci. USA 2003, 100, 16089–16094. [Google Scholar] [CrossRef]
- Saini, C.; Morf, J.; Stratmann, M.; Gos, P.; Schibler, U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012, 26, 567–580. [Google Scholar] [CrossRef]
- Balsalobre, A.; Damiola, F.; Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef]
- Mol, B.W.J.; Roberts, C.T.; Thangaratinam, S.; Magee, L.A.; De Groot, C.J.M.; Hofmeyr, G.J. Pre-eclampsia. Lancet 2016, 387, 999–1011. [Google Scholar] [CrossRef]
- Waddell, B.J.; Wharfe, M.D.; Crew, R.C.; Mark, P.J. A rhythmic placenta? Circadian variation, clock genes and placental function. Placenta 2012, 33, 533–539. [Google Scholar] [CrossRef]
- Papacleovoulou, G.; Nikolova, V.; Oduwole, O.; Chambers, J.; Vazquez-Lopez, M.; Jansen, E.; Nicolaides, K.; Parker, M.; Williamson, C. Gestational disruptions in metabolic rhythmicity of the liver, muscle, and placenta affect fetal size. FASEB J. 2017, 31, 1698–1708. [Google Scholar] [CrossRef]
- Wharfe, M.D.; Mark, P.J.; Waddell, B.J. Circadian variation in placental and hepatic clock genes in rat pregnancy. Endocrinology 2011, 152, 3552–3560. [Google Scholar] [CrossRef]
- Ratajczak, C.K.; Herzog, E.D.; Muglia, L.J. Clock gene expression in gravid uterus and extra-embryonic tissues during late gestation in the mouse. Reprod. Fertil. Dev. 2010, 22, 743–750. [Google Scholar] [CrossRef]
- Barr, M. Prenatal growth of Wistar rats: Circadian periodicity of fetal growth late in gestation. Teratology 1973, 7, 283–287. [Google Scholar] [CrossRef]
- Lellupitiyage Don, S.S.; Mas-Rosario, J.A.; Lin, H.-H.; Nguyen, E.M.; Taylor, S.R.; Farkas, M.E. Macrophage circadian rhythms are differentially affected based on stimuli. Integr. Biol. 2022, 14, 62–75. [Google Scholar] [CrossRef]
- Souza, E.S.; Santos, A.A.; Ribeiro-Paz, E.E.; Córdoba-Moreno, M.; Trevisan, I.L.; Caldeira, W.; Muxel, S.M.; Sousa, K.D.; Markus, R.P. Melatonin synthesized by activated microglia orchestrates the progression of microglia from a pro-inflammatory to a recovery/repair phenotype. Melatonin Res. 2022, 5, 55–67. [Google Scholar] [CrossRef]
- Maltepe, E.; Fisher, S.J. Placenta: The Forgotten Organ. Annu. Rev. Cell Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef]
- Besseau, L.; Benyassi, A.; Møller, M.; Coon, S.L.; Weller, J.L.; Boeuf, G.; Klein, D.C.; Falcón, J. Melatonin pathway: Breaking the ‘high-at-night’ rule in trout retina. Exp. Eye Res. 2006, 82, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Kivelä, A. Serum melatonin during human pregnancy. Acta Endocrinol. 1991, 124, 233–237. [Google Scholar] [CrossRef]
- Nakamura, Y.; Tamura, H.; Kashida, S.; Takayama, H.; Yamagata, Y.; Karube, A.; Sugino, N.; Kato, H. Changes of serum melatonin level and its relationship to feto-placental unit during pregnancy. J. Pineal Res. 2001, 30, 29–33. [Google Scholar] [CrossRef]
- Sagrillo-Fagundes, L.; Assunção Salustiano, E.M.; Ruano, R.; Markus, R.P.; Vaillancourt, C. Melatonin modulates autophagy and inflammation protecting human placental trophoblast from hypoxia/reoxygenation. J. Pineal Res. 2018, 65, e12520. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Winn, E.; Nguyen, D.; Kasten, E.P.; Petroff, M.G.; Hoffmann, H.M. Co-alterations of circadian clock gene transcripts in human placenta in preeclampsia. Sci. Rep. 2022, 12, 17856. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.B.; Chaves, I.; Herzog, E.M.; Willemsen, S.P.; van der Horst, G.T.J.; Steegers-Theunissen, R.P.M. Early- and late-onset preeclampsia and the DNA methylation of circadian clock and clock-controlled genes in placental and newborn tissues. Chronobiol. Int. 2017, 34, 921–932. [Google Scholar] [CrossRef]
- Milczarek, R.; Hallmann, A.; Sokołowska, E.; Kaletha, K.; Klimek, J. Melatonin enhances antioxidant action of α-tocopherol and ascorbate against NADPH- and iron-dependent lipid peroxidation in human placental mitochondria. J. Pineal Res. 2010, 49, 149–155. [Google Scholar] [CrossRef]
- Gozeri, E.; Celik, H.; Ozercan, I.; Gurates, B.; Polat, S.A.; Hanay, F. The effect of circadian rhythm changes on fetal and placental development (experimental study). Neuro Endocrinol. Lett. 2008, 29, 87–90. [Google Scholar]
- Grosse, J.; Davis, F.C. Transient entrainment of a circadian pacemaker during development by dopaminergic activation in Syrian hamsters. Brain Res. Bull. 1999, 48, 185–194. [Google Scholar] [CrossRef]
- Viswanathan, N.; Weaver, D.R.; Reppert, S.M.; Davis, F.C. Entrainment of the fetal hamster circadian pacemaker by prenatal injections of the dopamine agonist SKF 38393. J. Neurosci. 1994, 14, 5393–5398. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Schwartz, W.J. Maternal Endocrine Extirpations Do Not Abolish Maternal Coordination of the Fetal Circadian Clock. Endocrinology 1986, 119, 1763–1767. [Google Scholar] [CrossRef]
- Mezouar, S.; Ben Amara, A.; Chartier, C.; Gorvel, L.; Mege, J.-L. A fast and reliable method to isolate human placental macrophages. Curr. Protoc. Immunol. 2019, 125, e77. [Google Scholar] [CrossRef] [PubMed]
- Mezouar, S.; Benammar, I.; Boumaza, A.; Diallo, A.B.; Chartier, C.; Buffat, C.; Boudjarane, J.; Halfon, P.; Katsogiannou, M.; Mege, J.-L. Full-term human placental macrophages eliminate Coxiella burnetii through an IFN-γ autocrine loop. Front. Microbiol. 2019, 10, 2434. [Google Scholar] [CrossRef]
- Belhareth, R.; Mezouar, S.; Ben Amara, A.; Chartier, C.; Azzouz, E.B.; Chabrière, E.; Amri, M.; Mege, J.L. Cigarette smoke extract interferes with placenta macrophage functions: A new mechanism to compromise placenta functions? Reprod. Toxicol. 2018, 78, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Ben Amara, A.; Gorvel, L.; Baulan, K.; Derain-Court, J.; Buffat, C.; Vérollet, C.; Textoris, J.; Ghigo, E.; Bretelle, F.; Maridonneau-Parini, I.; et al. Placental Macrophages Are Impaired in Chorioamnionitis, an Infectious Pathology of the Placenta. J. Immunol. 2013, 191, 5501–5514. [Google Scholar] [CrossRef]
- Ghigo, E.; Capo, C.; Raoult, D.; Mege, J.L. Interleukin-10 stimulates Coxiella burnetii replication in human monocytes through tumor necrosis factor down-modulation: Role in microbicidal defect of Q fever. Infect. Immun. 2001, 69, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Ben Azzouz, E.; Boumaza, A.; Mezouar, S.; Bardou, M.; Carlini, F.; Picard, C.; Raoult, D.; Mège, J.-L.; Desnues, B. Tropheryma whipplei increases expression of human leukocyte antigen-G on monocytes to reduce tumor necrosis factor and promote bacterial replication. Gastroenterology 2018, 155, 1553–1563. [Google Scholar] [CrossRef]
- Holness, C.L.; Simmons, D.L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 1993, 81, 1607–1613. [Google Scholar] [CrossRef]
- Diallo, A.B.; Gay, L.; Coiffard, B.; Leone, M.; Mezouar, S.; Mege, J.-L. Daytime variation in SARS-CoV-2 infection and cytokine production. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Mezouar, S.; Vitte, J.; Gorvel, L.; Ben Amara, A.; Desnues, B.; Mege, J.-L. Mast cell cytonemes as a defense mechanism against Coxiella burnetii. mBio 2019, 10, e02669-18. [Google Scholar] [CrossRef] [PubMed]
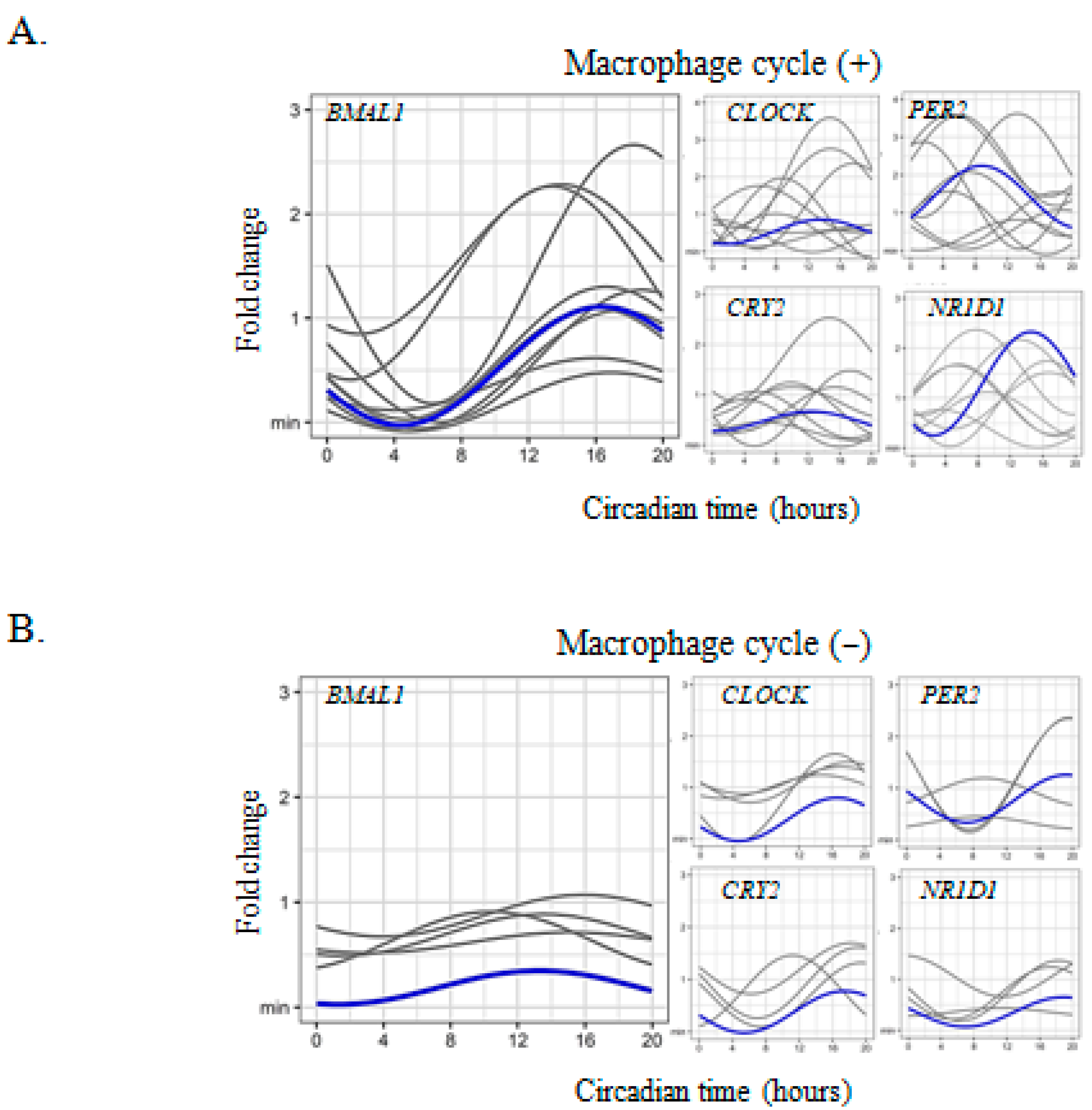
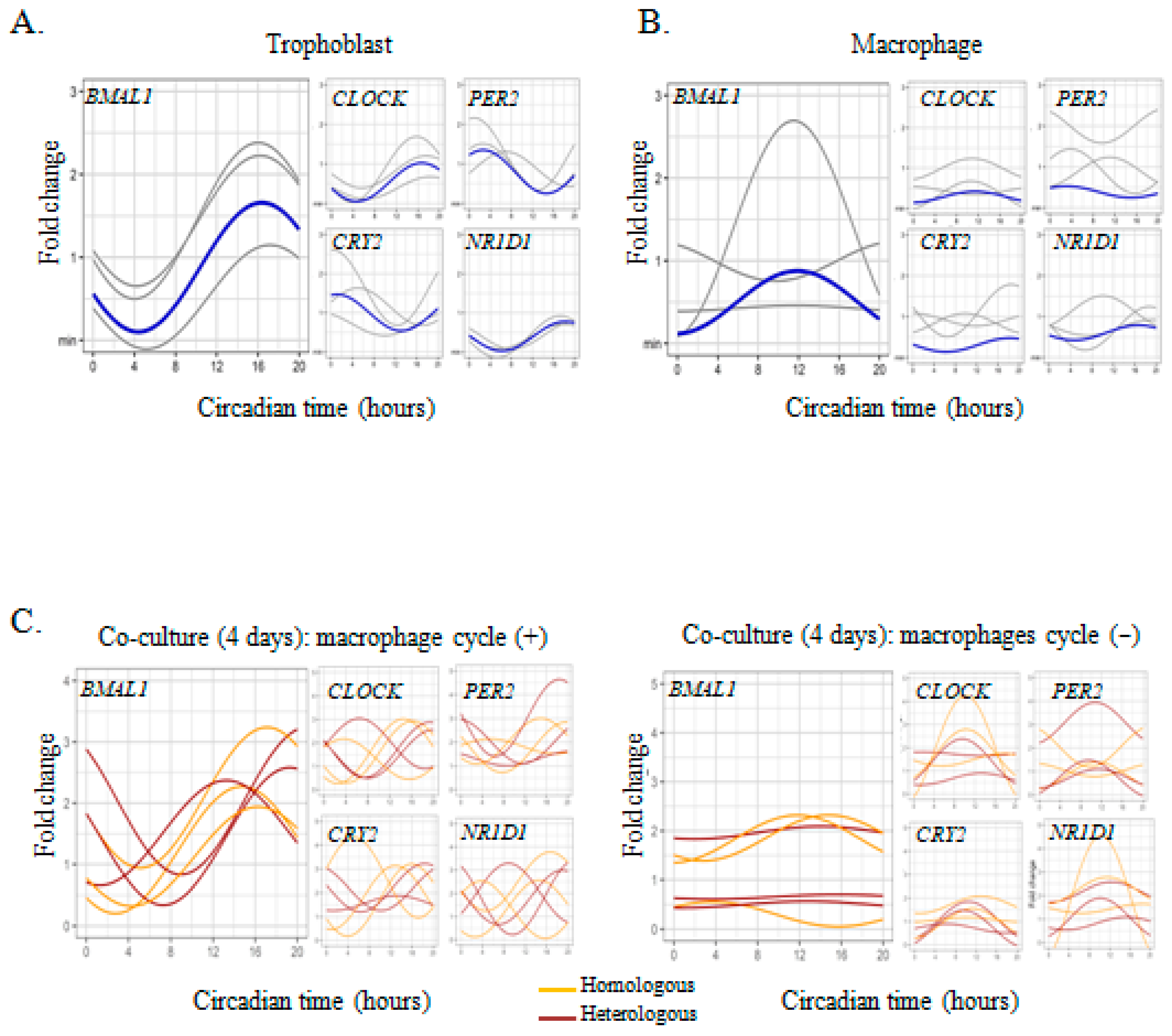

| Conditions | Gene | Cycle | Mesor | CI 95% | p | Amplitude | CI 95% | p | Acrophase | CI 95% | p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| / | BMAL1 | + | 0.80 | (0.62; 0.98) | <0.001 | 0.56 | (0.31; 0.83) | <0.001 | 16.38 | (14.63; 18.14) | <0.001 |
| − | 0.70 | (0.53; 0.88) | <0.001 | 0.16 | (−0.08; 0,41) | 0.20 | 13.25 | (7.39; 19.11) | 0.67 | ||
| CLOCK | + | 0.36 | (0.16; 0.55) | <0.001 | 0.23 | (−0.04; 0.51) | 0.09 | 17.11 | (12.56; 21.66) | 0.02 | |
| − | 1.21 | (0.66; 1.76) | <0.001 | 0.49 | (−0.2 9; 1.27) | 0.22 | 6.21 | (0.10; 12.32) | 0.06 | ||
| CRY2 | + | 0.73 | (0.50; 0.96) | <0.001 | 0.75 | (0.43; 1.07) | <0.001 | 17.27 | (15.63; 18.87) | <0.001 | |
| − | 0.93 | (0.10; 1.75) | 0.027 | 0.27 | (−0.90; 1.43) | 0.65 | 9.379 | (4.77; 13.99) | 0.75 | ||
| PER2 | + | 6.70 | (4.09; 9.31) | <0.001 | 2.74 | (−0.95; 6.43) | 0.14 | 8.46 | (3.32; 13.60) | 0.18 | |
| − | 1.14 | (0.47; 1.81) | <0.001 | 0.26 | (−0.68; 1.21 | 0.58 | 17.12 | (15.38; 18.86) | 0.46 | ||
| NR1D1 | + | 0.89 | (0.77; 1.01) | <0.001 | 0.85 | (0.68; 1.03) | <0.001 | 16.55 | (15.77; 17.32) | <0.001 | |
| − | 0.87 | (0.32; 1.42) | 0.002 | 0.19 | (−0.58; 0.97) | 0.61 | 15.93 | (13.00; 18.86) | 0.60 | ||
| 4 days of culture | BMAL1 | / | 0.93 | (0.51; 1.35) | <0.001 | 0.38 | (−0.21; 0.97) | 0.21 | 11.87 | (5.88; 17.85) | 0.96 |
| CLOCK | 0.42 | (0.07; 0.77) | 0.01 | 0.03 | (−0.46; 0.53) | 0.89 | 11.69 | / | 0.99 | ||
| CRY2 | 0.87 | (0.42; 1.33) | <0.001 | 0.36 | (−0.27; 0.10) | 0.26 | 11.02 | (4.30; 17.73) | 0.77 | ||
| PER2 | 0.40 | (0.14; 0.66) | 0.003 | 0.28 | (−0.09; 0.65) | 0.13 | 6.28 | (1.28; 11.28) | 0.02 | ||
| NR1D1 | 0.58 | (0.15; 1.01) | 0.008 | 0.22 | (−0.39; 0.82) | 0.48 | 21.43 | / | 0.63 | ||
| Co-culture with trophoblasts (4 days) | BMAL1 | + | 1.58 | (1.23; 1.92) | <0.001 | 0.8 | (0.31; 1.30) | 0.001 | 17.20 | (14.87; 19.55) | <0.001 |
| − | 1.21 | (0.72; 1.71) | <0.001 | 0.14 | (−0.56; 0.84) | 0.70 | 12.99 | (5.39; 20.60) | 0.92 | ||
| CLOCK | + | 1.70 | (0.89; 2.50) | <0.001 | 1.18 | (0.04; 2.33) | 0.04 | 7.36 | (3.68; 11.04) | 0.01 | |
| − | 1.46 | (−0.54; 3.45) | 0.15 | 0.20 | (−2.63; 3.03) | 0.89 | 6.44 | / | 0.84 | ||
| CRY2 | + | 4.69 | (2.47; 6.91) | <0.001 | 1.88 | (−1.26; 5.03) | 0.24 | 11.12 | (4.73; 17.51) | 0.79 | |
| − | 1.38 | (−0.06; 2.82) | 0.06 | 1.04 | (−0.99; 3.07) | 0.32 | 12.31 | (4.83; 19.79) | 0.93 | ||
| PER2 | + | 2.03 | (1.28; 2.79) | <0.001 | 0.93 | (−0.14; 1.99) | 0.09 | 11.58 | (7.18; 15.98) | 0.85 | |
| − | 1.07 | (−0.07; 2.21) | 0.06 | 0.28 | (−1.33; 1.89) | 0.73 | 10.65 | (0.72; 20.58) | 0.90 | ||
| NR1D1 | + | 1.82 | (1.06; 2.59) | <0.001 | 1.16 | (0.07; 2.25) | 0.03 | 6.99 | (3.41; 10.57) | 0.006 | |
| − | 1.48 | (−0.42; 3.38) | 0.12 | 0.27 | (−2.42; 2.96) | 0.12 | 17.57 | / | 0.77 |
| Gene | Cycle | Mesor | CI 95% | p | Amplitude | CI 95% | p | Acrophase | CI 95% | p |
|---|---|---|---|---|---|---|---|---|---|---|
| BMAL1 | Single | 1.10 | (0.80; 1.40) | <0.001 | 0.27 | (−0.15; 0.70) | 0.20 | 16.85 | (10.97; 22.73) | 0.10 |
| Co-culture | 2.03 | (1.30; 2.75) | <0.001 | 0.08 | (−0.95; 1.11) | 0.87 | 16.18 | / | 0.10 | |
| CLOCK | Single | 0.40 | (0.25; 0.55) | <0.001 | 0.51 | (0.30; 0.72) | <0.001 | 8.66 | (7.07; 10.24) | <0.001 |
| Co-culture | 0.79 | (0.16; 1.42) | 0.014 | 0.46 | (−0.43; 1.35) | 0.31 | 13.55 | (6.16; 20.95) | 0.68 | |
| CRY2 | Single | 0.77 | (0.40; 1.14) | <0.001 | 0.28 | (−0.25; 0.80) | 0.29 | 14.41 | (7.20; 21.63) | 0.51 |
| Co-culture | 4.12 | (1.22; 7.03) | 0.005 | 2.34 | (−1.77; 6.44) | 0.26 | 11.63 | (4.92; 18.35) | 0.91 | |
| PER2 | Single | 0.52 | (0.25; 0.79) | <0.001 | 0.44 | (0.05; 0.83) | 0.02 | 12.53 | (9.17; 15.88 | 0.75 |
| Co-culture | 1.31 | (0.58; 2.03) | <0.001 | 0.99 | (−0.04; 2.02 | 0.06 | 14.91 | (10.95; 18.88) | 0.14 | |
| NR1D1 | Single | 0.53 | (0.26; 0.81) | <0.001 | 0.36 | (−0.03; 0.75) | 0.07 | 13.53 | (9.37; 17.69) | 0.46 |
| Co-culture | 1.10 | (0.42; 1.79) | 0.002 | 0.66 | (−0.31; 1.63) | 0.18 | 10.07 | (4.49; 15.65) | 0.49 |
| Cells | Mesor | CI 95% | p | Amplitude | CI 95% | p | Acrophase | CI 95% | p |
|---|---|---|---|---|---|---|---|---|---|
| Trophoblast (healthy) | 113.35 | (96.92; 129.78) | <0.001 | 25.2816 | (2.04; 48.52) | 0.03 | 18.28 | (14.77; 21.79) | 0.001 |
| Macrophage (healthy) | 141.74 | (130.12; 153.36) | <0.001 | 23.9745 | (7.54; 40.41) | 0.004 | 15.58 | (12.96; 18.20) | 0.007 |
| Macrophage (preeclampsia) | 48.32 | (26.82; 69.82) | / | 24.0605 | (−6.34; 54.46) | 0.12 | 2.99 | / | 0.22 |
| Control (n = 20) | Preeclampsia (n = 9) | |
|---|---|---|
| Maternal age (year) | 32 [24–43] | 33 [24–44] |
| Gestational age (week) | 39 [36–41] | 36 [34–41] |
| BMI | 25.75 [17.3–37] | 28.75 [20.3–43.3] |
| Cesarean (n) | 0 | 4 |
| Weight at birth (g) | 3430 [2740–4160] | 2410 [1640–3260] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diallo, A.B.; Coiffard, B.; Desbriere, R.; Katsogiannou, M.; Donato, X.; Bretelle, F.; Mezouar, S.; Mege, J.-L. Disruption of the Expression of the Placental Clock and Melatonin Genes in Preeclampsia. Int. J. Mol. Sci. 2023, 24, 2363. https://doi.org/10.3390/ijms24032363
Diallo AB, Coiffard B, Desbriere R, Katsogiannou M, Donato X, Bretelle F, Mezouar S, Mege J-L. Disruption of the Expression of the Placental Clock and Melatonin Genes in Preeclampsia. International Journal of Molecular Sciences. 2023; 24(3):2363. https://doi.org/10.3390/ijms24032363
Chicago/Turabian StyleDiallo, Aïssatou Bailo, Benjamin Coiffard, Raoul Desbriere, Maria Katsogiannou, Xavier Donato, Florence Bretelle, Soraya Mezouar, and Jean-Louis Mege. 2023. "Disruption of the Expression of the Placental Clock and Melatonin Genes in Preeclampsia" International Journal of Molecular Sciences 24, no. 3: 2363. https://doi.org/10.3390/ijms24032363
APA StyleDiallo, A. B., Coiffard, B., Desbriere, R., Katsogiannou, M., Donato, X., Bretelle, F., Mezouar, S., & Mege, J.-L. (2023). Disruption of the Expression of the Placental Clock and Melatonin Genes in Preeclampsia. International Journal of Molecular Sciences, 24(3), 2363. https://doi.org/10.3390/ijms24032363







