Serum Levels of IFABP2 and Differences in Lactobacillus and Porphyromonas gingivalis Abundance on Gut Microbiota Are Associated with Poor Therapeutic Response in Rheumatoid Arthritis: A Pilot Study
Abstract
1. Introduction
2. Results
2.1. Demographic Data, Markers for Inflammation, Intestinal Bacteria Abundance, and Phenotype of Response to Therapy
2.2. Relation between Gut Microbiota’s Bacteria Abundance with the Dose of Drugs and Inflammation Markers
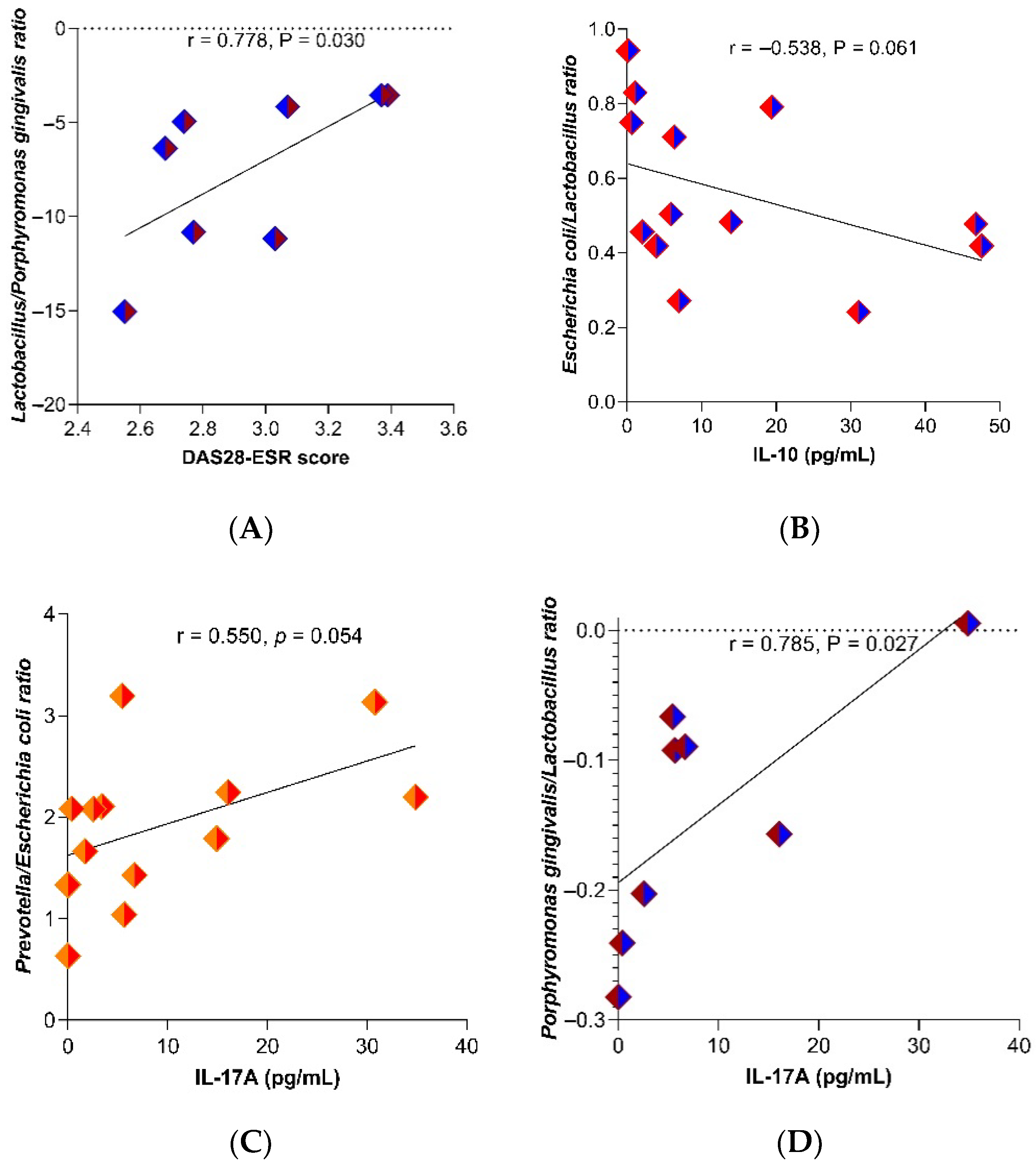
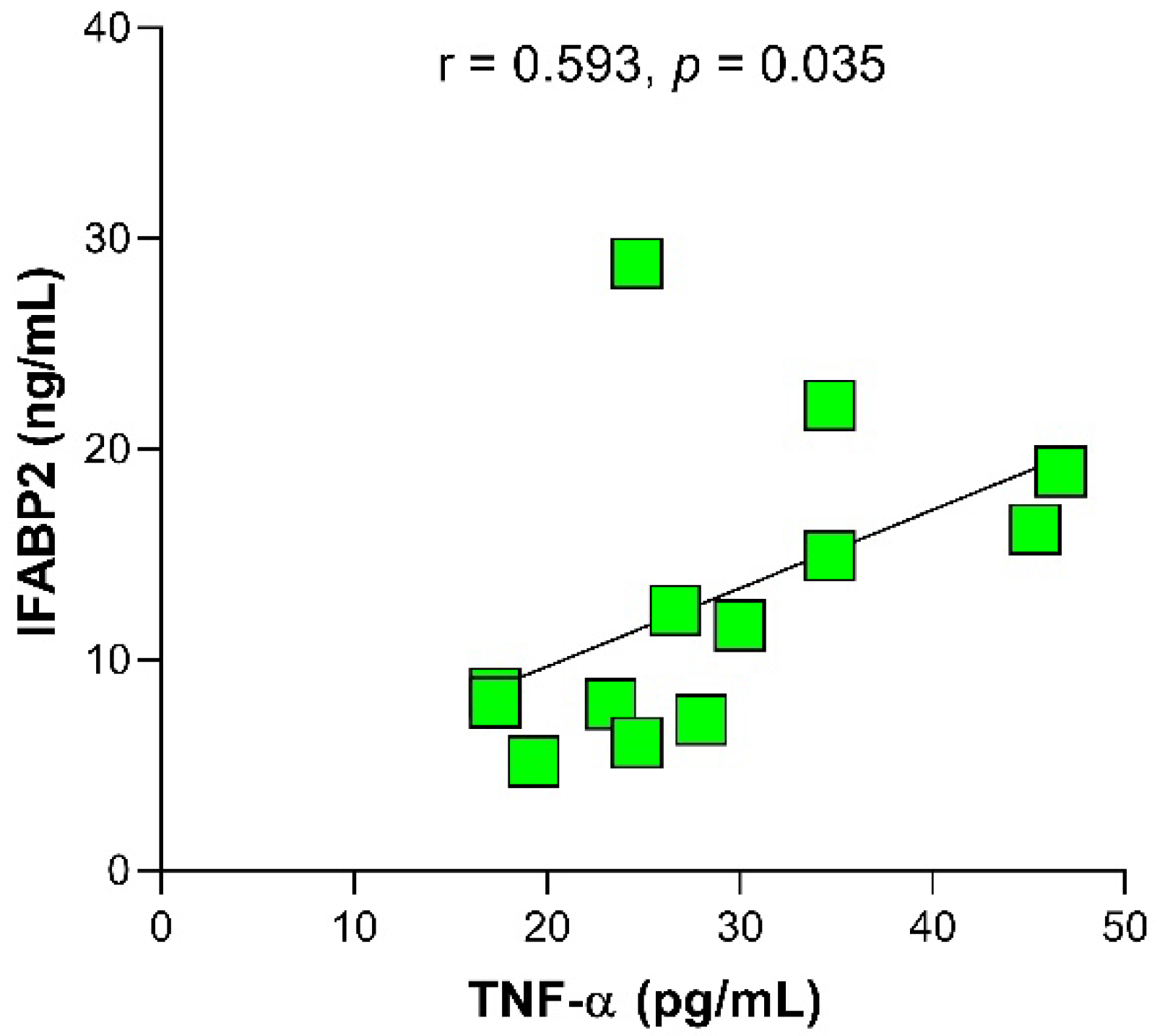
2.3. Relationship of IFABP2 Serum Levels and Inflammation Markers with Gut Microbiota’s Bacteria and Therapy Response Phenotype in RA
3. Discussion
4. Materials and Methods
4.1. Studied Population
4.2. Sample Collection
4.3. Clinical Assessment
4.4. DNA Extraction from Stool and qPCR to Estimation of Microbial Abundance
4.5. Cytokines (TNF-α, IL-10, and IL-17A) and Serum IFABP2 Levels Quantification
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACPAs | antibodies to citrullinated protein antigens |
| ACR | American College of Rheumatology |
| CD14 | cluster of differentiation 14 |
| CFU | colony-forming unit |
| CLQ | Chloroquine |
| CRP | C reactive protein |
| Cs | corticosteroids |
| CsDMARDs | conventional synthetic disease modifying anti-rheumatic drugs |
| DAS28 | Disease Activity Score 28 |
| DAS28-ESR | Disease Activity Score-28 with Erythrocyte Sedimentation Rate |
| DNA | deoxyribonucleic acid |
| ELISA | Enzyme-Linked ImmunoSorbent Assay |
| ESR | erythrocyte sedimentation rate |
| EULAR | European League Against Rheumatism |
| HAQ-DI | Health Assessment Questionnaire-Disability Index |
| IFABP2 | intestinal fatty-acid binding protein 2 |
| IFN-γ | interferon-gamma |
| IL-6 | interleukin-6 |
| IL-10 | interleukin-10 |
| IL-17A | interleukin-17A |
| IL-1β | interleukin-1 beta |
| IL-22 | interleukin-22 |
| IL-23 | interleukin-23 |
| LBP | lipopolysaccharide-binding protein |
| LPS | lipopolysaccharide |
| MLCK | myosin light chain kinase |
| MTX | methotrexate |
| NORA | new-onset RA |
| PDN | prednisone |
| qPCR | quantitative polymerase chain reaction |
| RA | rheumatoid arthritis |
| RF | rheumatoid factor |
| sCD14 | soluble cluster of differentiation 14 |
| TGF-β | transforming growth factor-β |
| Th1 | T helper 1 cells |
| Th17 | T helper 17 cells |
| TLR-2 | Toll-like receptor 2 |
| TNF-α | tumor necrosis factor alpha |
| ZO-1 | zonula occludens-1 |
References
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zou, Q.; Zeng, B.; Fang, Y.; Wei, H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 2013, 67, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Maresz, K.J.; Hellvard, A.; Sroka, A.; Adamowicz, K.; Bielecka, E.; Koziel, J.; Gawron, K.; Mizgalska, D.; Marcinska, K.A.; Benedyk, M.; et al. Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD). PLoS Pathog. 2013, 9, e1003627. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef]
- Block, K.E.; Zheng, Z.; Dent, A.L.; Kee, B.L.; Huang, H. Gut Microbiota Regulates K/BxN Autoimmune Arthritis through Follicular Helper T but Not Th17 Cells. J. Immunol. 2016, 196, 1550–1557. [Google Scholar] [CrossRef]
- Neumann, V.C.; Shinebaum, R.; Cooke, E.M.; Wright, V. Effects of sulphasalazine on faecal flora in patients with rheumatoid arthritis: A comparison with penicillamine. Br. J. Rheumatol. 1987, 26, 334–337. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef]
- Picchianti-Diamanti, A.; Panebianco, C.; Salemi, S.; Sorgi, M.L.; Di Rosa, R.; Tropea, A.; Sgrulletti, M.; Salerno, G.; Terracciano, F.; D´Amelio, R.; et al. Analysis of Gut Microbiota in Rheumatoid Arthritis Patients: Disease-Related Dysbiosis and Modifications Induced by Etanercept. Int. J. Mol. Sci. 2018, 19, 2938. [Google Scholar] [CrossRef]
- Rodrigues, G.S.P.; Cayres, L.C.F.; Gonçalves, F.P.; Takaoka, N.N.C.; Lengert, A.H.; Tansini, A.; Brisotti, J.L.; Sasdelli, C.B.G.; Oliveira, G.L.V. Detection of Increased Relative Expression Units of Bacteroides and Prevotella, and Decreased Clostridium leptum in Stool Samples from Brazilian Rheumatoid Arthritis Patients: A Pilot Study. Microorganisms 2019, 7, 413. [Google Scholar] [CrossRef]
- Chiang, H.I.; Li, J.R.; Liu, C.C.; Liu, P.Y.; Chen, H.H.; Chen, Y.M.; Lan, J.L.; Chen, D.Y. An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis. J. Clin. Med. 2019, 8, 1770. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, S.X.; Yin, X.F.; Zhang, M.X.; Qiao, J.; Xin, X.H.; Chang, M.J.; Gao, C.; Li, Y.F.; Li, X.F. The Gut Microbiota and Its Relevance to Peripheral Lymphocyte Subpopulations and Cytokines in Patients with Rheumatoid Arthritis. J. Immunol. Res. 2021, 2021, 6665563. [Google Scholar] [CrossRef]
- Gupta, V.K.; Cunningham, K.Y.; Hur, B.; Bakshi, U.; Huang, H.; Warrington, K.J.; Taneja, V.; Myasoedova, E.; Davis, J.M., 3rd; Sung, J. Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med. 2021, 13, 149. [Google Scholar] [CrossRef]
- Hammad, D.B.M.; Hider, S.L.; Liyanapathirana, V.C.; Tonge, D.P. Molecular Characterization of Circulating Microbiome Signatures in Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2020, 9, 440. [Google Scholar] [CrossRef]
- Guerreiro, C.S.; Calado, Â.; Sousa, J.; Fonseca, J.E. Diet, Microbiota, and Gut Permeability-The Unknown Triad in Rheumatoid Arthritis. Front. Med. 2018, 5, 349. [Google Scholar] [CrossRef]
- Matei, D.E.; Menon, M.; Alber, D.G.; Smith, A.M.; Nedjat-Shokouhi, B.; Fassano, A.; Magill, L.; Duhlin, A.; Bitoun, s.; Gleizes, A.; et al. Intestinal barrier dysfunction play an integral role in arthritis pathology and can be targeted to ameliorate disease. Med 2021, 2, 864–883.e9. [Google Scholar] [CrossRef]
- Audo, R.; Sanchez, P.; Rivière, B.; Mielle, J.; Tan, J.; Lukas, C.; Macia, L.; Morel, J.; Daien, C.I. Rheumatoid arthritis is associated with increased gut permeability and bacterial translocation wich are reversed by inflammation control. Rheumatology 2022, 2022, keac454. [Google Scholar] [CrossRef]
- Tajik, N.; Frech, M.; Schulz, O.; Schälter, F.; Lucas, S.; Azizov, V.; Dürholz, K.; Steffen, F.; Omata, Y.; Rings, A.; et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat. Commun. 2020, 11, 1995. [Google Scholar] [CrossRef]
- Scher, J.U.; Nayack, R.R.; Ubeda, C.; Turnbaugh, P.J.; Abramson, S.B. Pharmacomicrobiomics in inflammatory arthritis: Gut microbiome as modulator of therapeutic response. Nat. Rev. Rheumatol. 2020, 16, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza-García, O.; Castro-Alarcón, N.; Pérez-Rubio, G.; Guzmán-Guzmán, I.P. DMARDs-Gut Microbiota Feedback: Implications in the Response to Therapy. Biomolecules 2020, 10, 1479. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.R.; Alexander, M.; Stapleton-Grey, K.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.J. Perturbation of the human gut microbiome by a non-antibiotic drug contributes to the resolution of autoimmune disease. bioRxiv 2019. [Google Scholar] [CrossRef]
- Artacho, A.; Issac, S.; Nayak, R.; Flor-Duro, A.; Alexander, M.; Koo, I.; Manasson, J.; Smith, P.B.; Rosenthal, P.; Homsi, Y.; et al. The Pretreatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 931–942. [Google Scholar] [CrossRef]
- Nayak, R.R.; Alexander, M.; Deshpande, I.; Stapletom-Gray, K.; Rimal, B.; Patterson, A.D.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.T. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe 2021, 29, 362–377.e11. [Google Scholar] [CrossRef]
- Wu, T.; Yang, L.; Jiang, J.; Ni, Y.; Zhu, J.; Zheng, X.; Wang, Q.; Lu, X.; Fu, Z. Chronic glucocorticoid treatment induced circadian clock disorder leads to lipid metabolism and gut microbiota alterations in rats. Life Sci. 2018, 192, 173–182. [Google Scholar] [CrossRef]
- He, Z.; Kong, X.; Shao, T.; Zhang, Y.; Wen, C. Alterations of the Gut Microbiota Associated with Promoting Efficacy of Prednisone by Bromofuranone in MRL/lpr Mice. Front. Microbiol. 2019, 10, 978. [Google Scholar] [CrossRef]
- Huang, E.Y.; Inoue, T.; Leone, V.A.; Dalal, S.; Touw, K.; Wang, Y.; Musch, M.W.; Theriault, B.; Higuchi, K.; Donovan, S.; et al. Using corticosteroids to reshape the gut microbiome: Implications for inflammatory bowel diseases. Inflamm. Bowel Dis. 2015, 21, 963–972. [Google Scholar] [CrossRef]
- Alpizar-Rodríguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, c.; Finckh, A.; Strwing, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, S.; Zhang, M.; Chang, M.; Mao, X.; Wang, J.; Zhang, J.; Qiu, M.; Gao, C.; Li, X. Diversity Analysis of Intestinal Flora in Patients with Rheumatoid Arthritis. ACR Meeting Abstracts. Arthritis Rheumatol. 2019, 71 (Suppl. 10), 493. Available online: https://acrabstracts.org/abstract/diversity-analysis-of-intestinal-flora-in-patients-with-rheumatoid-arthritis/ (accessed on 24 September 2022).
- Tong, Y.; Tang, H.; Li, Y.; Su, L.C.; Wu, Y.; Bozec, A.; Zaiss, M.; Qing, P.; Zhao, H.; Tan, C.; et al. Gut Microbiota Dysbiosis in High-Risk Individuals For Rheumatoid Arthritis Triggers Mucosal Immunity Perturbation and Promotes Arthritis in Mice. Res. Square 2020. [Google Scholar] [CrossRef]
- Anis, R.H.; Dawa, G.A.; El-Korashi, L.A. Assessment of gut microbiota in rheumatoid arthritis patients. Microbes Infect. Dis. 2021, 2, 77–83. [Google Scholar] [CrossRef]
- Zhou, B.; Xia, X.; Wang, P.; Chen, S.; Yu, C.; Huang, R.; Zhang, R.; Wang, Y.; Lu, L.; Yuan, F.; et al. Induction and Amelioration of Methotrexate-Induced Gastrointestinal Toxicity are Related to Immune Response and Gut Microbiota. EBioMedicine 2018, 33, 122–133. [Google Scholar] [CrossRef]
- Tang, D.; Zeng, T.; Wang, Y.; Cui, H.; Wu, J.; Zou, B.; Tao, Z.; Zhang, L.; Garside, G.B.; Tao, S. Dietary restriction increases protective gut bacteria to rescue lethal methotrexate-induced intestinal toxicity. Gut Microbes 2020, 12, 1714401. [Google Scholar] [CrossRef]
- Shin, Y.S.; Buehring, K.U.; Stokstad, E.L. The metabolism of methotrexate in Lactobacillus casei and rat liver and the influence of methotrexate on metabolism of folic acid. J. Biol. Chem. 1974, 249, 5772–5777. [Google Scholar] [CrossRef]
- Levy, C.C.; Goldman, P. The enzymatic hydrolysis of methotrexate and folic acid. J. Biol. Chem. 1967, 242, 2933–2938. [Google Scholar] [CrossRef]
- Letertre, M.P.M.; Munjoma, N.; Wolfer, K.; Pechlivanis, A.; McDonald, J.A.K.; Hardwick, R.N.; Cherrington, N.J.; Coen, M.; Nicholson, J.K.; Hoyles, L.; et al. A Two-Way Interaction between Methotrexate and the Gut Microbiota of Male Sprague-Dawley Rats. J. Proteome Res. 2020, 19, 3326–3339. [Google Scholar] [CrossRef]
- Ceccarelli, F.; Orrú, G.; Pilloni, A.; Bartosiewicz, I.; Perricone, C.; Martino, E.; Lucchetti, R.; Fais, S.; Vomero, M.; Oliveiri, M.; et al. Porphyromonas gingivalis in the tongue biofilm is associated with clinical outcome in rheumatoid arthritis patients. Clin. Exp. Immunol. 2018, 194, 244–252. [Google Scholar] [CrossRef]
- de Aquino, S.G.; Talbot, J.; Sônego, F.; Turato, W.M.; Grespan, R.; Avila-Campos, M.J.; Cunha, F.Q.; Cirelli, J.A. The aggravation of arthritis by periodontitis is dependent of IL-17 receptor A activation. J. Clin. Periodontol. 2017, 44, 881–891. [Google Scholar] [CrossRef]
- de Aquino, S.G.; Abdollahi-Roodsaz, S.; Koenders, M.I.; van de Loo, F.A.; Prujin, G.; Marijnissen, R.J.; Walgreen, B.; Helsen, M.M.; van den Bersselaar, L.A.; de Molon, R.S.; et al. Periodontal pathogenesis directly promote autoimmune experimental arthritis by inducing a TLR2-and IL-1-driven Th17 response. J. Immunol. 2014, 192, 4103–4111. [Google Scholar] [CrossRef]
- Marchesan, J.T.; Gerow, E.A.; Schaff, R.; Taut, A.D.; Shin, S.Y.; Sugai, J.; Brand, D.; Burberry, A.; Jorns, J.; Lundy, S.K.; et al. Porphyromonas gingivalis oral infection exacerbates the development and severity of collagen-induced arthritis. Arthritis Res. Ther. 2013, 15, R186. [Google Scholar] [CrossRef]
- Scher, J.U.; Ubeda, C.; Equinda, M.; Khanin, R.; Buischi, Y.; Viale, A.; Lipuma, L.; Attur, M.; Pillinger, M.H.; Weissmann, G.; et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012, 64, 3083–3094. [Google Scholar] [CrossRef] [PubMed]
- Totaro, M.C.; Cattani, P.; Ria, F.; Tolusso, B.; Gremese, E.; Fedale, A.L.; DÓnghia, S.; Marchetti, S.; Di Sante, G.; Canestri, S.; et al. Porphyromonas gingivalis and the pathogenesis of rheumatoid arhritis of various compartments including the synovial tissue. Arthritis Res. Ther. 2013, 15, R66. [Google Scholar] [CrossRef] [PubMed]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence for Immune Relevance of Prevotella copri, a Gut Microbe, in Patients with Rheumatoid Arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Al-Saffar, A.K.; Meijer, C.H.; Gannavarapu, V.R.; Hall, G.; Li, Y.; Diaz Tartera, H.O.; Lördal, M.; Ljung, T.; Hellström, P.M.; Webb, D.L. Parallel Changes in Harvey-Bradshaw Index, TNF-α, and Intestinal Fatty Acid Binding Protein in Response to Infliximab in Crohn´s Disease. Gastroenterol. Res. Pract. 2017, 2017, 1745918. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, L.; Bathon, J.M.; England, B.R.; St Clair, E.W.; Arayssi, T.; Carandang, K.; Deane, K.D.; Genovese, M.; Huston, K.K.; Kerr, G.; et al. 2021 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 1108–1123. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Landewé, R.B.M.; Bijlsma, J.W.J.; Burmester, G.R.; Dougados, M.; Kerschbaumer, A.; McInnes, I.B.; Sepriano, A.; van Vollenhoven, R.F.; de Wit, M.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis. 2020, 79, 685–699. [Google Scholar] [CrossRef]
- Pelsers, M.M.; Hermens, W.T.; Glatz, J.F. Fatty acid-binding proteins as plasma markers of tissue injury. Clin. Chim. Acta 2005, 352, 15–35. [Google Scholar] [CrossRef]
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety of depression. Gut 2018, 67, 1555–1557. [Google Scholar] [CrossRef]
- Blaser, A.; Padar, M.; Tang, J.; Dulton, J.; Forbes, A. Citrulline and intestinal fatty acid-binding protein as biomarkers for gastrointestinal dysfunction in the critically ill. Anaesthesiol. Intensive Ther. 2019, 51, 230–239. [Google Scholar] [CrossRef]
- Ma, T.Y.; Iwamoto, G.K.; Hoa, N.T.; Akotia, V.; Pedram, A.; Boivin, M.A.; Said, H.M. TNF-alpha-induced increase in intestinal epithelial tight junction permeability requires NF-κB activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G367–G376. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R.; Ye, D.; Dokladny, K.; Ma, T.Y. Mechanism of IL-1β-Induced Increase in Intestinal Epithelial Tight Junction Permeability. J. Immunol. 2008, 180, 5653–5661. [Google Scholar] [CrossRef] [PubMed]
- He, W.Q.; Wang, J.; Sheng, J.Y.; Zha, J.M.; Graham, W.V.; Turner, J.R. Contributions of Myosin Light Chain Kinase to Regulation of Epithelial Paracellular Permeability and Mucosal Homeostasis. Int. J. Mol. Sci. 2020, 21, 933. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Graham, W.V.; Wang, Y.; Witkowski, E.D.; Schwarz, B.T.; Turner, J.R. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am. J. Pathol. 2005, 166, 409–419. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Guo, S.; Ye, D.; Rawat, M.; Ma, T.Y. TNF-α Modulation of Intestinal Tight Junction Permeability Is Mediated by NIK/IKK-α Axis Activation of the Canonical NF-κB Pathway. Am. J. Pathol. 2016, 186, 1151–1165. [Google Scholar] [CrossRef]
- Martinsson, K.; Dürholz, K.; Schett, G.; Zaiss, M.; Kastbom, A. Higher serum levels of short-chain fatty acids are associated with non-progression to arthritis in individuals at increased risk of RA. Ann. Rheum. Dis. 2022, 81, 445–447. [Google Scholar] [CrossRef]
- Dubé, N.; Delvin, E.; Yotov, W.; Garofalo, C.; Bendayan, M.; Veerkamp, J.H.; Levy, E. Modulation of intestinal and liver fatty acid-binding proteins in Caco-2 cells by lipids, hormones and cytokines. J. Cell. Biochem. 2001, 81, 613–620. [Google Scholar] [CrossRef]
- Maillefert, J.F.; Puéchal, X.; Falgarone, G.; Lizard, G.; Ornetti, P.; Solau, E.; Legré, V.; Lioté, F.; Sibilia, J.; Morel, J.; et al. Prediction of response to disease modifying antirheumatic drugs in rheumatoid arthritis. Jt. Bone Spine 2010, 77, 558–563. [Google Scholar] [CrossRef]
- Zhao, P.W.; Jiang, W.G.; Wang, L.; Jiang, Z.Y.; Shan, Y.X.; Jiang, Y.F. Plasma levels of IL-37 and correlation with TNF-α, IL-17A, and disease activity during DMARD treatment of rheumatoid arthritis. PLoS ONE 2014, 9, e95346. [Google Scholar] [CrossRef]
- Horta-Baas, G.; Saldoval-Cabrera, A.; Romero-Figueroa, M.D.S. Modification of Gut Microbiota in Inflammatory Arthritis: Highlights and future Challenges. Curr. Rheumatol. Rep. 2021, 23, 67. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., 3rd; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
- Vallejo-Yagüe, E.; Keystone, E.C.; Kandhasamy, S.; Micheroli, R.; Finck, A.; Burden, A.M. Primary and secondary non-response: In need of operational definitions in observational studies. Ann. Rheum. Dis. 2021, 80, 961–964. [Google Scholar] [CrossRef]
- Fu, C.J.; Carter, J.N.; Li, Y.; Porter, J.H.; Kerley, M.S. Comparison of agar plate and real-time PCR on enumeration of Lactobacillus, Clostridium perfringens and total anaerobic bacteria in dog faeces. Lett. Appl. Microbiol. 2006, 42, 490–494. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Miyamoto, Y.; Takada, T.; Matsumoto, K.; Oyaizu, H.; Tanaka, R. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 2002, 68, 5445–5451. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T.; Tanaka, R. Use of 16rNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef]
- Slots, J.; Ashimoto, A.; Flynn, M.J.; Li, G.; Chen, C. Detection of putative periodontal pathogens in subgingival specimens by 16S ribosomal DNA amplification with the polymerase chain reaction. Clin. Infect. Dis. 1995, 20, S304–S307. [Google Scholar] [CrossRef]
- Huijsdens, X.W.; Linskens, R.K.; Mak, M.; Meuwissen, S.G.; Vandenbroucke-Grauls, C.M.; Savelkoul, P.H. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 2002, 40, 4423–4427. [Google Scholar] [CrossRef]

| Characteristics | NORA (n = 10) | Treated (n = 13) | p-Value |
|---|---|---|---|
| Clinical assessment | |||
| Age, years a | 44.8 ± 10.11 | 45.69 ± 11.94 | 0.851 |
| Disease evolution, months b | 19.5 (4–192) | 60 (12–300) | 0.087 |
| ESR, mm/hr a | 36.90 ± 10.56 | 36.15 ± 15.30 | 0.896 |
| CRP, mg/Lb | 9.5 (4.99–46.9) | 20.1 (4.99–33.9) | 0.092 |
| DAS28-ESR, score a | 4.94 ± 1.56 | 3.28 ± 0.76 | 0.002 |
| HAQ-DI, score b | 0.57 (0–1.2) | 0.10 (0–1) | 0.134 |
| Rheumatoid factor, UI/mL b | 72 (8.59–984.3) | 103.5 (8.59–942.5) | 0.619 |
| Positive rheumatoid factor ≥ 20 UI/mL, n (%) c | 7 (70) | 11 (84.62) | 0.618 |
| Inflammatory markers | |||
| TNF-α, pg/mL b | 23.33 (13.33–65.33) | 26.66 (17.33–46.66) | 0.419 |
| IL-10, pg/mL b | 8.64 (0–55.78) | 6.36 (0.11–47.55) | 0.732 |
| IL-17A, pg/mL b | 3.105 (0–48.04) | 5.46 (0–34.86) | 0.707 |
| IFABP2, ng/mL b | 21.18 (7.46–29.16) | 11.64 (5.186–28.82) | 0.035 |
| Specific group bacterial | |||
| Lactobacillus, Log10 CFU/mL b | 6.26 (5.33–9.38) | 5.93 (4.32–8.59) | 0.192 |
| Bacteroides fragilis, Log10 CFU/mL b | 2.85 (1.38–4.45) | 2.86 (0.60–5.14) | 0.804 |
| Prevotella, Log10 CFU/mL b | 5.73 (1.72–6.88) | 6.35 (2.58–7.02) | 0.664 |
| Porphyromonas gingivalis, Log10 CFU/mL b | −0.56 (−0.80–0.03) | −0.71 (−1.61–0.03) | 0.340 |
| Escherichia coli, Log10 CFU/mL b | 3.45 (2.51–4.68) | 3.05 (2.07–5.01) | 0.385 |
| Current therapy scheme | |||
| MTX, n (%) c | - | 3 (23.08) | |
| MTX + PDN, n (%) c | - | 6 (46.15) | |
| MTX + CLQ + PDN, n (%) c | - | 4 (30.77) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaragoza-García, O.; Castro-Alarcón, N.; Pérez-Rubio, G.; Falfán-Valencia, R.; Briceño, O.; Navarro-Zarza, J.E.; Parra-Rojas, I.; Tello, M.; Guzmán-Guzmán, I.P. Serum Levels of IFABP2 and Differences in Lactobacillus and Porphyromonas gingivalis Abundance on Gut Microbiota Are Associated with Poor Therapeutic Response in Rheumatoid Arthritis: A Pilot Study. Int. J. Mol. Sci. 2023, 24, 1958. https://doi.org/10.3390/ijms24031958
Zaragoza-García O, Castro-Alarcón N, Pérez-Rubio G, Falfán-Valencia R, Briceño O, Navarro-Zarza JE, Parra-Rojas I, Tello M, Guzmán-Guzmán IP. Serum Levels of IFABP2 and Differences in Lactobacillus and Porphyromonas gingivalis Abundance on Gut Microbiota Are Associated with Poor Therapeutic Response in Rheumatoid Arthritis: A Pilot Study. International Journal of Molecular Sciences. 2023; 24(3):1958. https://doi.org/10.3390/ijms24031958
Chicago/Turabian StyleZaragoza-García, Oscar, Natividad Castro-Alarcón, Gloria Pérez-Rubio, Ramcés Falfán-Valencia, Olivia Briceño, José Eduardo Navarro-Zarza, Isela Parra-Rojas, Mario Tello, and Iris Paola Guzmán-Guzmán. 2023. "Serum Levels of IFABP2 and Differences in Lactobacillus and Porphyromonas gingivalis Abundance on Gut Microbiota Are Associated with Poor Therapeutic Response in Rheumatoid Arthritis: A Pilot Study" International Journal of Molecular Sciences 24, no. 3: 1958. https://doi.org/10.3390/ijms24031958
APA StyleZaragoza-García, O., Castro-Alarcón, N., Pérez-Rubio, G., Falfán-Valencia, R., Briceño, O., Navarro-Zarza, J. E., Parra-Rojas, I., Tello, M., & Guzmán-Guzmán, I. P. (2023). Serum Levels of IFABP2 and Differences in Lactobacillus and Porphyromonas gingivalis Abundance on Gut Microbiota Are Associated with Poor Therapeutic Response in Rheumatoid Arthritis: A Pilot Study. International Journal of Molecular Sciences, 24(3), 1958. https://doi.org/10.3390/ijms24031958









