Germline Mutations in Steroid Metabolizing Enzymes: A Focus on Steroid Transforming Aldo-Keto Reductases
Abstract
1. Introduction
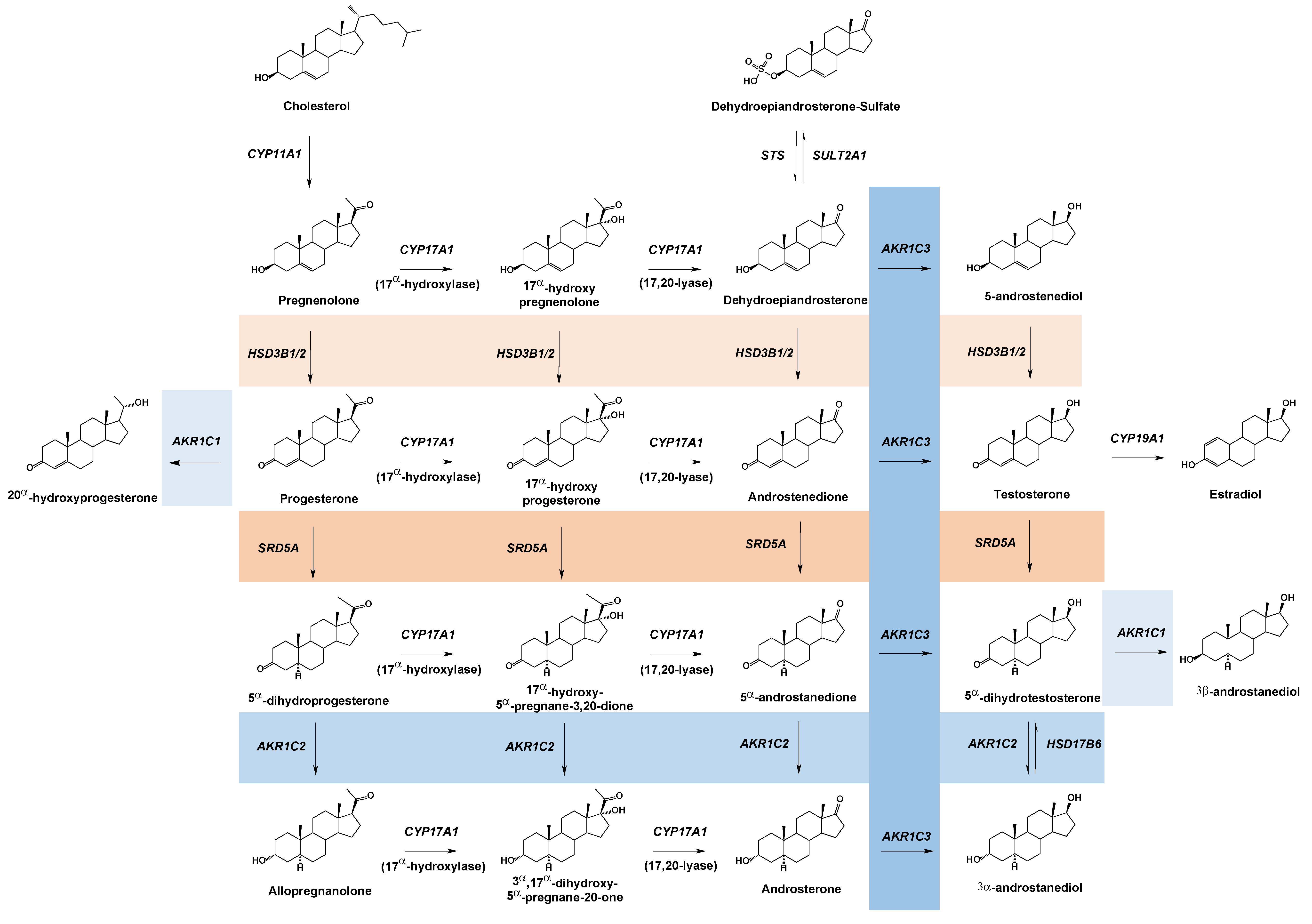
2. Aldo-Keto Reductases
3. Steroid Metabolizing AKR Enzymes
3.1. AKR1C1
3.2. AKR1C2
3.3. AKR1C3
3.4. AKR1C4
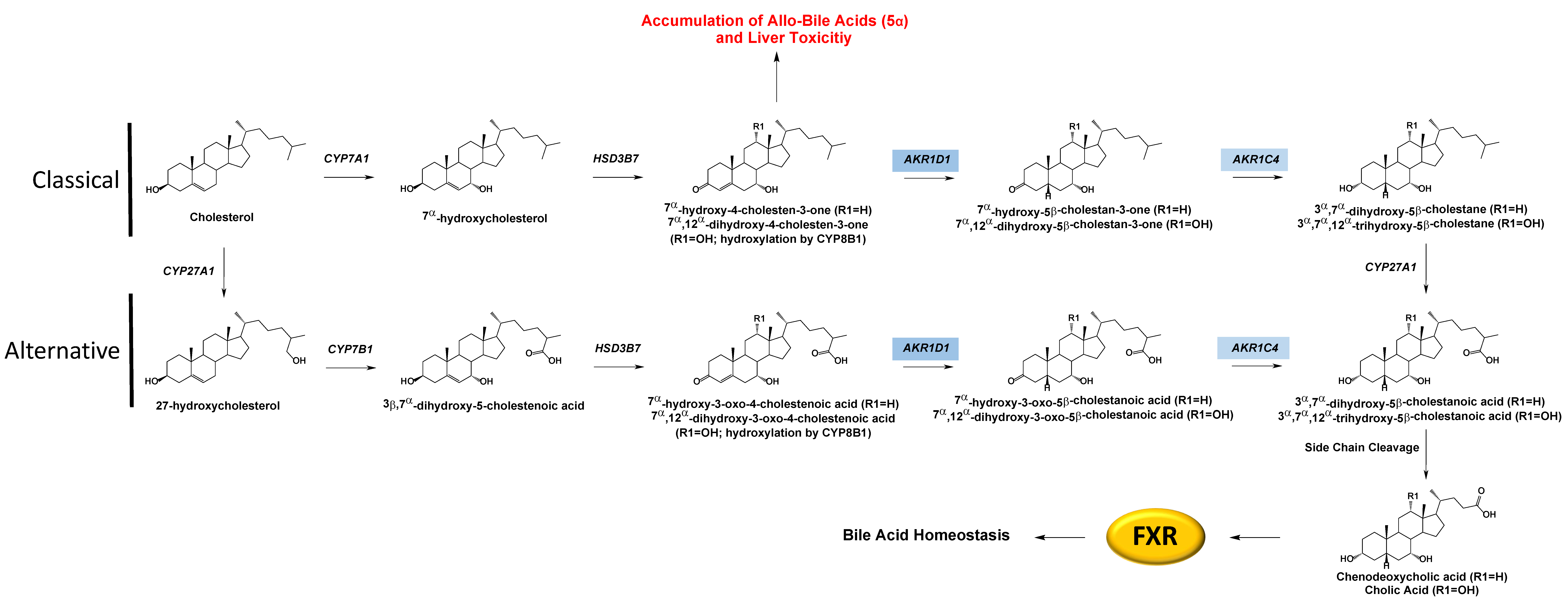
3.5. AKR1D1
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, J.; Zhang, Z.; Shen, W.J.; Azhar, S. Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutr. Metab. 2010, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.L.; Mendonca, B.B. The Molecular Basis of 5alpha-Reductase Type 2 Deficiency. Sex. Dev. 2022, 16, 171–183. [Google Scholar] [CrossRef] [PubMed]
- George, M.M.; New, M.I.; Ten, S.; Sultan, C.; Bhangoo, A. The clinical and molecular heterogeneity of 17betaHSD-3 enzyme deficiency. Horm. Res. Paediatr. 2010, 74, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Al Alawi, A.M.; Nordenstrom, A.; Falhammar, H. Clinical perspectives in congenital adrenal hyperplasia due to 3beta-hydroxysteroid dehydrogenase type 2 deficiency. Endocrine 2019, 63, 407–421. [Google Scholar] [CrossRef]
- Sahakitrungruang, T. Clinical and molecular review of atypical congenital adrenal hyperplasia. Ann. Pediatr. Endocrinol. Metab. 2015, 20, 1–7. [Google Scholar] [CrossRef]
- Chang, K.H.; Li, R.; Kuri, B.; Lotan, Y.; Roehrborn, C.G.; Liu, J.; Vessella, R.; Nelson, P.S.; Kapur, P.; Guo, X.; et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell 2013, 154, 1074–1084. [Google Scholar] [CrossRef]
- Penning, T.M. The Aldo-Keto Reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 176, 139–148. [Google Scholar] [CrossRef]
- Velica, P.; Davies, N.J.; Rocha, P.P.; Schrewe, H.; Ride, J.P.; Bunce, C.M. Lack of functional and expression homology between human and mouse aldo-keto reductase 1C enzymes: Implications for modelling human cancers. Mol. Cancer 2009, 8, 121. [Google Scholar] [CrossRef]
- Clarke, L.; Zheng-Bradley, X.; Smith, R.; Kulesha, E.; Xiao, C.; Toneva, I.; Vaughan, B.; Preuss, D.; Leinonen, R.; Shumway, M.; et al. The 1000 Genomes Project: Data management and community access. Nat. Methods 2012, 9, 459–462. [Google Scholar] [CrossRef]
- Detlefsen, A.J.; Wangtrakuldee, P.; Penning, T.M. Characterization of the major single nucleotide polymorphic variants of aldo-keto reductase 1C3 (type 5 17beta-hydroxysteroid dehydrogenase). J. Steroid Biochem. Mol. Biol. 2022, 221, 106121. [Google Scholar] [CrossRef]
- Halldorsson, B.V.; Eggertsson, H.P.; Moore, K.H.S.; Hauswedell, H.; Eiriksson, O.; Ulfarsson, M.O.; Palsson, G.; Hardarson, M.T.; Oddsson, A.; Jensson, B.O.; et al. The sequences of 150,119 genomes in the UK Biobank. Nature 2022, 607, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, E.T. Second Stand Up To Cancer Prostate Cancer Dream Team. Oncology Times 2012, 34, 38. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Cooper, W.C.; Jin, Y.; Penning, T.M. Elucidation of a complete kinetic mechanism for a mammalian hydroxysteroid dehydrogenase (HSD) and identification of all enzyme forms on the reaction coordinate: The example of rat liver 3alpha-HSD (AKR1C9). J. Biol. Chem. 2007, 282, 33484–33493. [Google Scholar] [CrossRef]
- Penning, T.M.; Wangtrakuldee, P.; Auchus, R.J. Structural and Functional Biology of Aldo-Keto Reductase Steroid-Transforming Enzymes. Endocr. Rev. 2019, 40, 447–475. [Google Scholar] [CrossRef]
- Paulukinas, R.D.; Mesaros, C.A.; Penning, T.M. Conversion of Classical and 11-Oxygenated Androgens by Insulin-Induced AKR1C3 in a Model of Human PCOS Adipocytes. Endocrinology 2022, 163, bqac068. [Google Scholar] [CrossRef]
- Piekorz, R.P.; Gingras, S.; Hoffmeyer, A.; Ihle, J.N.; Weinstein, Y. Regulation of progesterone levels during pregnancy and parturition by signal transducer and activator of transcription 5 and 20alpha-hydroxysteroid dehydrogenase. Mol. Endocrinol. 2005, 19, 431–440. [Google Scholar] [CrossRef]
- Michelini, S.; Chiurazzi, P.; Marino, V.; Dell’Orco, D.; Manara, E.; Baglivo, M.; Fiorentino, A.; Maltese, P.E.; Pinelli, M.; Herbst, K.L.; et al. Aldo-Keto Reductase 1C1 (AKR1C1) as the First Mutated Gene in a Family with Nonsyndromic Primary Lipedema. Int. J. Mol. Sci. 2020, 21, 6264. [Google Scholar] [CrossRef] [PubMed]
- Flück, C.E.; Meyer-Boni, M.; Pandey, A.V.; Kempna, P.; Miller, W.L.; Schoenle, E.J.; Biason-Lauber, A. Why boys will be boys: Two pathways of fetal testicular androgen biosynthesis are needed for male sexual differentiation. Am. J. Hum. Genet. 2011, 89, 201–218. [Google Scholar] [CrossRef]
- Lemonde, H.A.; Custard, E.J.; Bouquet, J.; Duran, M.; Overmars, H.; Scambler, P.J.; Clayton, P.T. Mutations in SRD5B1 (AKR1D1), the gene encoding delta(4)-3-oxosteroid 5beta-reductase, in hepatitis and liver failure in infancy. Gut 2003, 52, 1494–1499. [Google Scholar] [CrossRef]
- Drury, J.E.; Mindnich, R.; Penning, T.M. Characterization of disease-related 5beta-reductase (AKR1D1) mutations reveals their potential to cause bile acid deficiency. J. Biol. Chem. 2010, 285, 24529–24537. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, E.; Cresteil, D.; Baussan, C.; Dabadie, A.; Gerhardt, M.F.; Jacquemin, E. SRD5B1 (AKR1D1) gene analysis in delta(4)-3-oxosteroid 5beta-reductase deficiency: Evidence for primary genetic defect. J. Hepatol. 2004, 40, 716–718. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Wu, J.F.; Kimura, A.; Nittono, H.; Liou, B.Y.; Lee, C.S.; Chen, H.S.; Chiu, Y.C.; Ni, Y.H.; Peng, S.S.; et al. AKR1D1 and CYP7B1 mutations in patients with inborn errors of bile acid metabolism: Possibly underdiagnosed diseases. Pediatr. Neonatol. 2020, 61, 75–83. [Google Scholar] [CrossRef]
- Seki, Y.; Mizuochi, T.; Kimura, A.; Takahashi, T.; Ohtake, A.; Hayashi, S.; Morimura, T.; Ohno, Y.; Hoshina, T.; Ihara, K.; et al. Two neonatal cholestasis patients with mutations in the SRD5B1 (AKR1D1) gene: Diagnosis and bile acid profiles during chenodeoxycholic acid treatment. J. Inherit. Metab Dis. 2013, 36, 565–573. [Google Scholar] [CrossRef]
- Ueki, I.; Kimura, A.; Chen, H.L.; Yorifuji, T.; Mori, J.; Itoh, S.; Maruyama, K.; Ishige, T.; Takei, H.; Nittono, H.; et al. SRD5B1 gene analysis needed for the accurate diagnosis of primary 3-oxo-Delta4-steroid 5beta-reductase deficiency. J. Gastroenterol. Hepatol. 2009, 24, 776–785. [Google Scholar] [CrossRef]
- Taketani, Y.; Yamagishi, R.; Fujishiro, T.; Igarashi, M.; Sakata, R.; Aihara, M. Activation of the prostanoid FP receptor inhibits adipogenesis leading to deepening of the upper eyelid sulcus in prostaglandin-associated periorbitopathy. Invest. Ophthalmol. Vis. Sci. 2014, 55, 1269–1276. [Google Scholar] [CrossRef]
- Lepak, N.M.; Serrero, G. Prostaglandin F2 alpha stimulates transforming growth factor-alpha expression in adipocyte precursors. Endocrinology 1995, 136, 3222–3229. [Google Scholar] [CrossRef]
- Komoto, J.; Yamada, T.; Watanabe, K.; Takusagawa, F. Crystal structure of human prostaglandin F synthase (AKR1C3). Biochemistry 2004, 43, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.W.; Kempegowda, P.; Walsh, M.; Taylor, A.E.; Manolopoulos, K.N.; Allwood, J.W.; Semple, R.K.; Hebenstreit, D.; Dunn, W.B.; Tomlinson, J.W.; et al. AKR1C3-Mediated Adipose Androgen Generation Drives Lipotoxicity in Women With Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2017, 102, 3327–3339. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, J.V.; Esquenet, M.; Goossens, K.; Heyns, W.; Verhoeven, G. Androgens stimulate fatty acid synthase in the human prostate cancer cell line LNCaP. Cancer Res. 1997, 57, 1086–1090. [Google Scholar]
- Takahashi, R.H.; Grigliatti, T.A.; Reid, R.E.; Riggs, K.W. The effect of allelic variation in aldo-keto reductase 1C2 on the in vitro metabolism of dihydrotestosterone. J. Pharmacol. Exp. Ther. 2009, 329, 1032–1039. [Google Scholar] [CrossRef]
- Zachmann, M.; Vollmin, J.A.; Hamilton, W.; Prader, A. Steroid 17,20-desmolase deficiency: A new cause of male pseudohermaphroditism. Clin. Endocrinol. 1972, 1, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.W.; Reichardt, J.K. Modeling single nucleotide polymorphisms in the human AKR1C1 and AKR1C2 genes: Implications for functional and genotyping analyses. PLoS ONE 2010, 5, e15604. [Google Scholar] [CrossRef]
- Penning, T.M.; Detlefsen, A.J. Intracrinology-revisited and prostate cancer. J. Steroid Biochem. Mol. Biol. 2020, 196, 105499. [Google Scholar] [CrossRef]
- Karunasinghe, N.; Ambs, S.; Wang, A.; Tang, W.; Zhu, S.; Dorsey, T.H.; Goudie, M.; Masters, J.G.; Ferguson, L.R. Influence of lifestyle and genetic variants in the aldo-keto reductase 1C3 rs12529 polymorphism in high-risk prostate cancer detection variability assessed between US and New Zealand cohorts. PLoS ONE 2018, 13, e0199122. [Google Scholar] [CrossRef]
- Karunasinghe, N.; Symes, E.; Gamage, A.; Wang, A.; Murray, P.; Zhu, S.; Goudie, M.; Masters, J.; Ferguson, L.R. Interaction between leukocyte aldo-keto reductase 1C3 activity, genotypes, biological, lifestyle and clinical features in a prostate cancer cohort from New Zealand. PLoS ONE 2019, 14, e0217373. [Google Scholar] [CrossRef]
- Karunasinghe, N.; Zhu, Y.; Han, D.Y.; Lange, K.; Zhu, S.; Wang, A.; Ellett, S.; Masters, J.; Goudie, M.; Keogh, J.; et al. Quality of life effects of androgen deprivation therapy in a prostate cancer cohort in New Zealand: Can we minimize effects using a stratification based on the aldo-keto reductase family 1, member C3 rs12529 gene polymorphism? BMC Urol. 2016, 16, 48. [Google Scholar] [CrossRef]
- Shiota, M.; Endo, S.; Fujimoto, N.; Tsukahara, S.; Ushijima, M.; Kashiwagi, E.; Takeuchi, A.; Inokuchi, J.; Uchiumi, T.; Eto, M. Polymorphisms in androgen metabolism genes with serum testosterone levels and prognosis in androgen-deprivation therapy. Urol. Oncol. 2020, 38, 849.e11–849.e18. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, J.; Palonek, E.; Lorentzon, M.; Ohlsson, C.; Rane, A.; Ekstrom, L. A novel polymorphism in the 17beta-hydroxysteroid dehydrogenase type 5 (aldo-keto reductase 1C3) gene is associated with lower serum testosterone levels in caucasian men. Pharmacogenomics J. 2007, 7, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Ju, R.; Wu, W.; Fei, J.; Qin, Y.; Tang, Q.; Wu, D.; Xia, Y.; Wu, J.; Wang, X. Association analysis between the polymorphisms of HSD17B5 and HSD17B6 and risk of polycystic ovary syndrome in Chinese population. Eur. J. Endocrinol. 2015, 172, 227–233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roberts, R.O.; Bergstralh, E.J.; Farmer, S.A.; Jacobson, D.J.; Hebbring, S.J.; Cunningham, J.M.; Thibodeau, S.N.; Lieber, M.M.; Jacobsen, S.J. Polymorphisms in genes involved in sex hormone metabolism may increase risk of benign prostatic hyperplasia. Prostate 2006, 66, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Soderhall, C.; Korberg, I.B.; Thai, H.T.; Cao, J.; Chen, Y.; Zhang, X.; Shulu, Z.; van der Zanden, L.F.; van Rooij, I.A.; Frisen, L.; et al. Fine mapping analysis confirms and strengthens linkage of four chromosomal regions in familial hypospadias. Eur. J. Hum. Genet. 2015, 23, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Tiryakioglu, N.O.; Tunali, N.E. Association of AKR1C3 Polymorphisms with Bladder Cancer. Urol J 2016, 13, 2615–2621. [Google Scholar]
- Figueroa, J.D.; Malats, N.; Garcia-Closas, M.; Real, F.X.; Silverman, D.; Kogevinas, M.; Chanock, S.; Welch, R.; Dosemeci, M.; Lan, Q.; et al. Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis 2008, 29, 1955–1962. [Google Scholar] [CrossRef]
- Lan, Q.; Mumford, J.L.; Shen, M.; Demarini, D.M.; Bonner, M.R.; He, X.; Yeager, M.; Welch, R.; Chanock, S.; Tian, L.; et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis 2004, 25, 2177–2181. [Google Scholar] [CrossRef]
- Platt, A.; Xia, Z.; Liu, Y.; Chen, G.; Lazarus, P. Impact of nonsynonymous single nucleotide polymorphisms on in-vitro metabolism of exemestane by hepatic cytosolic reductases. Pharmacogenet Genomics 2016, 26, 370–380. [Google Scholar] [CrossRef]
- Andreen, L.; Nyberg, S.; Turkmen, S.; van Wingen, G.; Fernandez, G.; Backstrom, T. Sex steroid induced negative mood may be explained by the paradoxical effect mediated by GABAA modulators. Psychoneuroendocrinology 2009, 34, 1121–1132. [Google Scholar] [CrossRef]
- Liang, J.J.; Rasmusson, A.M. Overview of the Molecular Steps in Steroidogenesis of the GABAergic Neurosteroids Allopregnanolone and Pregnanolone. Chronic Stress 2018, 2, 2470547018818555. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.G.; Nikamo, P.; Schalling, M.; Landen, M. AKR1C4 gene variant associated with low euthymic serum progesterone and a history of mood irritability in males with bipolar disorder. J. Affect. Disord. 2011, 133, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.G.; Nikamo, P.; Schalling, M.; Landen, M. Polymorphisms in AKR1C4 and HSD3B2 and differences in serum DHEAS and progesterone are associated with paranoid ideation during mania or hypomania in bipolar disorder. Eur. Neuropsychopharmacol. 2012, 22, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Lord, S.J.; Mack, W.J.; Van Den Berg, D.; Pike, M.C.; Ingles, S.A.; Haiman, C.A.; Wang, W.; Parisky, Y.R.; Hodis, H.N.; Ursin, G. Polymorphisms in genes involved in estrogen and progesterone metabolism and mammographic density changes in women randomized to postmenopausal hormone therapy: Results from a pilot study. Breast Cancer Res. 2005, 7, R336–R344. [Google Scholar] [CrossRef] [PubMed]
- Kume, T.; Iwasa, H.; Shiraishi, H.; Yokoi, T.; Nagashima, K.; Otsuka, M.; Terada, T.; Takagi, T.; Hara, A.; Kamataki, T. Characterization of a novel variant (S145C/L311V) of 3alpha-hydroxysteroid/dihydrodiol dehydrogenase in human liver. Pharmacogenetics 1999, 9, 763–771. [Google Scholar] [CrossRef]
- Gathercole, L.L.; Nikolaou, N.; Harris, S.E.; Arvaniti, A.; Poolman, T.M.; Hazlehurst, J.M.; Kratschmar, D.V.; Todorcevic, M.; Moolla, A.; Dempster, N.; et al. AKR1D1 knockout mice develop a sex-dependent metabolic phenotype. J. Endocrinol. 2022, 253, 97–113. [Google Scholar] [CrossRef]
- Chen, M.; Jin, Y.; Penning, T.M. In-Depth Dissection of the P133R Mutation in Steroid 5beta-Reductase (AKR1D1): A Molecular Basis of Bile Acid Deficiency. Biochemistry 2015, 54, 6343–6351. [Google Scholar] [CrossRef]


| AKR Enzyme | Missense Mutation | References | Disorder |
|---|---|---|---|
| AKR1C1 | L213Q | Michelini et al., 2020 [21] | Nonsyndromic Primary Lipedema |
| AKR1C2 | I79V | Flück et al., 2011 [22] | 46,XY DSD |
| H90Q | |||
| N300T | |||
| H222Q | |||
| AKR1D1 | P198L | Lemonde et al., 2003, Drury et al., 2010 [23,24] | Bile Acid Deficiency |
| L106F | |||
| P133R | Gonzales et al., 2004, Drury et al. 2010, Chen et al., 2020 [24,25,26] | ||
| R261C | Gonzales et al., 2004, Seki et al., 2013, Drury et al., 2010 [24,25,27] | ||
| G223E | Ueki et al., 2009, Seki et al., 2013, Drury et al., 2010 [24,27,28] | ||
| R266Q | Seki et al., 2013, Chen et al., 2020 [26,27] | ||
| T25I | Chen et al., 2020 [26] |
| AKR Enzyme | Missense nsSNP | NCBI Identifier | MAF |
|---|---|---|---|
| AKR1C1 * | - | - | - |
| AKR1C2 | F46Y | rs2854482 | 0.0649 |
| AKR1C3 | H5Q | rs12529 | 0.4203 |
| K104D | rs12387 | 0.1518 | |
| E77G | rs11551177 | 0.0367 | |
| R258C | rs62621365 | 0.0325 | |
| R66Q | rs35961894 | 0.0230 | |
| AKR1C4 | C145S | rs3829125 | 0.1028 |
| L311V | rs17134592 | 0.1024 | |
| G135E | rs11253043 | 0.0270 | |
| AKR1D1 * | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detlefsen, A.J.; Paulukinas, R.D.; Penning, T.M. Germline Mutations in Steroid Metabolizing Enzymes: A Focus on Steroid Transforming Aldo-Keto Reductases. Int. J. Mol. Sci. 2023, 24, 1873. https://doi.org/10.3390/ijms24031873
Detlefsen AJ, Paulukinas RD, Penning TM. Germline Mutations in Steroid Metabolizing Enzymes: A Focus on Steroid Transforming Aldo-Keto Reductases. International Journal of Molecular Sciences. 2023; 24(3):1873. https://doi.org/10.3390/ijms24031873
Chicago/Turabian StyleDetlefsen, Andrea J., Ryan D. Paulukinas, and Trevor M. Penning. 2023. "Germline Mutations in Steroid Metabolizing Enzymes: A Focus on Steroid Transforming Aldo-Keto Reductases" International Journal of Molecular Sciences 24, no. 3: 1873. https://doi.org/10.3390/ijms24031873
APA StyleDetlefsen, A. J., Paulukinas, R. D., & Penning, T. M. (2023). Germline Mutations in Steroid Metabolizing Enzymes: A Focus on Steroid Transforming Aldo-Keto Reductases. International Journal of Molecular Sciences, 24(3), 1873. https://doi.org/10.3390/ijms24031873







