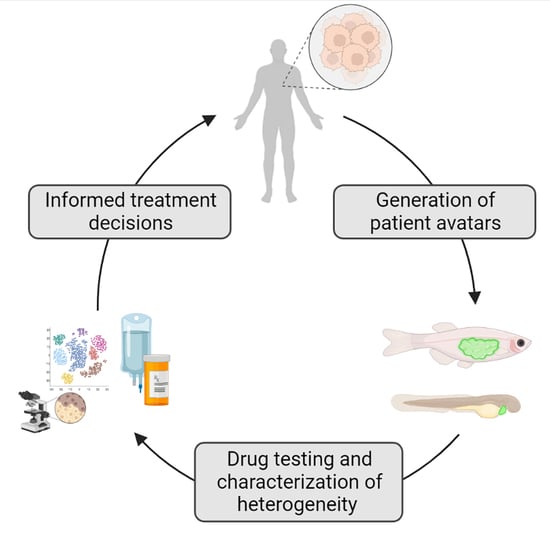
Zebrafish Cancer Avatars: A Translational Platform for Analyzing Tumor Heterogeneity and Predicting Patient Outcomes
Abstract
1. Introduction
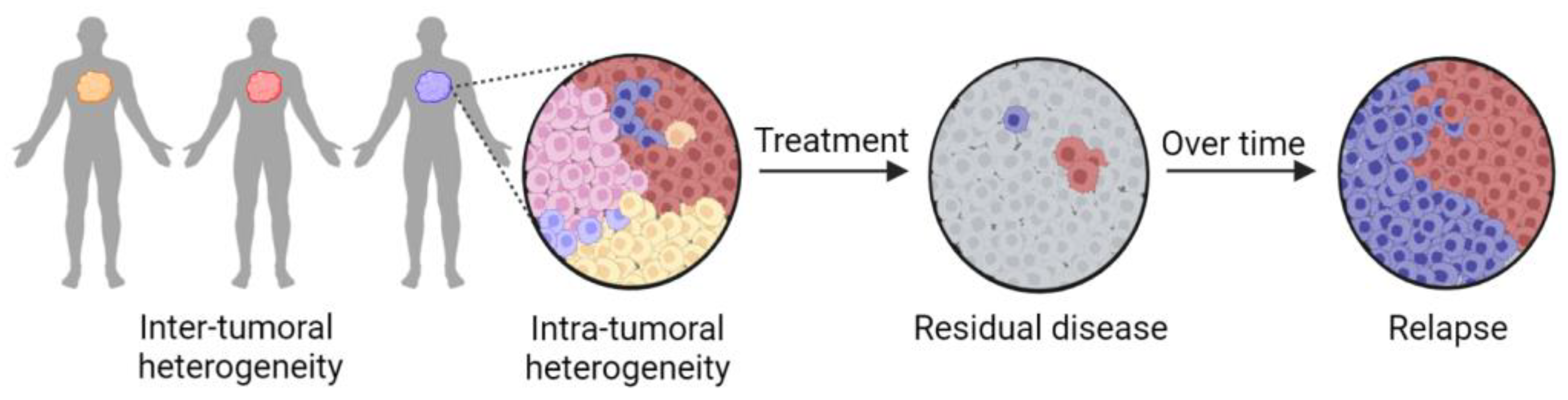
2. Tumor Heterogeneity Complicates Precision Medicine Approaches
3. Current Patient-Derived Avatar Models Are Not Suitable for Clinical Timelines
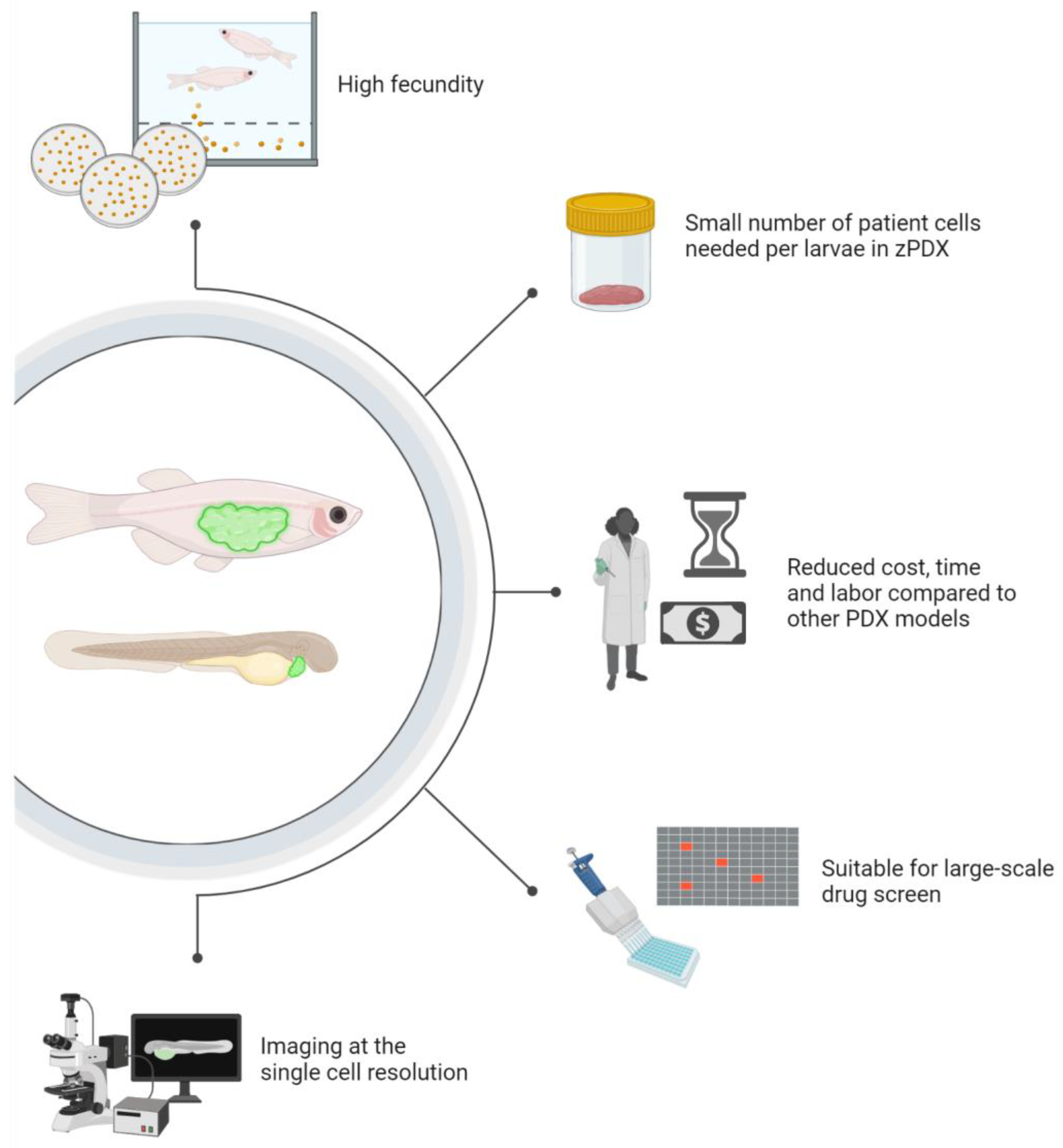
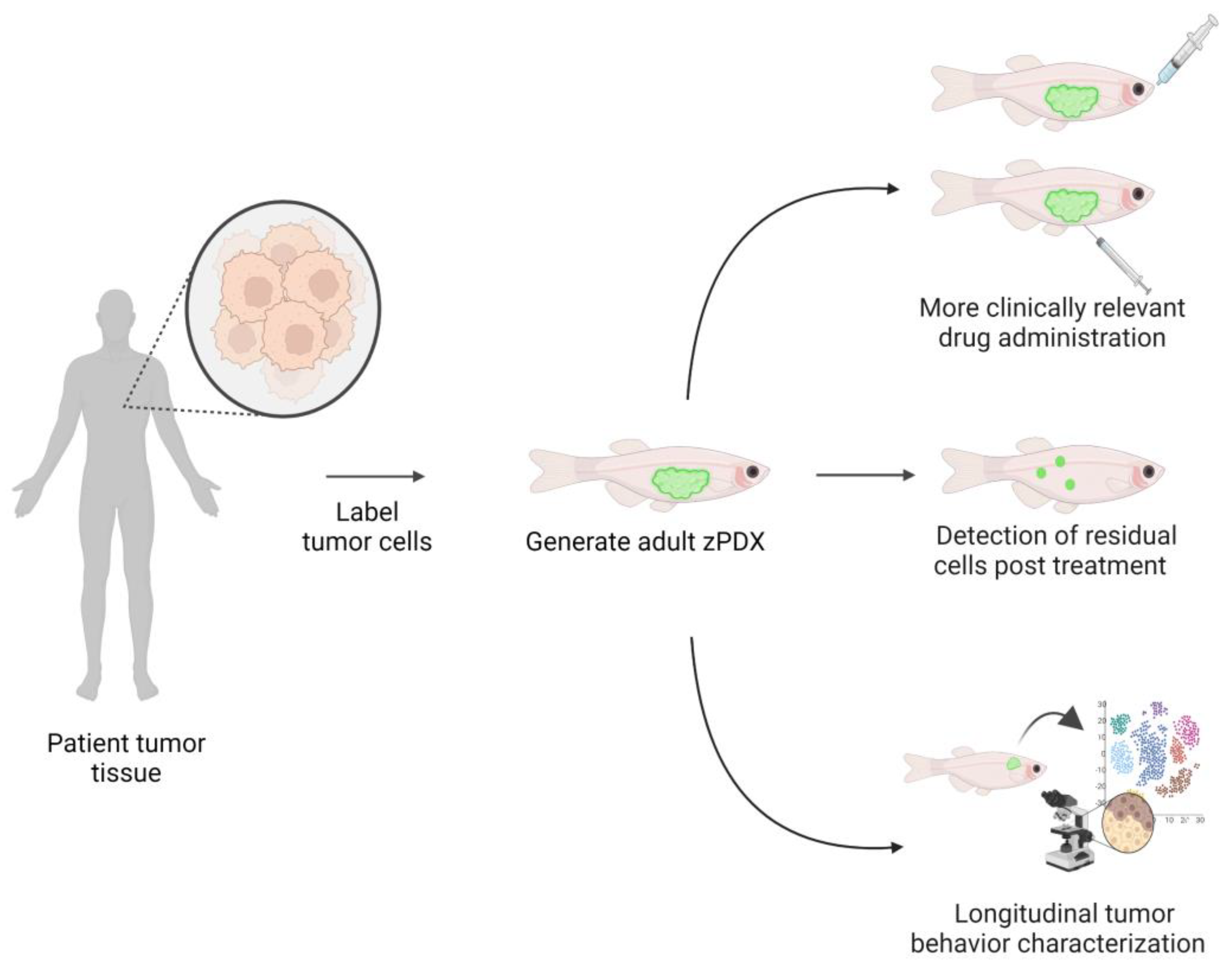
4. Zebrafish Patient-Derived Xenografts (zPDX) Fill Gaps in Current Model Systems
| Automated Step | Equipment and Software Involved | Application | Speed or Efficacy | Accuracy | Limitations | Ref. |
|---|---|---|---|---|---|---|
| Imaging and Image analysis | Automated microscopy platform, IncuCyte S3 and CNN for image analysis | Characterize the tumor growth in the xenografts in vivo | One image every 4–6 h over 4 days | 77% agreement with manual analysis | Difficulty of identification of early signs of death | [84] |
| Image analysis | Faster R-CNN algorithm, Inception Reset V2 feature extractor | Counting of tumor cells in zPDX | Not mentioned | 85% average precision | Larger datasets are considered for higher accuracy | [86] |
| Image analysis | SPIM for imaging, CellProfiler, CellTracker and R open source tools | Real time visualization and tracking of metastatic cancer cells in zPDX | Not mentioned | Differentiate the dissemination patterns between two types of cancers | Not mentioned | [81] |
| Imaging | VAST BioImager System, upright microscope, LP Sampler | Automate zebrafish larva handling, positioning, orientation and imaging | 100 and 75 larvae/h sampling from bulk and 96-well plate | 86% and 97% success rate imaging from bulk and 96-well plate | Applicable for 2–7 dpf larva, high storage space for obtained images | [79] |
| Imaging and treatment administration | Automated pipetting workstation, automated imaging system (ImagXpressMICRO) | Live imaging of zPDXand automated application of therapeutics | Not mentioned | Not mentioned | Not mentioned | [87] |
| Imaging and treatment administration | ImageXpress Micro System, JANUS automated workstation | Collect High-Content images for adult and embryo zPDX | Not mentioned | Not mentioned | Not mentioned | [88] |
| Injection | Motorized stage with paired controller, motorized micro-manipulator, “injectman II”, z-calibration unit | Semi-automate injection | 2000 embryos/h | 99% success rate | Automation is reported for injection into embryos and requires commercial injection needles | [78] |
| Injection and screening | Automated microinjection system and COPAS | Automate injection into zebrafish embryos and fluorescence quantification | 3000 embryos/15 min for sorting step | Not mentioned | Not mentioned | [89] |
| Imaging, image pre-processing and analysis | CLSM platform with movable stage, Image-Pro Software | Quantification of human cancer dissemination in zPDX | Less than 5 min per plate for imaging | Dissemination in zPDX significantly correlated with metastatic behavior in mouse model | Confocal microscopy is required | [80] |
5. Zebrafish Are Clinically Relevant Cancer Avatars Models
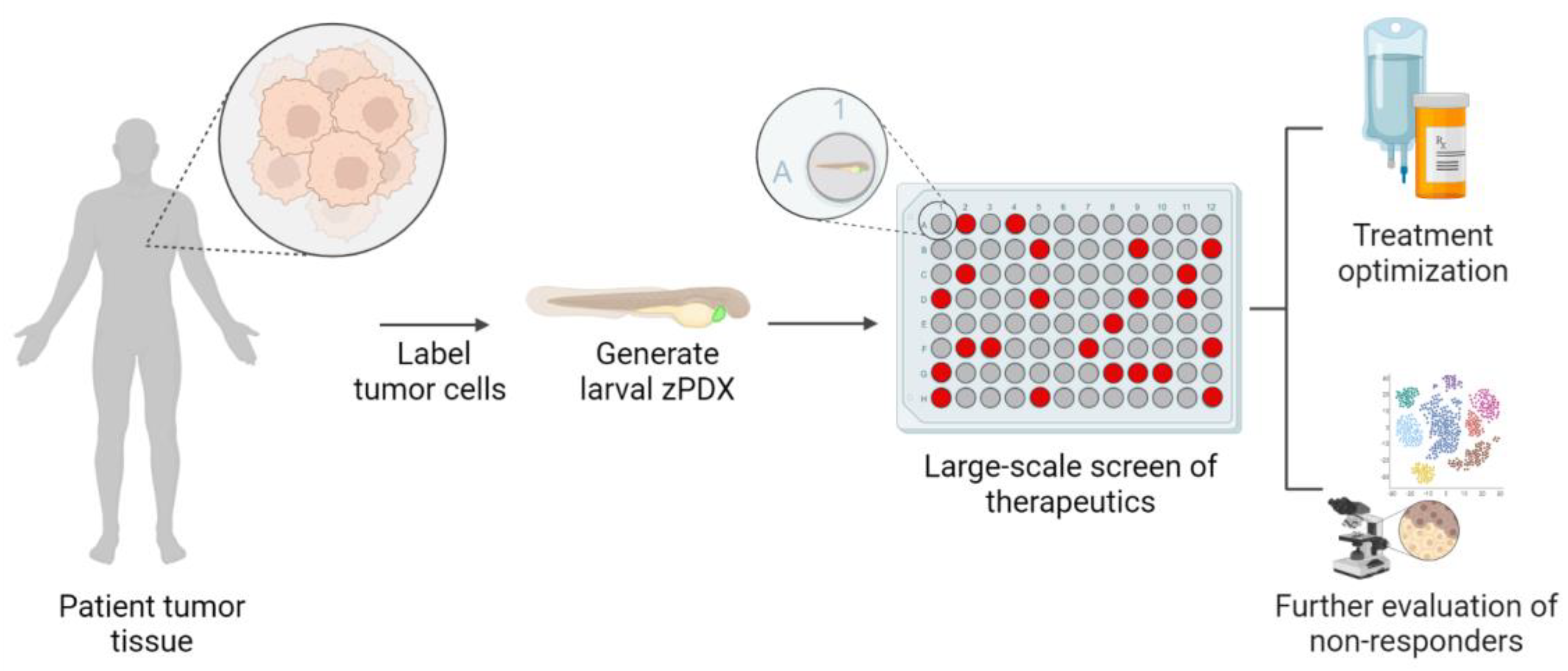
6. Zebrafish Bring Pharmacology Strengths into Precision Medicine Pipelines
7. Tank to Bedside: Zebrafish Avatars for the Prediction of Patient Outcomes
| Cancer | Zebrafish Age | Number of Patient Samples | Outcome Assessed | Therapeutic Regimen | Xenograft Characterization | Criteria for Zebrafish Response Evaluation | Criteria for Human Response Evaluation | Concordance 1 | Ref |
|---|---|---|---|---|---|---|---|---|---|
| CRC | 48 hpf | 36 | Chemotherapy response | 5-FU, FOLFOX, FOLFIRI and FOLFOXIRI | Engraftment success and change in equivalent tumor volume (%∆V) | Adaptive RECIST 2 Criteria | RECIST criteria | 75% | [119] |
| 48 hpf | 2 | Radiotherapy sensitivity | 25 Gy + 5-FU | Engraftment success, Tumor size and Apoptosis of tumor cells | Tumor size and cell death | MRI response and rectosigmoidoscopy | 100% | [124] | |
| 48 hpf | 5 | Chemotherapy response | FOLFIRI, FOLFOX and Cetumixab | Engraftment success and tumor size | Tumor size, angiogenesis potential, apoptosis and metastatic potential | Disease recurrence, CEA levels and treatment response | 80% | [73] | |
| NSCLC | 48 hpf | 4 | Chemotherapy response | Erlotinib and Paclitaxel | Engraftment success and tumor size | Relative tumor sizes, dissemination | Not mentioned | Not mentioned | [93] |
| 48 hpf | 21 | Chemotherapy response | Osimertinib, Pemetrexed, Docetaxel and Cisplatin | Engraftment success and tumor size | Tumor size | RECIST criteria | 76.9% | [125] | |
| PDAC | 48 hpf | 31 | Chemotherapy response | Gemcitabine, GEMOX, GEM/nab-P and FOLFOXIRI | Engraftment success and change in equivalent tumor volume (%∆V) | Adaptive WHO Criteria 3 | Adjuvant treatment administration, disease recurrence rate and DFS | 66.7% for cancer recurrence | [94] |
| 48 hpf | 15 | Chemotherapy response | Gemcitabine, GEMOX, GEM/nab-P and FOLFOXIR | Engraftment success, Larvae survival rate, Histology and Immunohistochemistry | Mean RTA | PFS, overall health and failure-free survival | Not mentioned | [118] | |
| TNBC and CRC 4 | 48 hpf | 2 | Bevacizumab response | Bevacizumab | Engraftment success | Apoptosis and metastatic potential | Bevacizumab treatment resistance | Resistance predication 5 | [126] |
| Pancreatic CRC and GC | 48 hpf | 24 | Chemotherapy response | 5-FU, FOLFOX, FOLFIRI, FOLFOXIRI, GEM, GEMOX, GEM/nab-P, FLOT and ECF | Engraftment success | Tumor size, Adaptive RECIST 2 criteria | Not mentioned | Not mentioned | [120] |
| GC | 48 hpf | 14 | Chemotherapy response and dissemination pattern | 5-FU, docetaxel and apatinib | Engraftment success and larvae survival rate | Cell proliferation rate | Clinical and histopathological characteristics | Not mentioned 6 | [92] |
8. Zebrafish Avatars Provide New Opportunities to Assess the Impact of Intra-Tumoral Heterogeneity in Precision Medicine
9. Conclusions
10. Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5-FU | Fluorouracil |
| ADME | Absorption, distribution, metabolism and excretion |
| BMP | Bone morphogenetic protein |
| CEA | Carcinoembryonic antigen |
| CHT | Caudal hematopoietic tissues |
| CLSM | Confocal Laser Scanning Microscope |
| CML | Chronic Myeloid Leukemia |
| CNN | Convolutional Neural Network |
| COPAS | Complex Object Parametric Analysis and Sorting |
| CR | Complete Response |
| CRC | Colorectal cancer |
| CRLM | Colorectal Liver Metastases |
| CROs | Contract Research Organizations |
| CYP | Human cytochrome P450 |
| DFS | Disease-free survival |
| dpf | Days post fertilization |
| ECF | 5-Fluorouracil + Cisplatin + Epirubicin |
| ER | Estrogen receptor |
| FGF | Fibroblast growth factor |
| FLOT | Fluorouracil, leucovorin, oxaliplatin and docetaxel |
| FOLFIRI | 5-Fluorouracil + Lederfolin + Irinotecan |
| FOLFOX | 5-Fluorouracil + Lederfolin + Oxaliplatin |
| FOLFOXIRI | 5-Fluorouracil + Folinic acid + Oxaliplatin + Irinotecan |
| FUCCI4 | Fluorescent, ubiquitination-based cell cycle indicator |
| GBM | Glioblastoma multiforme |
| GC | Gastric cancer |
| GEM | Gemcitabine |
| GEM/nab-P | Gemcitabine + nab-Paclitaxel |
| GEMOX | Gemcitabine + Oxaliplatin |
| GFP | Green Fluorescent Protein |
| Gy | Gray |
| jak3 | Janus kinase 3 |
| LP | Large Particle |
| mPDX | mouse patient-derived xenografts |
| MR | Minor Response |
| MRI | Magnetic resonance imaging |
| NCI MATCH | National Cancer Institute Molecular Analysis for Therapy Choice |
| NSCLC | Non-Small Cell Lung Cancer |
| PCA | Principal component analysis |
| PD | Progressive Disease |
| PDAC | Pancreatic ductal adenocarcinoma |
| PDO | Patient-derived organoids |
| PDX | Patient-derived xenografts |
| PFS | Progression-free survival |
| PR | Progesterone receptor |
| PR | Partial Response |
| R-CNN | Regions with Convolutional Neural Network |
| RECIST | Response Evaluation Criteria In Solid Tumors |
| RTA | Relative tumor area |
| SD | Stable Disease |
| SPIM | Selective Plane Illumination Microscopy |
| TNBC | Triple-Negative Breast Cancer |
| VAST | Vertebrate Automated Screening Technology |
| VEGF | Vascular endothelial growth factor |
| WHO | World Health Organization |
| zPDX | Zebrafish patient-derived xenografts |
References
- Hayes, D.F.; Schott, A.F. Personalized medicine: Genomics trials in oncology. Trans. Am. Clin. Climatol. Assoc. 2015, 126, 133. [Google Scholar] [PubMed]
- Yan, L.; Zhang, W. Precision medicine becomes reality—Tumor type-agnostic therapy. Cancer Commun. 2018, 38, 6. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Letai, A.; Bhola, P.; Welm, A.L. Functional precision oncology: Testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 2021, 40, 26–35. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.; Chen, A.; Li, S.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Mitchell, E.P.; Iafrate, A.J.; Sklar, J. The molecular analysis for therapy choice (NCI-MATCH) trial: Lessons for genomic trial design. JNCI J. Natl. Cancer Inst. 2020, 112, 1021–1029. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.J.; Chen, A.P.; Li, S.; McShane, L.M.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Iafrate, A.J.; Sklar, J. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J. Clin. Oncol. 2020, 38, 3883. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef]
- Stanta, G.; Bonin, S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef]
- Zellmer, V.R.; Zhang, S. Evolving concepts of tumor heterogeneity. Cell Biosci. 2014, 4, 69. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef]
- Malaney, P.; Nicosia, S.V.; Davé, V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett. 2014, 344, 1–12. [Google Scholar] [CrossRef]
- DeRose, Y.S.; Wang, G.; Lin, Y.-C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.; Factor, R.; Matsen, C.; Milash, B.A.; Nelson, E. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef]
- Topp, M.D.; Hartley, L.; Cook, M.; Heong, V.; Boehm, E.; McShane, L.; Pyman, J.; McNally, O.; Ananda, S.; Harrell, M. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol. Oncol. 2014, 8, 656–668. [Google Scholar] [CrossRef]
- Davies, A.; Hidalgo, M.; Stebbing, J.; Ciznadija, D.; Katz, A.; Sidransky, D. Mouse clinical trials of pancreatic cancer: Integration of PDX models with genomics to improve patient outcomes to chemotherapeutics. Ann. Oncol. 2016, 27, vi527. [Google Scholar] [CrossRef]
- Na, D.; Chae, J.; Cho, S.-Y.; Kang, W.; Lee, A.; Min, S.; Kang, J.; Kim, M.J.; Choi, J.; Lee, W. Predictive biomarkers for 5-fluorouracil and oxaliplatin-based chemotherapy in gastric cancers via profiling of patient-derived xenografts. Nat. Commun. 2021, 12, 4840. [Google Scholar] [CrossRef]
- Clohessy, J.G.; Pandolfi, P.P. Mouse hospital and co-clinical trial project—From bench to bedside. Nat. Rev. Clin. Oncol. 2015, 12, 491–498. [Google Scholar] [CrossRef]
- Costa, B.; Estrada, M.F.; Mendes, R.V.; Fior, R. Zebrafish avatars towards personalized medicine—A comparative review between avatar models. Cells 2020, 9, 293. [Google Scholar] [CrossRef]
- Jung, J.; Seol, H.S.; Chang, S. The generation and application of patient-derived xenograft model for cancer research. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer heterogeneity: Implications for targeted therapeutics. Br. J. Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef]
- Liu, J.; Dang, H.; Wang, X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018, 50, e416. [Google Scholar] [CrossRef] [PubMed]
- Saußele, S.; Krauß, M.-P.; Hehlmann, R.; Lauseker, M.; Proetel, U.; Kalmanti, L.; Hanfstein, B.; Fabarius, A.; Kraemer, D.; Berdel, W.E. Impact of comorbidities on overall survival in patients with chronic myeloid leukemia: Results of the randomized CML study IV. Blood J. Am. Soc. Hematol. 2015, 126, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.A.; Druker, B.J.; Guilhot, F.; O’Brien, S.G.; Riviere, G.J.; Krahnke, T.; Gathmann, I.; Wang, Y. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: A subanalysis of the IRIS study. Blood J. Am. Soc. Hematol. 2008, 111, 4022–4028. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Hochhaus, A.; Branford, S.; Müller, M.C.; Kaeda, J.S.; Foroni, L.; Druker, B.J.; Guilhot, F.; Larson, R.A.; O’Brien, S.G. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: An analysis from the International Randomized Study of Interferon and STI571 (IRIS). Blood J. Am. Soc. Hematol. 2010, 116, 3758–3765. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019, 20, 404–416. [Google Scholar] [CrossRef]
- Vaupel, P. Oxygen supply to malignant tumors. In Tumor Blood Circulation: Angiogenesis, Vascular Morphology and Blood Flow of Experimental and Human Tumors; CRC Press: Boca Raton, FL, USA, 2020; pp. 143–168. [Google Scholar]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef]
- Zamarron, B.F.; Chen, W. Dual roles of immune cells and their factors in cancer development and progression. Int. J. Biol. Sci. 2011, 7, 651. [Google Scholar] [CrossRef]
- Bozic, I.; Antal, T.; Ohtsuki, H.; Carter, H.; Kim, D.; Chen, S.; Karchin, R.; Kinzler, K.W.; Vogelstein, B.; Nowak, M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 18545–18550. [Google Scholar] [CrossRef]
- Vitale, I.; Shema, E.; Loi, S.; Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021, 27, 212–224. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 1–9. [Google Scholar] [CrossRef]
- Bell, C.C.; Gilan, O. Principles and mechanisms of non-genetic resistance in cancer. Br. J. Cancer 2020, 122, 465–472. [Google Scholar] [CrossRef]
- Longo, D.L. Tumor heterogeneity and personalized medicine. N. Engl. J. Med. 2012, 366, 956–957. [Google Scholar] [CrossRef]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Hua, X.; Zhao, W.; Pesatori, A.C.; Consonni, D.; Caporaso, N.E.; Zhang, T.; Zhu, B.; Wang, M.; Jones, K.; Hicks, B. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat. Commun. 2020, 11, 2459. [Google Scholar] [CrossRef]
- Magill, S.T.; Vasudevan, H.N.; Seo, K.; Villanueva-Meyer, J.E.; Choudhury, A.; John Liu, S.; Pekmezci, M.; Findakly, S.; Hilz, S.; Lastella, S. Multiplatform genomic profiling and magnetic resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Rye, I.H.; Trinh, A.; Sætersdal, A.B.; Nebdal, D.; Lingjærde, O.C.; Almendro, V.; Polyak, K.; Børresen-Dale, A.L.; Helland, Å.; Markowetz, F. Intratumor heterogeneity defines treatment-resistant HER 2+ breast tumors. Mol. Oncol. 2018, 12, 1838–1855. [Google Scholar] [CrossRef] [PubMed]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B. Inter-and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021, 12, 1426. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B.-F.; Chen, C.-Y.; Zhou, J.-Y.; Chen, Y.-S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.-S. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S. A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell 2020, 180, 188–204.e122. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Niida, A.; Uchi, R.; Hirata, H.; Komatsu, H.; Sakimura, S.; Hayashi, S.; Nambara, S.; Kuroda, Y.; Ito, S. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Ho, D.W.-H.; Tsui, Y.-M.; Sze, K.M.-F.; Chan, L.-K.; Cheung, T.-T.; Lee, E.; Sham, P.-C.; Tsui, S.K.-W.; Lee, T.K.-W.; Ng, I.O.-L. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett. 2019, 459, 176–185. [Google Scholar] [CrossRef]
- Araf, S.; Wang, J.; Korfi, K.; Pangault, C.; Kotsiou, E.; Rio-Machin, A.; Rahim, T.; Heward, J.; Clear, A.; Iqbal, S. Genomic profiling reveals spatial intra-tumor heterogeneity in follicular lymphoma. Leukemia 2018, 32, 1261–1265. [Google Scholar] [CrossRef]
- Raynaud, F.; Mina, M.; Tavernari, D.; Ciriello, G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet. 2018, 14, e1007669. [Google Scholar] [CrossRef]
- Dentro, S.C.; Leshchiner, I.; Haase, K.; Tarabichi, M.; Wintersinger, J.; Deshwar, A.G.; Yu, K.; Rubanova, Y.; Macintyre, G.; Demeulemeester, J. Characterizing genetic intra-tumor heterogeneity across 2658 human cancer genomes. Cell 2021, 184, 2239–2254.e39. [Google Scholar] [CrossRef]
- Gould, S.E.; Junttila, M.R.; de Sauvage, F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015, 21, 431–439. [Google Scholar] [CrossRef]
- Cho, S.-Y. Patient-derived xenografts as compatible models for precision oncology. Lab. Anim. Res. 2020, 36, 14. [Google Scholar] [CrossRef]
- Park, H.; Kim, Y.; Sul, J.W.; Jeong, I.G.; Yi, H.J.; Ahn, J.B.; Kang, J.S.; Yun, J.; Hwang, J.J.; Kim, C.S. Synergistic anti-cancer efficacy of MEK inhibition and dual PI3K/mTOR inhibition in castration-resistant prostate cancer. Prostate 2015, 75, 1747–1759. [Google Scholar] [CrossRef]
- Zhang, R.; Jin, S.; Rao, W.; Song, F.; Yin, Q.; Wang, Y.; Wang, L.; Xi, Y.; Zhang, X.; Wang, M. OVA12, a novel tumor antigen, promotes cancer cell growth and inhibits 5-fluorouracil-induced apoptosis. Cancer Lett. 2015, 357, 141–151. [Google Scholar] [CrossRef]
- Garralda, E.; Paz, K.; López-Casas, P.P.; Jones, S.; Katz, A.; Kann, L.M.; López-Rios, F.; Sarno, F.; Al-Shahrour, F.; Vasquez, D. Integrated Next-Generation Sequencing and Avatar Mouse Models for Personalized Cancer TreatmentPersonalized Cancer Treatment using Genomics and Avatar Models. Clin. Cancer Res. 2014, 20, 2476–2484. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Servant, R.; Garioni, M.; Vlajnic, T.; Blind, M.; Pueschel, H.; Müller, D.C.; Zellweger, T.; Templeton, A.J.; Garofoli, A.; Maletti, S. Prostate cancer patient-derived organoids: Detailed outcome from a prospective cohort of 81 clinical specimens. J. Pathol. 2021, 254, 543–555. [Google Scholar] [CrossRef]
- Green, S.; Dam, M.S.; Svendsen, M.N. Mouse avatars of human cancers: The temporality of translation in precision oncology. Hist. Philos. Life Sci. 2021, 43, 1–22. [Google Scholar] [CrossRef]
- Cone, E.B.; Marchese, M.; Paciotti, M.; Nguyen, D.-D.; Nabi, J.; Cole, A.P.; Molina, G.; Molina, R.L.; Minami, C.A.; Mucci, L.A. Assessment of time-to-treatment initiation and survival in a cohort of patients with common cancers. JAMA Netw. Open 2020, 3, e2030072. [Google Scholar] [CrossRef]
- Lin, D.; Wyatt, A.W.; Xue, H.; Wang, Y.; Dong, X.; Haegert, A.; Wu, R.; Brahmbhatt, S.; Mo, F.; Jong, L. High Fidelity Patient-Derived Xenografts for Accelerating Prostate Cancer Discovery and Drug Development Next-Generation Models of Prostate Cancer. Cancer Res. 2014, 74, 1272–1283. [Google Scholar] [CrossRef]
- Mathew, R.K.; Rutka, J.T. Diffuse intrinsic pontine glioma: Clinical features, molecular genetics, and novel targeted therapeutics. J. Korean Neurosurg. Soc. 2018, 61, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Moore, J.C.; Ignatius, M.S.; Tenente, I.M.; Hayes, M.N.; Garcia, E.G.; Torres Yordán, N.; Bourque, C.; He, S.; Blackburn, J.S. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 2016, 7, 10358. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Brunson, D.C.; Tang, Q.; Do, D.; Iftimia, N.A.; Moore, J.C.; Hayes, M.N.; Welker, A.M.; Garcia, E.G.; Dubash, T.D. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 2019, 177, 1903–1914.e14. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, M.; Póvoa, V.; Fior, R. Generation of zebrafish larval xenografts and tumor behavior analysis. JoVE J. Vis. Exp. 2021, 172, e62373. [Google Scholar]
- Yan, C.; Do, D.; Yang, Q.; Brunson, D.C.; Rawls, J.F.; Langenau, D.M. Single-cell imaging of human cancer xenografts using adult immunodeficient zebrafish. Nat. Protoc. 2020, 15, 3105–3128. [Google Scholar] [CrossRef]
- Saland, E.; Boutzen, H.; Castellano, R.; Pouyet, L.; Griessinger, E.; Larrue, C.; De Toni, F.; Scotland, S.; David, M.; Danet-Desnoyers, G. A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J. 2015, 5, e297. [Google Scholar] [CrossRef]
- Gamble, J.T.; Elson, D.J.; Greenwood, J.A.; Tanguay, R.L.; Kolluri, S.K. The zebrafish xenograft models for investigating cancer and cancer therapeutics. Biology 2021, 10, 252. [Google Scholar] [CrossRef]
- Mendonça-Gomes, J.M.; Valverde, T.M.; da Mata Martins, T.M.; Charlie-Silva, I.; Padovani, B.N.; Fénero, C.M.; da Silva, E.M.; Domingues, R.Z.; Melo-Hoyos, D.C.; Corrêa-Junior, J.D. Long-term dexamethasone treatment increases the engraftment efficiency of human breast cancer cells in adult zebrafish. Fish Shellfish. Immunol. Rep. 2021, 2, 100007. [Google Scholar] [CrossRef]
- Traver, D.; Winzeler, A.; Stern, H.M.; Mayhall, E.A.; Langenau, D.M.; Kutok, J.L.; Look, A.T.; Zon, L.I. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 2004, 104, 1298–1305. [Google Scholar] [CrossRef]
- Stoletov, K.; Montel, V.; Lester, R.D.; Gonias, S.L.; Klemke, R. High-resolution imaging of the dynamic tumor cell–vascular interface in transparent zebrafish. Proc. Natl. Acad. Sci. USA 2007, 104, 17406–17411. [Google Scholar] [CrossRef]
- Moore, J.C.; Tang, Q.; Yordán, N.T.; Moore, F.E.; Garcia, E.G.; Lobbardi, R.; Ramakrishnan, A.; Marvin, D.L.; Anselmo, A.; Sadreyev, R.I. Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J. Exp. Med. 2016, 213, 2575–2589. [Google Scholar] [CrossRef]
- Fior, R.; Póvoa, V.; Mendes, R.V.; Carvalho, T.; Gomes, A.; Figueiredo, N.; Ferreira, M.G. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl. Acad. Sci. USA 2017, 114, E8234–E8243. [Google Scholar] [CrossRef]
- Costa, B.; Estrada, M.F.; Barroso, M.T.; Fior, R. Zebrafish Patient-Derived Avatars from Digestive Cancers for Anti-cancer Therapy Screening. Curr. Protoc. 2022, 2, e415. [Google Scholar] [CrossRef]
- Weintraub, A. All eyes on zebrafish. Lab. Anim. 2017, 46, 323–326. [Google Scholar] [CrossRef]
- Aparicio, S.; Hidalgo, M.; Kung, A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 2015, 15, 311–316. [Google Scholar] [CrossRef]
- Dockins, S.R.; Blackburn, J.S.; Haney, M.G.; Wilson, B. Optimization of human cancer cell xenografts into zebrafish larvae for high-throughput drug screening. Cancer Res. 2019, 79, 3697. [Google Scholar]
- Spaink, H.P.; Cui, C.; Wiweger, M.I.; Jansen, H.J.; Veneman, W.J.; Marín-Juez, R.; de Sonneville, J.; Ordas, A.; Torraca, V.; van der Ent, W. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 2013, 62, 246–254. [Google Scholar] [CrossRef]
- Pulak, R. Tools for automating the imaging of zebrafish larvae. Methods 2016, 96, 118–126. [Google Scholar] [CrossRef]
- Ghotra, V.P.; He, S.; De Bont, H.; van Der Ent, W.; Spaink, H.P.; van De Water, B.; Snaar-Jagalska, B.E.; Danen, E.H. Automated whole animal bio-imaging assay for human cancer dissemination. PLoS ONE 2012, 7, e31281. [Google Scholar] [CrossRef]
- Asokan, N.; Daetwyler, S.; Bernas, S.N.; Schmied, C.; Vogler, S.; Lambert, K.; Wobus, M.; Wermke, M.; Kempermann, G.; Huisken, J. Long-term in vivo imaging reveals tumor-specific dissemination and captures host tumor interaction in zebrafish xenografts. Sci. Rep. 2020, 10, 13254. [Google Scholar]
- Haney, M.G.; Moore, L.H.; Blackburn, J.S. Drug screening of primary patient derived tumor xenografts in zebrafish. JoVE J. Vis. Exp. 2020, e60996. [Google Scholar]
- Mercatali, L.; La Manna, F.; Groenewoud, A.; Casadei, R.; Recine, F.; Miserocchi, G.; Pieri, F.; Liverani, C.; Bongiovanni, A.; Spadazzi, C. Development of a patient-derived xenograft (PDX) of breast cancer bone metastasis in a zebrafish model. Int. J. Mol. Sci. 2016, 17, 1375. [Google Scholar] [CrossRef] [PubMed]
- Almstedt, E.; Rosén, E.; Gloger, M.; Stockgard, R.; Hekmati, N.; Koltowska, K.; Krona, C.; Nelander, S. Real-time evaluation of glioblastoma growth in patient-specific zebrafish xenografts. Neuro-Oncology 2022, 24, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Phase I Study of Olaparib and Temozolomide for Ewings Sarcoma or Rhabdoomyosarcoma. Available online: https://clinicaltrials.gov/ct2/show/record/NCT01858168 (accessed on 15 December 2022).
- Albuquerque, C.; Vanneschi, L.; Henriques, R.; Castelli, M.; Póvoa, V.; Fior, R.; Papanikolaou, N. Object detection for automatic cancer cell counting in zebrafish xenografts. PLoS ONE 2021, 16, e0260609. [Google Scholar] [CrossRef]
- Zhang, B.; Shimada, Y.; Kuroyanagi, J.; Umemoto, N.; Nishimura, Y.; Tanaka, T. Quantitative phenotyping-based in vivo chemical screening in a zebrafish model of leukemia stem cell xenotransplantation. PLoS ONE 2014, 9, e85439. [Google Scholar] [CrossRef]
- Zhang, B.; Shimada, Y.; Kuroyanagi, J.; Nishimura, Y.; Umemoto, N.; Nomoto, T.; Shintou, T.; Miyazaki, T.; Tanaka, T. Zebrafish xenotransplantation model for cancer stem-like cell study and high-throughput screening of inhibitors. Tumor Biol. 2014, 35, 11861–11869. [Google Scholar] [CrossRef]
- Veneman, W.J.; Stockhammer, O.W.; De Boer, L.; Zaat, S.A.; Meijer, A.H.; Spaink, H.P. A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genom. 2013, 14, 255. [Google Scholar] [CrossRef]
- Haldi, M.; Ton, C.; Seng, W.L.; McGrath, P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis 2006, 9, 139–151. [Google Scholar] [CrossRef]
- Nicoli, S.; Ribatti, D.; Cotelli, F.; Presta, M. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res. 2007, 67, 2927–2931. [Google Scholar] [CrossRef]
- Wu, J.-Q.; Zhai, J.; Li, C.-Y.; Tan, A.-M.; Wei, P.; Shen, L.-Z.; He, M.-F. Patient-derived xenograft in zebrafish embryos: A new platform for translational research in gastric cancer. J. Exp. Clin. Cancer Res. 2017, 36, 160. [Google Scholar] [CrossRef]
- Ali, Z.; Vildevall, M.; Rodriguez, G.V.; Tandiono, D.; Vamvakaris, I.; Evangelou, G.; Lolas, G.; Syrigos, K.N.; Villanueva, A.; Wick, M. Zebrafish patient-derived xenograft models predict lymph node involvement and treatment outcome in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2022, 41, 58. [Google Scholar] [CrossRef]
- Usai, A.; Di Franco, G.; Piccardi, M.; Cateni, P.; Pollina, L.E.; Vivaldi, C.; Vasile, E.; Funel, N.; Palmeri, M.; Dente, L. Zebrafish patient-derived xenografts identify chemo-response in pancreatic ductal adenocarcinoma patients. Cancers 2021, 13, 4131. [Google Scholar] [CrossRef]
- Ai, X.; Ye, Z.; Xiao, C.; Zhong, J.; Lancman, J.J.; Chen, X.; Pan, X.; Yang, Y.; Zhou, L.; Wang, X. Clinically relevant orthotopic xenograft models of patient-derived glioblastoma in zebrafish. Dis. Model. Mech. 2022, 15, dmm049109. [Google Scholar] [CrossRef]
- Tulotta, C.; Stefanescu, C.; Beletkaia, E.; Bussmann, J.; Tarbashevich, K.; Schmidt, T.; Snaar-Jagalska, B.E. Inhibition of signaling between human CXCR4 and zebrafish ligands by the small molecule IT1t impairs the formation of triple-negative breast cancer early metastases in a zebrafish xenograft model. Dis. Model. Mech. 2016, 9, 141–153. [Google Scholar]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Stachura, D.L.; Reyes, J.R.; Bartunek, P.; Paw, B.H.; Zon, L.I.; Traver, D. Zebrafish kidney stromal cell lines support multilineage hematopoiesis. Blood J. Am. Soc. Hematol. 2009, 114, 279–289. [Google Scholar] [CrossRef]
- Rajan, V.; Melong, N.; Wong, W.H.; King, B.; Tong, R.S.; Mahajan, N.; Gaston, D.; Lund, T.; Rittenberg, D.; Dellaire, G. Humanized zebrafish enhance human hematopoietic stem cell survival and promote acute myeloid leukemia clonal diversity. Haematologica 2020, 105, 2391. [Google Scholar] [CrossRef]
- Pype, C.; Verbueken, E.; Saad, M.A.; Casteleyn, C.R.; Van Ginneken, C.J.; Knapen, D.; Van Cruchten, S.J. Incubation at 32.5 C and above causes malformations in the zebrafish embryo. Reprod. Toxicol. 2015, 56, 56–63. [Google Scholar] [CrossRef]
- Astin, J.; Keerthisinghe, P.; Du, L.; Sanderson, L.; Crosier, K.; Crosier, P.; Hall, C. Innate immune cells and bacterial infection in zebrafish. Methods Cell Biol. 2017, 138, 31–60. [Google Scholar]
- Cabezas-Sáinz, P.; Pensado-López, A.; Sáinz, B., Jr.; Sánchez, L. Modeling cancer using zebrafish xenografts: Drawbacks for mimicking the human microenvironment. Cells 2020, 9, 1978. [Google Scholar] [CrossRef]
- Lal, S.; La Du, J.; Tanguay, R.L.; Greenwood, J.A. Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J. Neurosci. Res. 2012, 90, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Farin, A.; Suzuki, S.O.; Weiker, M.; Goldman, J.E.; Bruce, J.N.; Canoll, P. Transplanted glioma cells migrate and proliferate on host brain vasculature: A dynamic analysis. Glia 2006, 53, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Rennekamp, A.J.; Peterson, R.T. 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 2015, 24, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.J.; Xu, Y.Q.; He, J.H.; Yu, H.P.; Huang, C.J.; Gao, J.M.; Dong, Q.X.; Xuan, Y.X.; Li, C.Q. Human cardiotoxic drugs delivered by soaking and microinjection induce cardiovascular toxicity in zebrafish. J. Appl. Toxicol. 2014, 34, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Saneja, A. Zebrafish as a powerful alternative model organism for pre-clinical investigation of nanomedicines. Drug Discov. Today 2022, 27, 1513–1522. [Google Scholar]
- de Souza Anselmo, C.; Sardela, V.F.; de Sousa, V.P.; Pereira, H.M.G. Zebrafish (Danio rerio): A valuable tool for predicting the metabolism of xenobiotics in humans? Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 212, 34–46. [Google Scholar] [CrossRef]
- Guarin, M.; Faelens, R.; Giusti, A.; De Croze, N.; Léonard, M.; Cabooter, D.; Annaert, P.; de Witte, P.; Ny, A. Spatiotemporal imaging and pharmacokinetics of fluorescent compounds in zebrafish eleuthero-embryos after different routes of administration. Sci. Rep. 2021, 11, 12229. [Google Scholar] [CrossRef]
- Goldstone, J.V.; McArthur, A.G.; Kubota, A.; Zanette, J.; Parente, T.; Jönsson, M.E.; Nelson, D.R.; Stegeman, J.J. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genom. 2010, 11, 643. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish disease models in drug discovery: From pre-clinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef]
- Cully, M. Zebrafish earn their drug discovery stripes. Nat. Rev. Drug Discov. 2019, 18, 811–814. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef]
- Fleming, A.; Alderton, W. Zebrafish in pharmaceutical industry research: Finding the best fit. Drug Discov. Today Dis. Model. 2013, 10, e43–e50. [Google Scholar] [CrossRef]
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005, 4, 35–44. [Google Scholar] [CrossRef]
- Fazio, M.; Ablain, J.; Chuan, Y.; Langenau, D.M.; Zon, L.I. Zebrafish patient avatars in cancer biology and precision cancer therapy. Nat. Rev. Cancer 2020, 20, 263–273. [Google Scholar] [CrossRef]
- Gatzweiler, C.; Ridinger, J.; Herter, S.; Gerloff, X.F.; ElHarouni, D.; Berker, Y.; Imle, R.; Schmitt, L.; Kreth, S.; Stainczyk, S. Functional therapeutic target validation using pediatric zebrafish xenograft models. Cancers 2022, 14, 849. [Google Scholar] [CrossRef]
- Di Franco, G.; Usai, A.; Funel, N.; Palmeri, M.; Montesanti, I.E.R.; Bianchini, M.; Gianardi, D.; Furbetta, N.; Guadagni, S.; Vasile, E. Use of zebrafish embryos as avatar of patients with pancreatic cancer: A new xenotransplantation model towards personalized medicine. World J. Gastroenterol. 2020, 26, 2792. [Google Scholar] [CrossRef]
- Di Franco, G.; Usai, A.; Piccardi, M.; Cateni, P.; Palmeri, M.; Pollina, L.E.; Gaeta, R.; Marmorino, F.; Cremolini, C.; Dente, L. Zebrafish Patient-Derived Xenograft Model to Predict Treatment Outcomes of Colorectal Cancer Patients. Biomedicines 2022, 10, 1474. [Google Scholar] [CrossRef]
- Usai, A.; Di Franco, G.; Colucci, P.; Pollina, L.E.; Vasile, E.; Funel, N.; Palmeri, M.; Dente, L.; Falcone, A.; Morelli, L. A model of a zebrafish avatar for co-clinical trials. Cancers 2020, 12, 677. [Google Scholar] [CrossRef]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018, 172, 373–386.e310. [Google Scholar] [CrossRef]
- Bose, S.; Clevers, H.; Shen, X. Promises and challenges of organoid-guided precision medicine. Med 2021, 2, 1011–1026. [Google Scholar] [CrossRef]
- Worst, B.C.; van Tilburg, C.M.; Balasubramanian, G.P.; Fiesel, P.; Witt, R.; Freitag, A.; Boudalil, M.; Previti, C.; Wolf, S.; Schmidt, S. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur. J. Cancer 2016, 65, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.; Ferreira, S.; Póvoa, V.; Cardoso, M.J.; Vieira, S.; Stroom, J.; Fidalgo, P.; Rio-Tinto, R.; Figueiredo, N.; Parés, O. Developments in zebrafish avatars as radiotherapy sensitivity reporters—Towards personalized medicine. EBioMedicine 2020, 51, 102578. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Wu, X.; Xu, K.; Zhan, P.; Liu, H.; Zhang, F.; Lv, T.; Song, Y. Zebrafish patient-derived xenografts accurately and quickly reproduce treatment outcomes in non-small cell lung cancer patients. Exp. Biol. Med. 2022, 15353702221142612. [Google Scholar] [CrossRef] [PubMed]
- Rebelo de Almeida, C.; Mendes, R.V.; Pezzarossa, A.; Gago, J.; Carvalho, C.; Alves, A.; Nunes, V.; Brito, M.J.; Cardoso, M.J.; Ribeiro, J. Zebrafish xenografts as a fast screening platform for bevacizumab cancer therapy. Commun. Biol. 2020, 3, 299. [Google Scholar] [CrossRef] [PubMed]
- Lampreht Tratar, U.; Horvat, S.; Cemazar, M. Transgenic mouse models in cancer research. Front. Oncol. 2018, 8, 268. [Google Scholar] [CrossRef]
- Consortium, E.P. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J.; Cai, S.; Clarke, R.B. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573. [Google Scholar] [CrossRef]
- Pearson, A.T.; Finkel, K.A.; Warner, K.A.; Nör, F.; Tice, D.; Martins, M.D.; Jackson, T.L.; Nör, J.E. Patient-derived xenograft (PDX) tumors increase growth rate with time. Oncotarget 2016, 7, 7993. [Google Scholar] [CrossRef]
- Perez, D.R.; Nickl, C.K.; Waller, A.; Delgado-Martin, C.; Woods, T.; Sharma, N.D.; Hermiston, M.L.; Loh, M.L.; Hunger, S.P.; Winter, S.S. High-throughput flow cytometry identifies small-molecule inhibitors for drug repurposing in T-alL. SLAS DISCOVERY Adv. Life Sci. RD 2018, 23, 732–741. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.-Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef]
- Belderbos, M.E.; Koster, T.; Ausema, B.; Jacobs, S.; Sowdagar, S.; Zwart, E.; de Bont, E.; de Haan, G.; Bystrykh, L.V. Clonal selection and asymmetric distribution of human leukemia in murine xenografts revealed by cellular barcoding. Blood J. Am. Soc. Hematol. 2017, 129, 3210–3220. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Cox, C.L.; Eirew, P.; Knapp, D.J.; Pellacani, D.; Kannan, N.; Carles, A.; Moksa, M.; Balani, S.; Shah, S. DNA barcoding reveals diverse growth kinetics of human breast tumour subclones in serially passaged xenografts. Nat. Commun. 2014, 5, 5871. [Google Scholar] [CrossRef]
- Shi, J.; Li, Y.; Jia, R.; Fan, X. The fidelity of cancer cells in PDX models: Characteristics, mechanism and clinical significance. Int. J. Cancer 2020, 146, 2078–2088. [Google Scholar] [CrossRef]
- Greaves, M. Evolutionary determinants of cancer. Cancer Discov. 2015, 5, 806–820. [Google Scholar] [CrossRef]
- Seidlitz, T.; Koo, B.-K.; Stange, D.E. Gastric organoids—An in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ. 2021, 28, 68–83. [Google Scholar] [CrossRef]
- Blackburn, J.S.; Langenau, D.M. Zebrafish as a model to assess cancer heterogeneity, progression and relapse. Dis. Model. Mech. 2014, 7, 755–762. [Google Scholar] [CrossRef]
- Taylor, K.L.; Grant, N.J.; Temperley, N.D.; Patton, E.E. Small molecule screening in zebrafish: An in vivo approach to identifying new chemical tools and drug leads. Cell Commun. Signal. 2010, 8, 11. [Google Scholar] [CrossRef]
- Phillips, J.B.; Westerfield, M. Zebrafish models in translational research: Tipping the scales toward advancements in human health. Dis. Model. Mech. 2014, 7, 739–743. [Google Scholar] [CrossRef]
- Yoganantharjah, P.; Gibert, Y. The use of the zebrafish model to aid in drug discovery and target validation. Curr. Top. Med. Chem. 2017, 17, 2041–2055. [Google Scholar] [CrossRef]
- Lam, P.-Y.; Peterson, R.T. Developing zebrafish disease models for in vivo small molecule screens. Curr. Opin. Chem. Biol. 2019, 50, 37–44. [Google Scholar] [CrossRef]
| Model | Current Status | Advantages | Disadvantages |
|---|---|---|---|
| PDO | PDO are used less frequently in terms of predicting patient outcomes compared to animal models. |
|
|
| Mouse PDX | Large number of models are established and well-characterized for different cancers with high predictive ability of patient response. However, applications for the first line of therapy determination are limited due to long engraftment timelines. |
|
|
| Zebrafish PDX | Larval and adult zPDX models are established for several cancers and can predict patient treatment response. Many steps of the PDX process can be automated allowing for large-scale drug screens. |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Hamaly, M.A.; Turner, L.T.; Rivera-Martinez, A.; Rodriguez, A.; Blackburn, J.S. Zebrafish Cancer Avatars: A Translational Platform for Analyzing Tumor Heterogeneity and Predicting Patient Outcomes. Int. J. Mol. Sci. 2023, 24, 2288. https://doi.org/10.3390/ijms24032288
Al-Hamaly MA, Turner LT, Rivera-Martinez A, Rodriguez A, Blackburn JS. Zebrafish Cancer Avatars: A Translational Platform for Analyzing Tumor Heterogeneity and Predicting Patient Outcomes. International Journal of Molecular Sciences. 2023; 24(3):2288. https://doi.org/10.3390/ijms24032288
Chicago/Turabian StyleAl-Hamaly, Majd A., Logan T. Turner, Angelica Rivera-Martinez, Analiz Rodriguez, and Jessica S. Blackburn. 2023. "Zebrafish Cancer Avatars: A Translational Platform for Analyzing Tumor Heterogeneity and Predicting Patient Outcomes" International Journal of Molecular Sciences 24, no. 3: 2288. https://doi.org/10.3390/ijms24032288
APA StyleAl-Hamaly, M. A., Turner, L. T., Rivera-Martinez, A., Rodriguez, A., & Blackburn, J. S. (2023). Zebrafish Cancer Avatars: A Translational Platform for Analyzing Tumor Heterogeneity and Predicting Patient Outcomes. International Journal of Molecular Sciences, 24(3), 2288. https://doi.org/10.3390/ijms24032288