Abstract
Micromelalopha troglodyta (Graeser) (Lepidoptera: Notodontidae) is a notorious pest of poplar. Coevolution with poplars rich in plant secondary metabolites prompts M. troglodyta to expand effective detoxification mechanisms against toxic plant secondary metabolites. Although glutathione S-transferases (GSTs) play an important role in xenobiotic detoxification in M. troglodyta, it is unclear how GSTs act in response to toxic secondary metabolites in poplar. In this study, five GST gene core promoters were accurately identified by a 5’ loss luciferase reporter assay, and the core promoters were significantly induced by two plant secondary metabolites in vitro. Two transcription factors, cap ‘n’ collar C (CncC) and aryl hydrocarbon receptor nuclear translocator (ARNT), were cloned in M. troglodyta. MtCncC and MtARNT clustered well with other insect CncCs and ARNTs, respectively. In addition, MtCncC and MtARNT could bind the MtGSTt1 promoter and strongly improve transcriptional activity, respectively. However, MtCncC and MtARNT had no regulatory function on the MtGSTz1 promoter. Our findings revealed the molecular mechanisms of the transcription factors MtCncC and MtARNT in regulating the GST genes of M. troglodyta. These results provide useful information for the control of M. troglodyta.
1. Introduction
Plants are the main food source for animals, while insects serve as important consumers of plants. They constantly fight against each other. From a plant perspective, many plants produce defensive toxins or inhibitors to repel insects, including secondary metabolites such as isoflavones, furanocoumarins, terpenoids, alkaloids and cyanogenic glycosides [1]. From an insect perspective, insects are protected from plant secondary metabolites mainly through increased physiological tolerance, metabolic capacity of the detoxification system, or behavioral avoidance [2]. As insects attempt to increase new host plant species, these mechanisms will continue to evolve [3].
Micromelalopha troglodyta (Graeser), which is mainly found in China, is an important leaf-feeding pest of poplar trees and can be widely spread causing heavy losses to the forestry industry [4,5]. Poplar secondary metabolites such as tannic acid and quercetin, as toxic natural products, have a toxic effect on M. troglodyta [6,7]. It is well known that the powerful detoxification metabolism mechanism of insects is an important way to overcome plant chemicals [8]. There are several enzymes involved in metabolizing heterologous substances and converting them into less toxic hydrophilic compounds. Major enzyme superfamilies include glutathione S-transferases (GSTs), cytochrome P450 monooxygenases (P450s), ATP-binding cassette (ABC) transporters, UDP-glycosyltransferases (UGTs), and carboxylesterases (CarEs) [9]. GSTs are important detoxifying enzymes in M. troglodyta that are involved in the metabolism of plant secondary metabolites [6,8]. Mounting evidence suggests that GSTs can detoxify secondary metabolites produced by plants during the insect feeding process [10,11]. GST activity of Myzus persicae could be induced by different host plants of Brassica species, suggesting that GSTs might play roles in host plant adaptation in M. persicase [12]. Multiple poplar secondary metabolites could induce the expression of GST genes in Lymantria dispar, and the adaptation of L. dispar to plant secondary metabolites was reduced after interference with GST genes [13]. Red palm weevils could excrete and degrade a variety of toxic plant secondary metabolites by upregulating GST genes [14]. Hence, these findings demonstrated that GSTs play an important role in participating in the metabolism of xenobiotics in pests.
The detoxification genes are transcriptionally activated by a common mechanism of transcription factors [15]. Transcription factors are a class of DNA-binding proteins, including cis- and trans-acting factors, that can enhance or repress the initiation of gene transcription by binding to specific promoter sequences [16]. Cap ‘n’ collar C (CncC) and aryl hydrocarbon receptor nuclear translocator (ARNT) are two important transcription factors in insects, with an essential role in the regulation of detoxification enzymes [15,16,17,18]. Mammalian nuclear factor erythroid 2-related Factor 2 (Nrf2), a homolog of CncC, is able to induce the expression of detoxification genes in response to stress from xenobiotic substances [19,20]. CncC regulates the expression of many detoxification genes (GSTs, P450s and esterases) involved in insect metabolism and detoxification [20,21]. Spodoptera litura Nrf2 acted as a cis-regulatory element that activated the excretion and degradation of indole-3-methanol (I3C) and chlorpyrifos by upregulating GSTe1, which could improve the response of S. litura to phytochemicals and insecticides [22]. In Leptinotarsa decemlineata, several imidacloprid-resistant GST genes were regulated by the transcription factors CncC and Maf [23]. The aryl hydrocarbon receptor (AhR) and ARNT are members of the bHLH-PAS family of transcription factors. They are ligand-activated transcription factors involved in the perception and delivery of stimuli in response to endogenous and xenobiotic stress [24,25]. Enhancer sequences of phase I and phase II genes (such as P450s and GSTs) can be recognized and activated by the AhR-ARNT complex [26]. The Spodoptera exigua transcription factors AhR and ARNT coordinately regulated the expression of multiple GSTs conferring insecticide resistance [27]. In locusts, AhR could improve tolerance to chlorpyrifos by increasing the expression of LmGSTd7 [28]. Combining the information above, CncC and ARNT are important transcriptional regulators of insect GST genes, and they play an important role in governing the expression of GSTs. However, the mode of regulation of GST genes by the two transcription factors in M. troglodyta is not clear.
We cloned five GST promoters of M. troglodyta and demonstrated that tannic acid could induce the activity of these five MtGST promoters [4]. To date, it has not been reported at home or abroad how the expression of GST in M. troglodyta is regulated to adapt to plant secondary metabolite stress. In this study, we focused on the following three questions: (1) Where is the core region of the MtGST promoter located? (2) Will the core region of the MtGST promoter be responsive to plant secondary metabolites? and (3) Do MtCncC and MtARNT interact with the promoters of GST? This study elucidated the role of MtCncC and MtARNT in regulating GST metabolism of plant secondary metabolites, which would be helpful to find new target genes to control M. troglodyta.
2. Results
2.1. MtGST Promoter Activity Analysis
In our previous study, we cloned five MtGST promoters. To determine the regions that are essential for GST expression, we constructed several deletion structures containing 5’ loss fragments and inserted them into the pGL4.10-Basic vector (Figure 1).
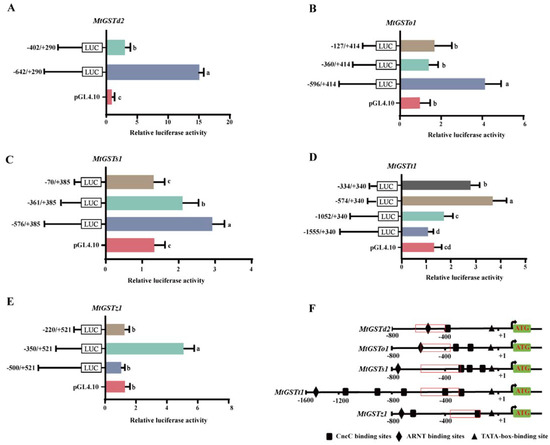
Figure 1.
Relative luciferase activities of the 5’ loss fragment of MtGST promoters. (A): MtGSTd2 promoter constructs; (B): MtGSTo1 promoter constructs; (C): MtGSTs1 promoter constructs; (D): MtGSTt1 promoter constructs; (E): MtGSTz1 promoter constructs; F: Prediction analysis of the promoter regions of five MtGST genes. Each value is presented as the mean ± SD of three replicates, and different lowercase letters show significant differences (p < 0.05). (F): The core regions deduced from five MtGST promoters. The location of nucleotides was marked relative to the transcription start site indicated by +1 and the translation start site indicated by ATG. The red box represents the core region of each promoter.
The pGL4.10-(−642/+290) vector of MtGSTd2 promoter had relatively high luciferase expression compared with that of pGL4.10-(−402/+290), which showed that sequences were essential for the transcription of MtGSTd2 located in the 240 bp region between −642 and −402 bp (Figure 1A). The pGL4.10-(−596/+414) vector of MtGSTo1 promoter exhibited relatively high luciferase expression compared with that of pGL4.10-(−360/+414) and pGL4.10-(−127/+414), which suggested that −596 to −360 region was critical for MtGSTo1 transcription (Figure 1B). The pGL4.10-(−576/+385) vector of MtGSTs1 promoter displayed relatively high luciferase expression than that of the pGL4.10-(−361/+385), and pGL4.10-(−70/+385) of MtGSTs1 had low luciferase expression compared to pGL4.10-(−361/+385), which suggested that sequences were essential for the transcription of MtGSTs1 located in the 215 bp region between −576 and −361 bp (Figure 1C). The pGL4.10-(−574/+340) vector of MtGSTt1 promoter had relatively high luciferase expression compared with that of pGL4.10-(−334/+340), and the pGL4.10-(−334/+340) vector of MtGSTt1 had high luciferase expression levels compared to pGL4.10-(−1052/+340) and pGL4.10-(−1555/+340), which showed that the 240 bp transcription region between −574 and −334 was critical for MtGSTt1 expression (Figure 1D). The pGL4.10-(−350/+521) vector of MtGSTz1 promoter had relatively high luciferase expression compared with that of pGL4.10-(−500/+521) and pGL4.10-(−220/+521), which suggested that sequences between −350 and −220 were critical for MtGSTz1 expression (Figure 1E). The main regulatory regions of the five MtGST promoters were shown in Figure 1F.
2.2. Induction Effect of Tannic Acid and Quercetin on the Core Region of Two Promoters
Due to the high transcriptional activity of MtGSTt1 (−574/+340) and MtGSTz1 (−350/+521) promoters, we used them as examples to explore the promoter response to tannic acid and quercetin (Figure 2 and Figure 3). The promoter activity of MtGSTt1 (−574/+340) was significantly induced by tannic acid at a low concentration (0.56 mg/L), while it was inhibited by tannic acid at 2.8, 14 and 70 mg/L (Figure 2A). The MtGSTt1 (−574/+340) promoter showed an increasing and then decreasing trend under quercetin stress, and the highest promoter activity was observed when the quercetin concentration was 7 mg/L (Figure 2B). These data showed that the promoter activity of the core region of MtGSTt1 (−574/+340) could be induced by tannic acid and quercetin. For MtGSTz1 (−350/+521) promoter, the promoter activity was strongly induced by 2.8 mg/L tannic acid (Figure 3A), while it was repressed at 70 mg/L tannic acid (Figure 3A). We also observed that the promoter activity of MtGSTz1 (−350/+521) was notably increased when Sf9 cells were treated with quercetin at 0.28, 1.4 and 7 mg/L and sharply repressed at 35 mg/L (Figure 3B). These data suggested that tannic acid and quercetin could influence the core region of MtGSTz1 promoter.
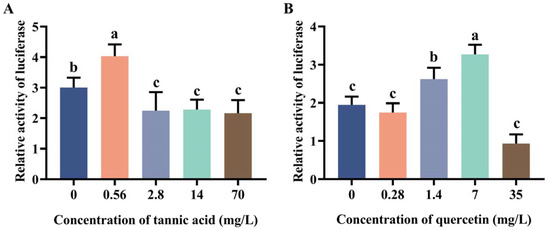
Figure 2.
Promoter activity of MtGSTt1 (−574/+340) under tannic acid (A) and quercetin (B) stress. Each value is presented as the mean ± SD of three replicates, and different lowercase letters show significant differences (p < 0.05).
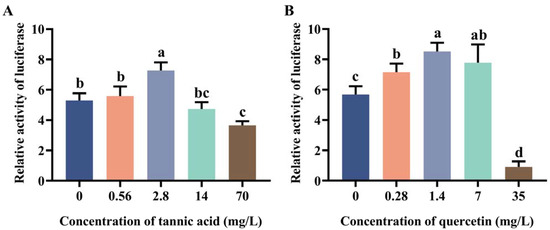
Figure 3.
Promoter activity of MtGSTz1 (−350/+521) under tannic acid (A) and quercetin (B) stress. Each value is presented as the mean ± SD of three replicates, and different lowercase letters show significant differences (p < 0.05).
2.3. Cloning and Phylogenetic Analysis of MtCncC
To explore the regulatory mode of promoters by transcription factors. The CncC gene of M. troglodyta was obtained from the transcriptome database and confirmed by cloning and resequencing. MtCncC is a 1671 bp open reading frame (ORF) sequence and encodes 556 AA residues. The amino acid sequence of MtCncC is listed in Supplemental Material S1. Twenty-six representative insect CncC amino acid sequences were selected for constructing the phylogenetic tree using the maximum likelihood (ML) method of MEGA X based on the multiple alignment built with Clustal W. The phylogenetic analysis showed that CncCs from different species were classified into six order clusters, and MtCncC was clearly classified into Lepidoptera subclusters, which suggested that MtCncC had a high identity with other lepidopteran insects CncCs (Figure 4).
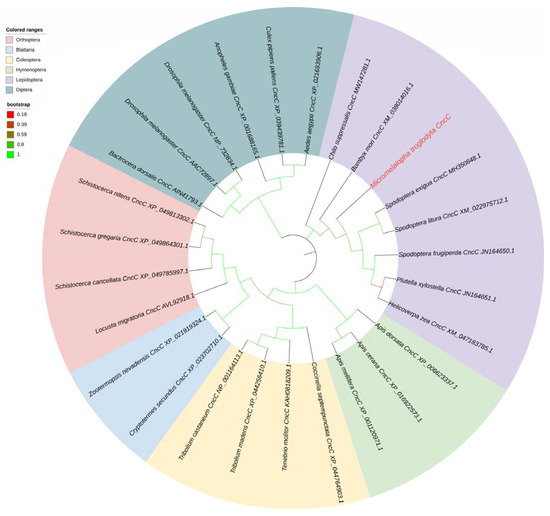
Figure 4.
Phylogenetic analysis of the CncC sequences of several insect species. The CncC gene name was shown as the Latin name of the species, CncC and the NCBI gene accession number, and the M. troglodyta CncC was marked in red and bold. The branches for Lepidoptera, Diptera, Blattodea, Coleoptera, Hymenoptera, and Orthoptera CncCs were shaded in different colors, and branch colors represented bootstrap values.
2.4. Cloning and Phylogenetic Analysis of MtARNT
Another transcription factor ARNT was also identified from the transcriptome database and confirmed by cloning and resequencing. MtARNT is a 1440 bp ORF sequence and encodes 479 AA residues. The amino acid sequence of MtARNT is listed in Supplemental Material S1. The phylogenetic tree of MtARNT was built according to the above method. Phylogenetic analysis showed that MtARNT was grouped into Lepidoptera subclusters and had a high similarity with ARNTs from other insects (Figure 5).
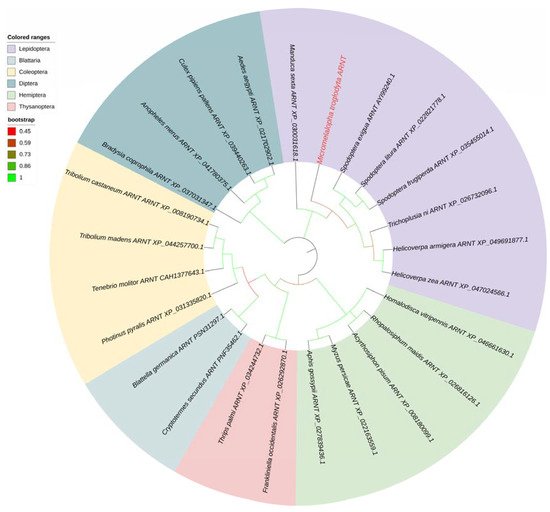
Figure 5.
Phylogenetic analysis of the ARNT sequences of several insect species. The ARNT gene name was shown as the Latin name of the species, ARNT and the NCBI gene accession number, and the M. troglodyta ARNT was marked in red and bold. The branches for Lepidoptera, Diptera, Blattodea, Coleoptera, Hymenoptera, and Thysanoptera ARNTs were shaded in different colors, and branch colors represented bootstrap values.
2.5. Analysis of the Transcriptional Activity of MtGST Promoters Regulated by MtCncC/MtARNT
To further determine the interaction relationship between the MtCncC/MtARNT and MtGST promoters, we selected the MtGSTt1 (−574/+340) and MtGSTz1 (−350/+521) promoters with high transcription activity to verify the regulatory role of ARNT and CncC. pGL4.10-MtGSTt1 (−574/+340) and pGL4.10-MtGSTz1 (−350/+521) promoters were transfected into Sf9 cells. MtCncC and MtARNT sequences were constructed into pAC-V5 basic expression vector and named pAC-CncC and pAC-ARNT, respectively. The pAC-V5 basic expression vector was used as a negative control. The transcription factors MtCncC/MtARNT and the promoter constructs were cotransfected into Sf9 cells. As shown in Figure 6A, the luciferase activity of pGL4.10-MtGSTt1 (−574/+340) promoter strongly increased when cotransfected with pAC-CncC or pAC-ARNT. These results showed that the expression of MtCncC/MtARNT in Sf9 cells facilitates MtGSTt1 (−574/+340) promoter transcription, but there was no significant change when cotransfected with pAC-CncC and pAC-ARNT. When pAC-CncC/pAC-ARNT was cotransfected with pGL4.10-MtGSTz1 (−350/+521) promoter, the transcriptional activity of the MtGSTz1 (−350/+521) promoter was not increased (Figure 6B).
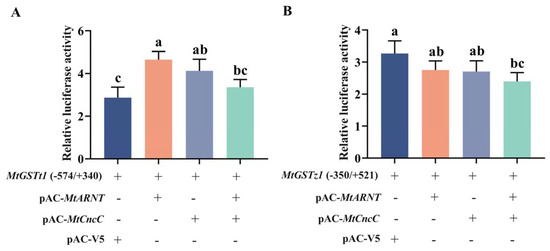
Figure 6.
Relative luciferase activity in cells transfected with the MtGST promoters, MtCncC and MtARNT. (A) MtGSTt1 (−574/+340) promoter; (B) MtGSTz1 (−350/+521) promoter. Each promoter activity was measured in the absence of protein (promoter + empty pAC-V5 basic vector), in the presence of a protein (MtARNT or MtCncC) or in the presence of two proteins (MtCncC and MtARNT). Each value is presented as the mean ± SD of three replicates, and lowercase letters show significant differences (p < 0.05).
3. Discussion
GSTs are broadly distributed and important detoxifying enzymes in aerobic organisms that catalyze glutathione (GSH) binding to endogenous and exogenous compounds and excrete them outside cells, thus reducing their damage to the cells [29,30]. In insects, GSTs are a family of multifunctional enzymes involved in the detoxification of toxic compounds, including plant secondary metabolites [4,31,32]. There are six cytoplasmic GST gene families, including epsilon, omega, delta, theta, sigma, and zeta [33]. In a previous study, we demonstrated that all five GST genes of M. troglodyta could be significantly induced by tannic acid, which belong to the omega, delta, theta, sigma, and zeta families. Subsequently, we cloned the 5’ flanking promoter sequences of these five GST genes and found that they could be induced by tannic acid [4].
The promoter is a very important regulatory element in gene transcription that determines the pattern and intensity of gene expression [34]. Many inducible promoters have been identified from insects, plants and pathogens to explore the in-depth mechanisms of their regulation [35,36,37]. The promoter of Drosophila heat shock protein (Hsp70) could enhance the expression of Hsp70 more than 200-fold after heat stimulation treatment [36]. Adding a stress-inducible promoter before the DREB1A gene in plants could enhance the drought, high salt and low temperature resistance of transgenic plants [37]. Bombyx mori nucleopolyhedrovirus (BmNPV)-inducible promoters were applied for gene therapy [38]. Currently, the promoters of GST have been reported in a few insects. In S. litura, the GST promoter acted as an important element for upregulating the expression of GST and improved S. litura tolerance to insecticides [22]. It was reported that the promoters of S. exigua GSTs were coregulated by two transcription factors, which enhanced the resistance of insects to xenobiotic stress [27]. Although we previously obtained five MtGST promoters, their regulatory mechanisms for MtGST genes are not clear. In the present study, we further identified the core regions of the five MtGST promoters in vitro by a 5’ loss fragment assay. The activity of the core regions of MtGSTt1 (−574/+340) and MtGSTz1 (−350/+521) promoters were also well induced by low concentrations of tannic acid and quercetin, which suggested that the core sequences of MtGST promoters have significant activity in response to plant secondary metabolites. However, we also found that higher concentrations of tannic acid and quercetin would decrease the activity of luciferase despite that they were not toxic to Sf9 cells. Based on a previous study, when human cells were exposed to xenobiotic stress, low concentrations of xenobiotics induced the promoter activity of pituitary adenylate cyclase-activating polypeptide (PACAP) receptor 1 (PAC1-R) while high concentrations of xenobiotics inhibited PAC1-R promoter activity [39]. Thus, we hypothesized that the relationship between GST promoter activity and plant secondary metabolites (tannic acid and quercetin) also presented a dose-dependent way. Once the concentrations of plant secondary metabolites exceeded the range causing induction, they might inhibit the activity of promoters. These findings provided useful information for understanding the mechanism of GST transcriptional regulation in M. troglodyta.
The transcription factor Nrf2, which is a member of the basic leucine-zipper family, is an oxygen-sensitive transcription factor and a vital physiological stress response mechanism in organisms [40]. Under oxidative stress, Nrf2 can translocate into the nucleus to bind to antioxidant response elements (AREs) and heterodimerize with MafK to regulate the expression of detoxification genes [41]. The mutation of Nrf2 in mice makes it more sensitive to xenobiotic stress [42]. Nrf2 can recognize specific DNA sequences in the presence of nuclear factor-erythroid 2 [43]. CncC in insects is homologous to Nrf2 and is an important transcription factor for regulating detoxification genes. In silkworm, both CncC and detoxification genes (including GSTs and P450s) regulated by CncC were upregulated after phoxim treatment [44]. The CncC-mediated detoxification pathway was associated with oxidative stress in Drosophila, and it was found that CncC could upregulate GSTd expression to enhance the ability to resist oxidative stress [45]. Nrf2 was able to regulate the detoxification enzyme gene CYP6A2 and increase resistance to DTT in Drosophila [46]. In Tribolium castaneum, the transcription factors CncC and Maf could regulate the expression of the CYP6BQ gene and increase resistance to deltamethrin [47]. Based on the results of phylogenetic analysis in this study, MtCncC was highly similar to CncCs from other insects. Therefore, we speculated that MtCncC is relatively conserved and has similar characteristics to other CncCs. In our study, by cotransfecting constructs containing MtCncC sequences and MtGST promoter sequences, we observed a significant induction of the MtGSTt1 (−574/+340) promoter by MtCncC, which suggests that MtCncC acts as a transcription factor responsible for the activity of the MtGSTt1 (−574/+340) promoter.
ARNT is also a regulatory element of xenobiotic stress response genes and a member of the bHLH-PAS transcription factor superfamily [25]. AhR is another bHLH-PAS protein family member that is a ligand-activated transcription factor [48]. In vertebrates, AhR has two isoforms, AhR1 and AhR2. AhR1 is found in all vertebrates, while AhR2 is present in some vertebrates [49]. AhR and ARNT can form heterodimers to bind enhancer DNA sequences and activate antioxidant and xenobiotic metabolic genes such as GSTs and P450s [26,50]. In mammals, some GSTs were regulated by AhR/ARNT [51,52]. In insects, AhR/ARNT was associated with the regulation of Aphis gossypii Glover CYP450 to improve its tolerance to spirotetramat [53]. In M. persicae, AhR/ARNT could upregulate the expression levels of CYP450 to confer resistance to pesticides [54]. NlARNT could bind to the CarE7 promoter and strongly induce transcriptional activity to enhance resistance to xenogenic stress in Nilaparvata lugens [55]. In this study, the phylogenetic relationship of MtARNT was closely related to that of other insect ARNTs. We hypothesized that MtARNT is highly similar to other ARNTs and has similar functions to other ARNTs. In this study, by cotransfecting constructs containing MtARNT sequences and MtGST promoter sequences, the MtGSTt1 (−574/+340) promoter was significantly induced by MtARNT, which suggests that MtARNT acts as an important cis-regulatory element responsible for the transcriptional activity of MtGSTt1 (−574/+340). CncC and ARNT coordinately regulated the expression of GST in S. exigua [27]. In mammals, the interaction between AhR and Nrf2 may be achieved through multiple mechanisms, including Nrf2 as a target gene of AhR, indirect activation of Nrf2 via CYP1A1-generated reactive oxygen species, and direct cross-interaction of AhR/XRE and Nrf2/ARE signaling [56]. According to our results, MtCncC and MtARNT did not coregulate the MtGST promoters and even appeared to reduce the transcriptional activity of the promoters. Thus, we speculated that MtCncC and MtARNT regulate the GST genes of M. troglodyta in a complex process.
In summary, this study identified the core regions of the five MtGST promoters and demonstrated their involvement in the response to tannic acid and quercetin stress. Furthermore, we identified two important transcription factors MtCncC and MtARNT involved in the regulation of the GST gene promoter. These results suggested that transcription factors regulate the expression of GSTs conferring resistance to plant secondary metabolites in M. troglodyta, and provided useful information for a better understanding of the regulatory mechanism between transcription factors and GSTs in M. troglodyta. Future studies will need to examine the mechanism of posttranscriptional regulation of GSTs in M. troglodyta.
4. Materials and Methods
4.1. Insect Rearing and Cell Culture
M. troglodyta larvae were gathered from poplar (Populus × euramericana ‘Nanlin 895’) trees in Nanjing, Jiangsu Province, China. The larvae were fed fresh poplar leaves with a photoperiod of 16 h:8 h (light: dark), a temperature of 26 ± 1 °C and a relative humidity of 70–80%. Third-instar larvae were used for subsequent experiments. Sf9 cells were routinely maintained with SF-900 II serum-free medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (HyClone, Logan, UT, USA), 50 mg/mL streptomycin and 50 mg/mL penicillin (Invitrogen) at 28 °C. Sf9 cells were cultured for 3 days and then used for transfection experiments.
4.2. Cloning and Sequencing 5’ Loss Fragments of GST Promoters
First, the transcription factor-binding sites for all full-length MtGST promoter sequences were predicted on the website http://alggen.lsi.upc.es (accessed on 10 November 2022) to avoid disrupting the integrity of the binding sites when the promoters of different fragments were cloned. The 5’ loss fragments of each MtGST promoter were amplified from the full-length sequences of the MtGST promoters using TaKaRa Ex Premier™ DNA Polymerase (Takara, Dalian, Liaoning, China). All primers were designed by Primer 5 software and were listed in Table 1. Each forward primer sequence and reverse primer sequence were added with Nhe I and Xho I restriction enzyme cleavage sites, respectively. Each 5’ loss fragment of MtGST promoter was ligated to a TA clone vector pMD-19T (Takara, Dalian, Liaoning, China), and the correct clone product was obtained by sequencing. The PGL4.10-Basic vector and the pMD-19T with the 5’ loss fragment were digested with Nhe I and Xho I. Then, the 5’ loss fragment of MtGST promoter was ligated to the PGL4.10-Basic vector using T4 DNA ligase (Takara, Dalian, Liaoning, China), and the ligation product was transformed into E. coli cells. The plasmid DNA was purified from E. coli cells for subsequent cell transfection experiments.

Table 1.
The primers used for this study.
4.3. Promoter Activity Analysis by Luciferase Reporter Assays
Sf9 cells (2.0 × 106 /well) were cultured in a 24-well culture plate, and each 5’ loss fragment promoter plasmid (700 ng/well) and pRL-TK (interference renilla luciferase reporter plasmid, Promega, Madison, WI, USA) (70 ng/well) were cotransfected using 2 µL/well Cellfectin II reagent (Invitrogen) in accordance with our previous method (Tang et al., 2020). After 48 h, Sf9 cells were harvested and lysed in 1 × passive lysis buffer (Promega), and the renilla and firefly luciferase activities were measured using the Dual-Luciferase® Reporter Assay System kit (Promega) on an FLx800TM fluorescence microplate reader (BioTek, Winooski, VT, USA). The promoter activity was calculated by normalizing the relative activity of firefly luciferase with that of renilla luciferase. Three replicates were performed for each treatment independently.
Tannic acid was initially solubilized in a small volume of acetone and then diluted in sterilized water to 70, 14, 2.8 and 0.56 mg/L, and sterilized water was used as a control. Quercetin was serially diluted in acetone to 35, 7, 1.4 and 0.28 mg/L, and acetone was used as a control. The concentrations of tannic acid and quercetin were determined according to previous study [4,57]. Sf9 cells (2.0 × 106 /well) were cultured in a 24-well culture plate, and then each 5’ loss fragment promoter plasmid (700 ng/well) and pRL-TK (interference plasmid) (70 ng/well) were cotransfected using 2 µL/well Cellfectin II reagent (Invitrogen). At 5 h posttransfection, we changed the transfection solution to cell culture medium containing 10 µL of tannic acid or quercetin with serum and double antibiotics. After 48 h, we measured luciferase activity using a Dual-Luciferase® Reporter Assay System kit on a microplate reader. The luciferase activity was calculated according to the above method.
4.4. Cloning the Sequences of MtCncC and MtARNT Genes
Total RNA was extracted from third-instar larvae using TRIzol Reagent (Takara, Dalian, Liaoning, China) according to the protocol. The quality and integrity of RNA were examined by a NanoDrop spectrophotometer and agarose gel electrophoresis, respectively. M. troglodyta RNA was reverse transcribed using the PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara, Dalian, Liaoning, China), and the cDNA was used for cloning MtCncC and MtARNT. The primers for cloning MtCncC and MtARNT were designed according to the transcriptome database and were listed in Table 1. Polymerase chain reaction (PCR) was performed using Premix Ex Taq™ (Takara, Dalian, Liaoning, China). The PCR program was as follows: 98 °C for 3 min; 35 cycles of 98 °C for 10 s, approximately 60 °C for 30 s and 72 °C for 90 s; an extension cycle of 72 °C for 5 min. The MtCncC and MtARNT DNA were ligated to the pMD-19T clone vector. The constructs were transformed into E. coli cells and sequenced by Sangon Biotech (Shanghai) Co., Ltd. The amino acid (AA) sequences of MtCncC and MtARNT were deduced from the NCBI Open Reading Frame (ORF) finder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 10 November 2022).
4.5. Phylogenetic Analysis of MtCncC and MtARNT
A phylogenetic tree was constructed to investigate the relationship between MtCncC and other insect CncC, and we picked Cap ‘n’ Collar as a keyword to query the nonredundant database (https://www.ncbi.nlm.nih.gov/ (accessed on 10 November 2022)). Multiple AA sequence alignment analysis was carried out using MEGA X (version 10.1) and Clustal X software (version 2.1). The phylogenetic tree was inferred by the maximum likelihood (ML) method in MEGA X with 1000 bootstrap replicates. The phylogenetic tree of MtARNT was inferred using the same methods.
4.6. Cotransfection of MtCncC and MtARNT with MtGSTt1 (−574/+340) or MtGSTz1 (−350/+521) Promoter
Using two primer pairs pAC-V5-CncC and pAC-V5-ARNT (Table 1), the MtCncC and MtARNT were amplified, respectively. Then MtCncC and MtARNT were cloned into pAC-V5 (Invitrogen). Sf9 cells (2.0 × 106/well) were cultured in a 24-well culture plate, and 350 ng of the promoter plasmid and 350 ng of the pAC-V5, pAC-V5-MtARNT, pAC-V5-MtCncC or pAC-V5-MtARNT and pAC-V5-MtCncC expression plasmids were cotransfected using 2 µL/well Cellfectin II reagent (Invitrogen). After 48 h induction, Sf9 cells were harvested and lysed to measure the renilla and firefly luciferase activities.
4.7. Statistical Analysis
ANOVA of the data collected from these experiments was performed using InStat software (GraphPad, San Diego, CA, USA). The significant differences of all two samples were evaluated using Student’s t test (two-tailed unpaired t test). The statistical significance of multisample comparisons was assessed with one-way ANOVA followed by Tukey’s multiple comparisons. A value of p < 0.05 was considered significantly different.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032190/s1.
Author Contributions
Conceptualization, F.T.; methodology, Z.W., X.S., F.T., X.G. and P.L.; software, Z.W., X.S. and Y.Z.; validation, Z.W., X.S. and Y.Z.; formal analysis, Z.W., X.S. and F.T.; investigation, Z.W. and X.S.; resources, Z.W. and X.S.; data curation, Z.W., X.S. and Y.Z.; writing—original draft preparation, Z.W. and X.S.; writing—review and editing, F.T.; visualization, Z.W. and X.S.; supervision, F.T.; project administration, F.T.; funding acquisition, F.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Contract No. 31370652, 30600476 and 30972376) and the Priority Academic Program Development Fund of Jiangsu Higher Education Institutions.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Krieger, R.I.; Feeny, P.P.; Wilkinson, C.F. Detoxication enzymes in the guts of caterpillars: An evolutionary answer to plant defenses? Science 1971, 172, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411–1419. [Google Scholar] [CrossRef]
- Tang, F.; Tu, H.; Shang, Q.; Gao, X.; Liang, P. Molecular Cloning and Characterization of Five Glutathione S-Transferase Genes and Promoters from Micromelalopha troglodyta (Graeser) (Lepidoptera: Notodontidae) and Their Response to Tannic Acid Stress. Insects 2020, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, F.; Xu, M.; Shen, T. Exploring miRNA-mRNA regulatory modules responding to tannic acid stress in Micromelalopha troglodyta (Graeser) (Lepidoptera: Notodontidae) via small RNA sequencing. Bull. Entomol. Res. 2022, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Zhang, X.; Liu, Y.; Gao, X.; Liu, N. In vitro inhibition of glutathione S-transferases by several insecticides and allelochemicals in two moth species. Int. J. Pest Manag. 2014, 60, 33–38. [Google Scholar] [CrossRef]
- Cheng, H.; Tang, F.; Li, W.; Xu, M. Tannic acid induction of a glutathione S-transferase in Micromelalopha troglodyta (Lepidoptera: Notodontidae) larvae. J. Entomol. Sci. 2015, 50, 350–362. [Google Scholar] [CrossRef]
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef]
- Koirala, B.K.S.; Moural, T.; Zhu, F. Functional and Structural Diversity of Insect Glutathione S-transferases in Xenobiotic Adaptation. Int. J. Biol. Sci. 2022, 18, 5713–5723. [Google Scholar] [CrossRef]
- Lee, K. Glutathione S-transferase activities in phytophagous insects: Induction and inhibition by plant phototoxins and phenols. Insect Biochem. 1991, 21, 353–361. [Google Scholar] [CrossRef]
- Gao, X.W.; Dong, X.L.; Zheng, B.Z.; Chen, Q. Glutathiones S-transferase (GSTs) of cotton bollworm: Induction of pesticides and plant secondary substances and metabolism of GSTs to pesticides. Acta Entomol. Sin. 1997, 40, 122–127. [Google Scholar]
- Francis, F.; Vanhaelen, N.; Haubruge, E. Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Arch. Insect Biochem. Physiol. 2005, 58, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sun, L.; Zhao, H.; Wang, Z.; Zou, L.; Cao, C. Functional identification and characterization of GST genes in the Asian gypsy moth in response to poplar secondary metabolites. Pestic. Biochem. Physiol. 2021, 176, 104860. [Google Scholar] [CrossRef]
- AlJabr, A.M.; Hussain, A.; Rizwan-Ul-Haq, M.; Al-Ayedh, H. Toxicity of Plant Secondary Metabolites Modulating Detoxification Genes Expression for Natural Red Palm Weevil Pesticide Development. Molecules 2017, 22, 169. [Google Scholar] [CrossRef]
- Palli, S.R. CncC/Maf-mediated xenobiotic response pathway in insects. Arch. Insect Biochem. Physiol. 2020, 104, e21674. [Google Scholar] [CrossRef]
- Li, X.; Deng, Z.; Chen, X. Regulation of insect P450s in response to phytochemicals. Curr. Opin. Insect Sci. 2021, 43, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Song, Y.; Zeng, R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect Sci. 2021, 43, 103–107. [Google Scholar] [CrossRef]
- Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Misra, J.R.; Horner, M.A.; Lam, G.; Thummel, C.S. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes. Dev. 2011, 25, 1796–1806. [Google Scholar] [CrossRef]
- Deng, H.; Kerppola, T.K. Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS Genet. 2013, 9, e1003263. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lu, M.; Zhang, N.; Zou, X.; Mo, M.; Zheng, S. Nuclear factor erythroid-derived 2-related factor 2 activates glutathione S-transferase expression in the midgut of Spodoptera litura (Lepidoptera: Noctuidae) in response to phytochemicals and insecticides. Insect Mol. Biol. 2018, 27, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, M.; Palli, S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 2017, 83, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell. Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Fernandez-Salguero, P.M. New Trends in Aryl Hydrocarbon Receptor Biology. Front. Cell. Dev. Biol. 2016, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Kann, S.; Huang, M.Y.; Estes, C.; Reichard, J.F.; Sartor, M.A.; Xia, Y.; Puga, A. Arsenite-induced aryl hydrocarbon receptor nuclear translocation results in additive induction of phase I genes and synergistic induction of phase II genes. Mol. Pharmacol. 2005, 68, 336–346. [Google Scholar] [CrossRef]
- Hu, B.; Huang, H.; Wei, Q.; Ren, M.; Mburu, D.K.; Tian, X.; Su, J. Transcription factors CncC/Maf and AhR/ARNT coordinately regulate the expression of multiple GSTs conferring resistance to chlorpyrifos and cypermethrin in Spodoptera exigua. Pest Manag. Sci. 2019, 75, 2009–2019. [Google Scholar] [CrossRef]
- Zhang, X.; Jie, D.; Liu, J.; Zhang, J.; Zhang, T.; Zhang, J.; Ma, E. Aryl hydrocarbon receptor regulates the expression of LmGSTd7 and is associated with chlorpyrifos susceptibility in Locusta migratoria. Pest Manag. Sci. 2019, 75, 2916–2924. [Google Scholar] [CrossRef]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef]
- Pavlidi, N.; Vontas, J.; Van Leeuwen, T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef]
- Samra, A.I.; Kamita, S.G.; Yao, H.W.; Cornel, A.J.; Hammock, B.D. Cloning and characterization of two glutathione S-transferases from pyrethroid-resistant Culex pipiens. Pest Manag. Sci. 2012, 68, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.A.; Liu, J.Y.; Rashid, M.; Wang, D.; Zhang, Y.L. Cantharidin Impedes Activity of Glutathione S-Transferase in the Midgut of Helicoverpa armigera Hubner. Int. J. Mol. Sci. 2013, 14, 5482–5500. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Jia, M.; Liu, T.; Xuan, T.; Yan, Z.K.; Guo, Y.; Ma, E.; Zhang, J. Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen). Pest Manag. Sci. 2011, 67, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Luo, Q.H.; Wang, J.M.; Li, X.F.; Yang, Y. Functional characterization and analysis of the Arabidopsis UGT71C5 promoter region. Genet. Mol. Res. 2015, 14, 19173–19183. [Google Scholar] [CrossRef] [PubMed]
- Kluge, J.; Terfehr, D.; Kuck, U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 6357–6372. [Google Scholar] [CrossRef]
- Concha, C.; Edman, R.M.; Belikoff, E.J.; Schiemann, A.H.; Carey, B.; Scott, M.J. Organization and expression of the Australian sheep blowfly (Lucilia cuprina) hsp23, hsp24, hsp70 and hsp83 genes. Insect Mol. Biol. 2012, 21, 169–180. [Google Scholar] [CrossRef]
- Pellegrineschi, A.; Reynolds, M.; Pacheco, M.; Brito, R.M.; Almeraya, R.; Yamaguchi-Shinozaki, K.; Hoisington, D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2004, 47, 493–500. [Google Scholar] [CrossRef]
- Cao, M.Y.; Kuang, X.X.; Li, H.Q.; Lei, X.J.; Xiao, W.F.; Dong, Z.Q.; Zhang, J.; Hu, N.; Chen, T.T.; Lu, C.; et al. Screening and optimization of an efficient Bombyx mori nucleopolyhedrovirus inducible promoter. J. Biotechnol. 2016, 231, 72–80. [Google Scholar] [CrossRef]
- Huang, X.; Li, J.; Song, S.; Wang, L.; Lin, Z.; Ouyang, Z.; Yu, R. Hormesis effect of hydrogen peroxide on the promoter activity of neuropeptide receptor PAC1-R. J. Food Biochem. 2019, 43, e12877. [Google Scholar] [CrossRef]
- Motohashi, H.; Yamamoto, M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004, 10, 549–557. [Google Scholar] [CrossRef]
- Nakata, K.; Tanaka, Y.; Nakano, T.; Adachi, T.; Tanaka, H.; Kaminuma, T.; Ishikawa, T. Nuclear receptor-mediated transcriptional regulation in Phase I, II, and III xenobiotic metabolizing systems. Drug Metab. Pharm. 2006, 21, 437–457. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; O’Connor, T.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 2002, 294, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yamamoto, M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme. Regul. 2006, 46, 113–140. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, J.; Wang, H.; Mao, T.; Li, J.; Cheng, X.; Hu, J.; Xue, B.; Li, B. Cloning and Functional Analysis of CncC and Keap1 Genes in Silkworm. J. Agric. Food Chem. 2018, 66, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Sykiotis, G.P.; Bohmann, D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 2008, 14, 76–85. [Google Scholar] [CrossRef]
- Wan, H.; Liu, Y.; Li, M.; Zhu, S.; Li, X.; Pittendrigh, B.R.; Qiu, X. Nrf2/Maf-binding-site-containing functional Cyp6a2 allele is associated with DDT resistance in Drosophila melanogaster. Pest Manag. Sci. 2014, 70, 1048–1058. [Google Scholar] [CrossRef]
- Kalsi, M.; Palli, S.R. Transcription factors, CncC and Maf, regulate expression of CYP6BQ genes responsible for deltamethrin resistance in Tribolium castaneum. Insect Biochem. Mol. Biol. 2015, 65, 47–56. [Google Scholar] [CrossRef]
- Vorrink, S.U.; Domann, F.E. Regulatory crosstalk and interference between the xenobiotic and hypoxia sensing pathways at the AhR-ARNT-HIF1alpha signaling node. Chem. Biol. Interact. 2014, 218, 82–88. [Google Scholar] [CrossRef]
- Jin, H.; Ji, C.; Ren, F.; Aniagu, S.; Tong, J.; Jiang, Y.; Chen, T. AHR-mediated oxidative stress contributes to the cardiac developmental toxicity of trichloroethylene in zebrafish embryos. J. Hazard. Mater. 2020, 385, 121521. [Google Scholar] [CrossRef]
- Wang, L.; He, X.; Szklarz, G.D.; Bi, Y.; Rojanasakul, Y.; Ma, Q. The aryl hydrocarbon receptor interacts with nuclear factor erythroid 2-related factor 2 to mediate induction of NAD(P)H:quinoneoxidoreductase 1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch. Biochem. Biophys. 2013, 537, 31–38. [Google Scholar] [CrossRef]
- Rushmore, T.H.; King, R.G.; Paulson, K.E.; Pickett, C.B. Regulation of glutathione S-transferase Ya subunit gene expression: Identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc. Natl. Acad. Sci. USA 1990, 87, 3826–3830. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, C. Antioxidant Functions of the Aryl Hydrocarbon Receptor. Stem Cells Int. 2016, 2016, 7943495. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Chen, X.; Pan, Y.; Zheng, Z.; Wei, X.; Xi, J.; Zhang, J.; Gao, X.; Shang, Q. Transcription factor aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator is involved in regulation of the xenobiotic tolerance-related cytochrome P450 CYP6DA2 in Aphis gossypii Glover. Insect Mol. Biol. 2017, 26, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Peng, T.; Xu, P.; Zeng, X.; Tian, F.; Song, J.; Shang, Q. Transcription Factors AhR/ARNT Regulate the Expression of CYP6CY3 and CYP6CY4 Switch Conferring Nicotine Adaptation. Int. J. Mol. Sci. 2019, 20, 4521, Published 12 September 2019. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, R.; Liu, C.; Gao, Y.; Deng, X.; Wan, H.; Li, J. Functional characterization of the transcription factors AhR and ARNT in Nilaparvata lugens. Pestic. Biochem. Physiol. 2021, 176, 104875. [Google Scholar] [CrossRef]
- Kohle, C.; Bock, K.W. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem. Pharmacol. 2007, 73, 1853–1862. [Google Scholar] [CrossRef]
- Li, F.; Ma, K.; Chen, X.; Zhou, J.J.; Gao, X. The regulation of three new members of the cytochrome P450 CYP6 family and their promoters in the cotton aphid Aphis gossypii by plant allelochemicals. Pest Manag. Sci. 2019, 75, 152–159. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).