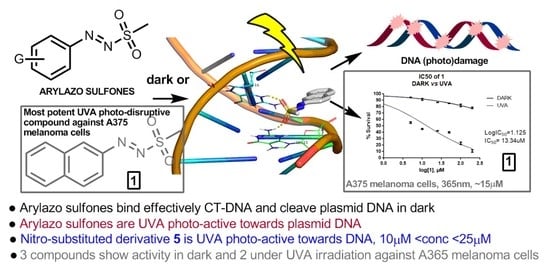
Effect of Arylazo Sulfones on DNA: Binding, Cleavage, Photocleavage, Molecular Docking Studies and Interaction with A375 Melanoma and Non-Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
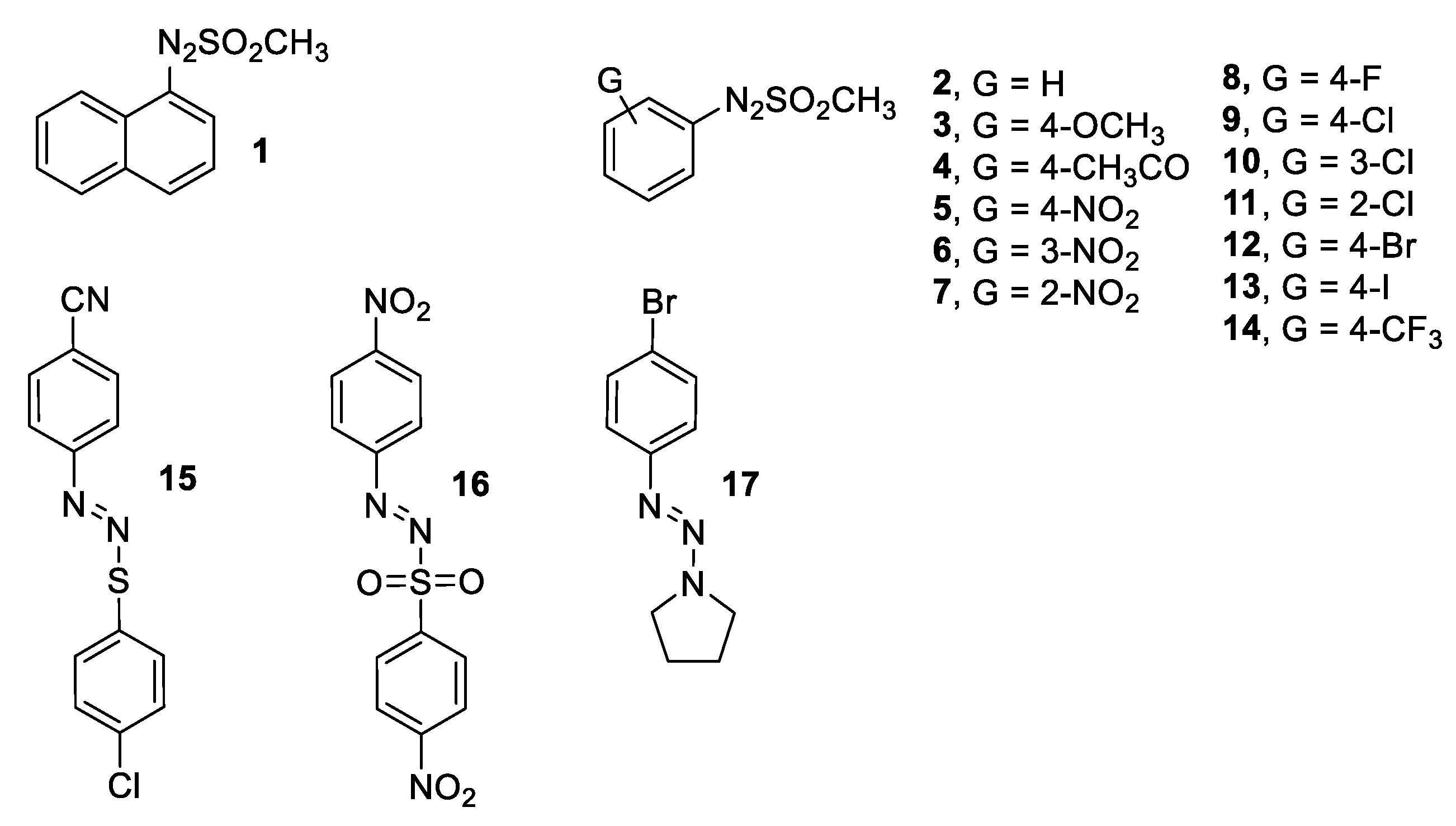
2.1. Synthesis and Characterization of Arylazo Sulfones
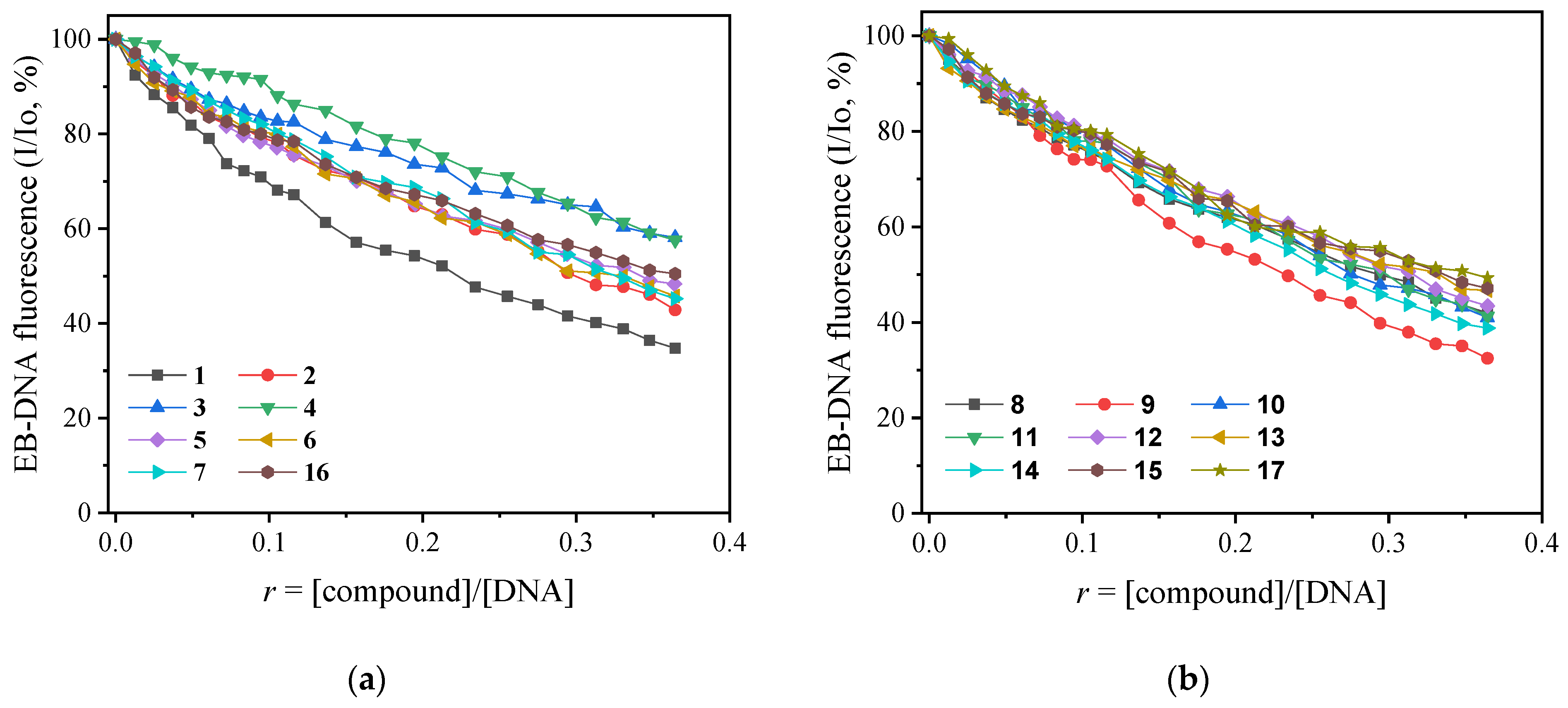
2.2. CT DNA Binding Studies of Arylazo Sulfones
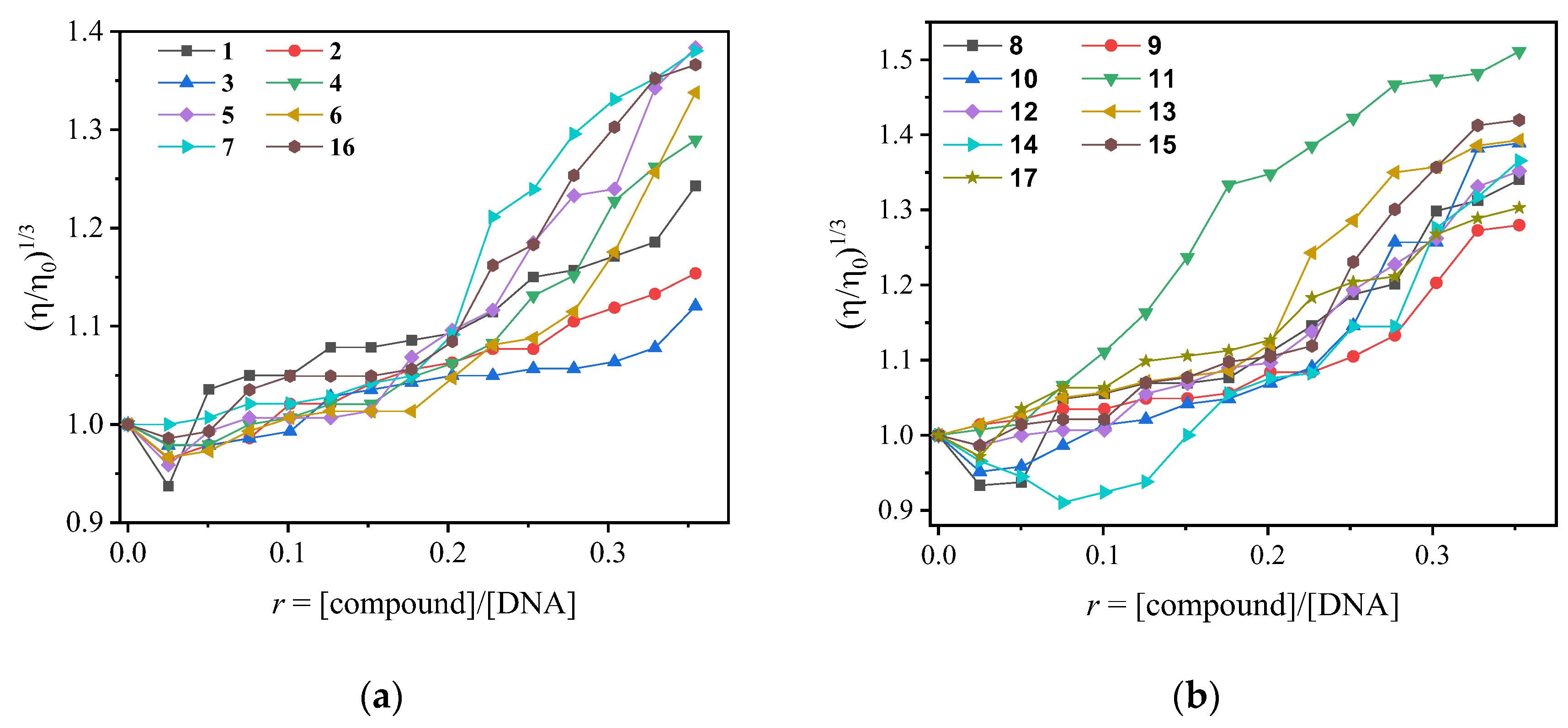
2.3. DNA Interactions of Arylazo Sulfones with Plasmid DNA pBluescript SK II
2.3.1. DNA Cleavage Experiments
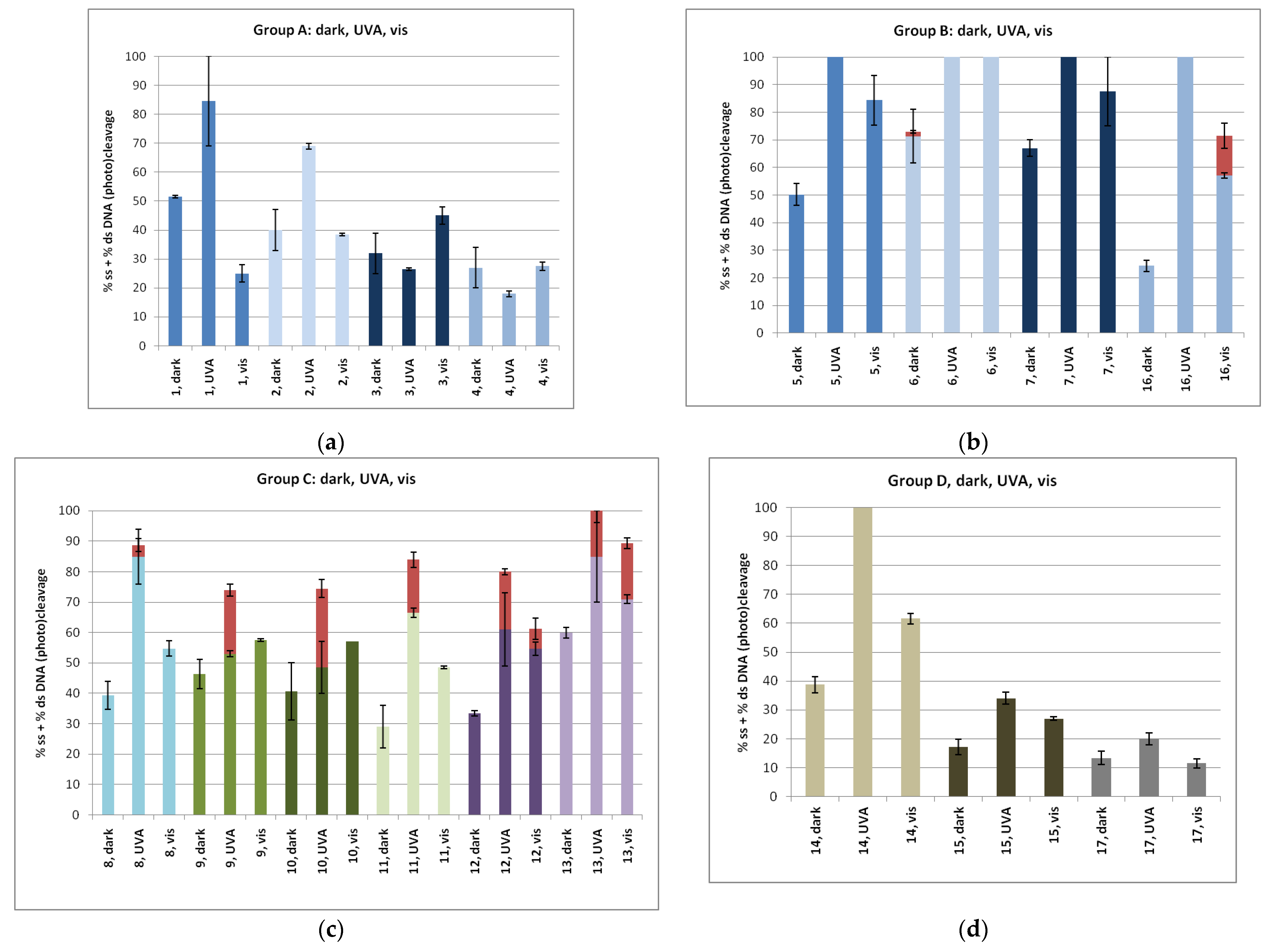
2.3.2. DNA Photo-Cleavage Experiments
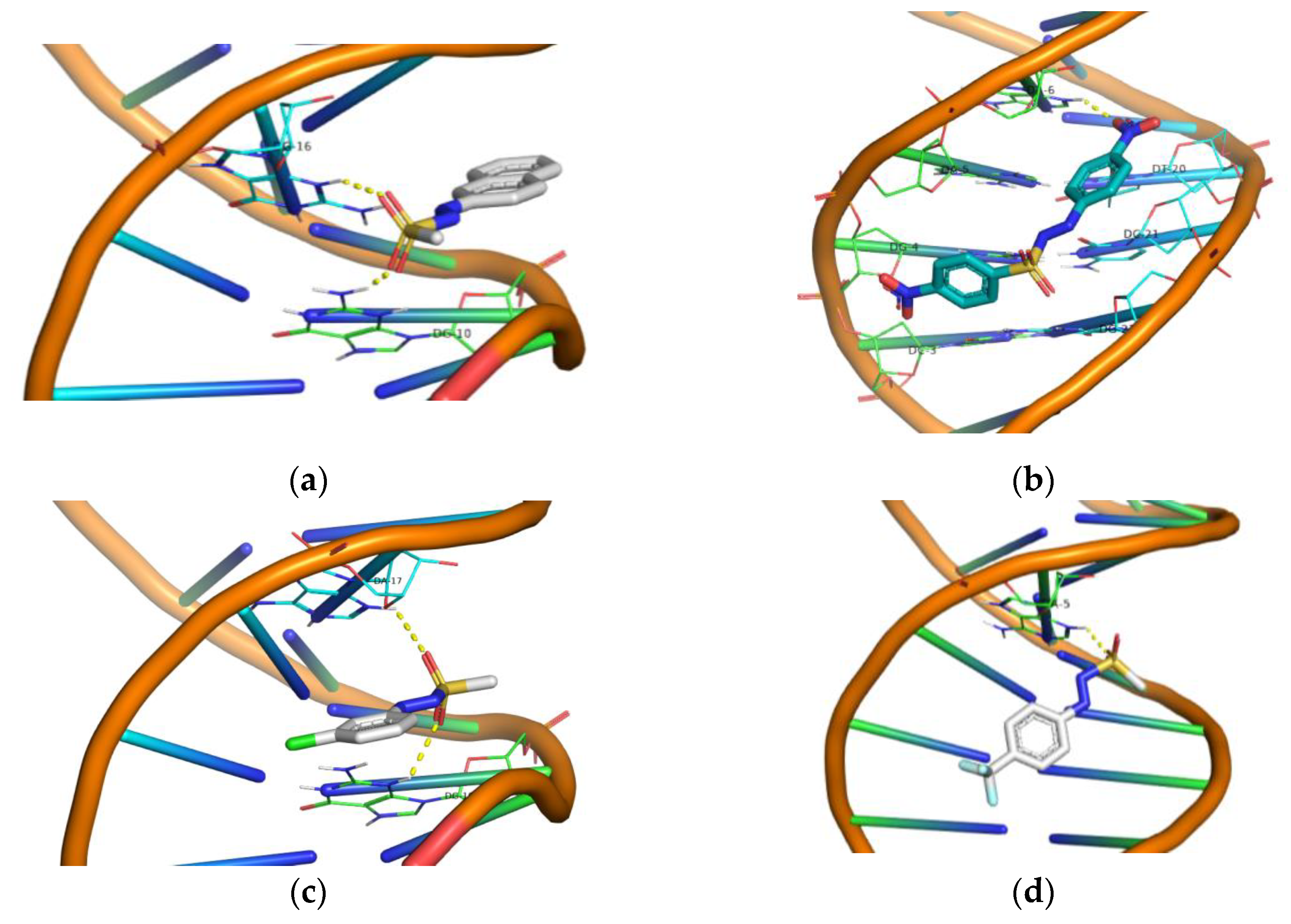
2.4. Molecular Docking “In Silico” Calculations of DNA/Arylazo Sulfones
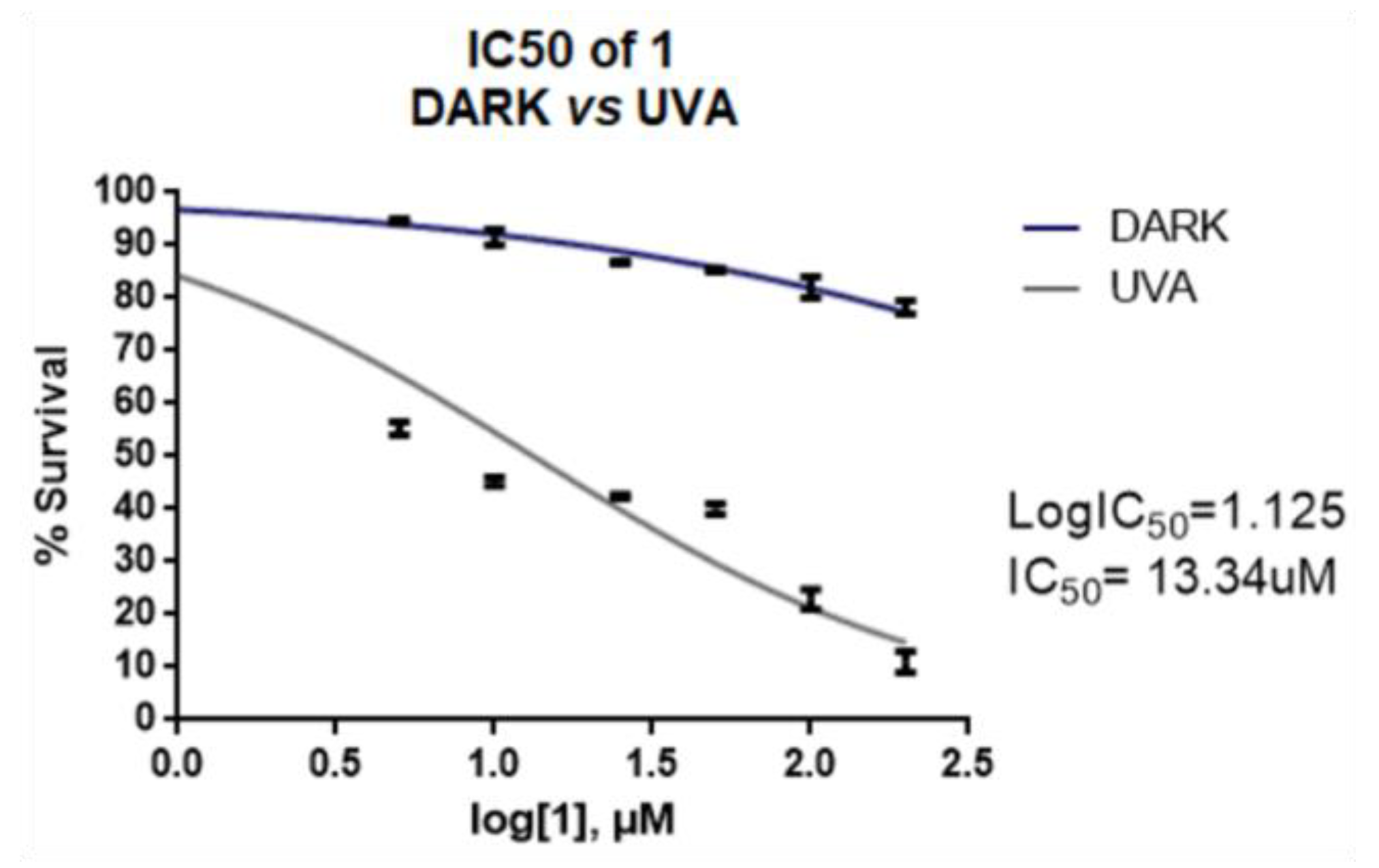
2.5. Cell Culture Experiments of Arylazo Sulfones with Melanoma Cell Lines
2.6. Cell Culture Experiments of the Photoactive Arylazo Sulfones 1, 14 and 17 with Non-Cancer Cell Lines
3. Materials and Methods
3.1. Interaction with CT DNA
3.2. DNA Cleavage and Photo-Cleavage Experiments
3.3. Molecular Docking Studies
3.4. Cell Culture Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, A.; Bhattacharya, S. DNA binders in clinical trials and chemotherapy. Bioorg. Med. Chem. 2014, 22, 4506–4521. [Google Scholar] [CrossRef] [PubMed]
- Portugal, J. Challenging transcription by DNA-binding antitumor drugs. Biochem. Pharmacol. 2018, 155, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Bhattacharya, S. Chemistry and biology of DNA-binding small molecules. Curr. Sci. 2012, 102, 212–231. [Google Scholar]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.; Fischer, F.L.; Ribeiro, C.W.; Silveira, G.P.; Sá, M.M.; Nome, F.; Terenzi, H. Metal-free artificial nucleases based on simple oxime and hydroxylamine scaffolds. Bioorg. Med. Chem. Lett. 2008, 18, 4499–4502. [Google Scholar] [CrossRef]
- Reddy, D.S.; Kongot, M.; Netalkar, S.P.; Kurjogi, M.M.; Kumar, R.; Avecilla, F.; Kumar, A. Synthesis and evaluation of novel coumarin-oxime ethers as potential anti-tubercular agents: Their DNA cleavage ability and BSA interaction study. Eur. J. Med. Chem. 2018, 150, 864–875. [Google Scholar] [CrossRef]
- Obalı, A.Y.; Akçaalan, S.; Arslan, E.; Obalı, İ. Antibacterial activities and DNA-cleavage properties of novel fluorescent imidazo-phenanthroline derivatives. Bioorg. Chem. 2020, 100, 103885. [Google Scholar] [CrossRef]
- Gratal, P.; Arias-Pérez, M.S.; Gude, L. 1H-imidazo[4,5-f][1,10]phenanthroline carbohydrate conjugates: Synthesis, DNA interactions and cytotoxic activity. Bioorg. Chem. 2022, 125, 105851. [Google Scholar] [CrossRef]
- Bhat, R.; Begum, N.S. Synthesis, characterization and molecular docking studies of new indol(1H-3-yl)pyrimidine derivatives: Insights into their role in DNA interaction. Nucleosides Nucleotides Nucleic Acids 2021, 40, 619–634. [Google Scholar] [CrossRef]
- Eryılmaz, S.; Türk Çelikoğlu, E.; İdil, Ö.; İnkaya, E.; Kozak, Z.; Mısır, E.; Gül, M. Derivatives of pyridine and thiazole hybrid: Synthesis, DFT, biological evaluation via antimicrobial and DNA cleavage activity. Bioorg. Chem. 2020, 95, 103476. [Google Scholar] [CrossRef]
- Doğan, A.; Özdemir, S.; Yalçin, M.S.; Sari, H.; Nural, Y. Naphthoquinone-thiazole hybrids bearing adamantane: Synthesis, antimicrobial, DNA cleavage, antioxidant activity, acid dissociation constant, and drug-likeness. J. Res. Pharm. 2021, 25, 292–304. [Google Scholar] [CrossRef]
- Mishra, V.R.; Ghanavatkar, C.W.; Mali, S.N.; Chaudhari, H.K.; Sekar, N. Schiff base clubbed benzothiazole: Synthesis, potent antimicrobial and MCF-7 anticancer activity, DNA cleavage and computational study. J. Biomol. Struct. Dyn. 2020, 38, 1772–1785. [Google Scholar] [CrossRef]
- Katrahalli, U.; Chanabasappa Yallur, B.; Manjunatha, D.H.; Krishna, P.M. BSA interaction and DNA cleavage studies of anti-bacterial benzothiazol-2-yl-malonaldehyde. J. Mol. Struct. 2019, 1196, 96–104. [Google Scholar] [CrossRef]
- Nural, Y.; Ozdemir, S.; Yalcin, M.S.; Demir, B.; Atabey, H.; Seferoglu, Z.; Ece, A. New bis- and tetrakis-1,2,3-triazole derivatives: Synthesis, DNA cleavage, molecular docking, antimicrobial, antioxidant activity and acid dissociation constants. Bioorg. Med. Chem. Lett. 2022, 55, 128453. [Google Scholar] [CrossRef]
- Caron, C.; Duong, X.N.T.; Guillot, R.; Bombard, S.; Granzhan, A. Interaction of Functionalized Naphthalenophanes with Abasic Sites in DNA: DNA Cleavage, DNA Cleavage Inhibition, and Formation of Ligand–DNA Adducts. Chem.—Eur. J. 2019, 25, 1949–1962. [Google Scholar] [CrossRef]
- Rafique, J.; Farias, G.; Saba, S.; Zapp, E.; Bellettini, I.C.; Momoli Salla, C.A.; Bechtold, I.H.; Scheide, M.R.; Santos Neto, J.S.; Monteiro de Souza Junior, D.; et al. Selenylated-oxadiazoles as promising DNA intercalators: Synthesis, electronic structure, DNA interaction and cleavage. Dye. Pigment. 2020, 180, 108519. [Google Scholar] [CrossRef]
- Wei, X.W.; Yuan, J.M.; Huang, W.Y.; Chen, N.Y.; Li, X.J.; Pan, C.X.; Mo, D.L.; Su, G.F. 2-Styryl-4-aminoquinazoline derivatives as potent DNA-cleavage, p53-activation and in vivo effective anticancer agents. Eur. J. Med. Chem. 2020, 186, 111851. [Google Scholar] [CrossRef]
- Ozkan, S.C.; Aksakal, F.; Yilmaz, A. Synthesis of novel calix[4]arene: P -benzazole derivatives and investigation of their DNA binding and cleavage activities with molecular docking and experimental studies. RSC Adv. 2020, 10, 38695–38708. [Google Scholar] [CrossRef]
- Rathod, A.S.; Biradar, J.S. Synthesis of Some Indolyl Derivatives under Solvent Free Conditions, Their Cytotoxicity, and DNA Cleavage Studies. Russ. J. Gen. Chem. 2020, 90, 135–141. [Google Scholar] [CrossRef]
- Danilkina, N.A.; Rumyantsev, A.M.; Lyapunova, A.L.; D’Yachenko, A.S.; Khlebnikov, A.F.; Balova, I.A. 10-Membered Azaenediyne Fused to a Benzothiophene through the Nicholas Macrocyclization: Synthesis and DNA Cleavage Ability. Synlett 2019, 30, 161–166. [Google Scholar] [CrossRef]
- Zhang, H.; Li, R.; Ba, S.; Lu, Z.; Pitsinos, E.N.; Li, T.; Nicolaou, K.C. DNA Binding and Cleavage Modes of Shishijimicin A. J. Am. Chem. Soc. 2019, 141, 7842–7852. [Google Scholar] [CrossRef] [PubMed]
- Marrani, E.; Foeldvari, I.; Lopez, J.A.; Cimaz, R.; Simonini, G. Comparing ultraviolet light A photo(chemo)therapy with Methotrexate protocol in childhood localized scleroderma: Evidence from systematic review and meta-analysis approach. Semin. Arthritis Rheum. 2018, 48, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Garritsen, F.M.; Brouwer, M.W.D.; Limpens, J.; Spuls, P.I. Photo(chemo)therapy in the management of atopic dermatitis: An updated systematic review with implications for practice and research. Br. J. Dermatol. 2014, 170, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.N.; Mascharak, P.K. Light-assisted and remote delivery of carbon monoxide to malignant cells and tissues: Photochemotherapy in the spotlight. J. Photochem. Photobiol. C Photochem. Rev. 2020, 42, 100341. [Google Scholar] [CrossRef]
- de Souza da Fonseca, A.; de Paoli, F.; Mencalha, A.L. Photodynamic therapy for treatment of infected burns. Photodiagnosis Photodyn. Ther. 2022, 38, 102831. [Google Scholar] [CrossRef]
- Yaqoob, M.D.; Xu, L.; Li, C.; Leong, M.M.L.; Xu, D.D. Targeting mitochondria for cancer photodynamic therapy. Photodiagnosis Photodyn. Ther. 2022, 38, 102830. [Google Scholar] [CrossRef]
- Heerfordt, I.M.; Philipsen, P.A.; Wulf, H.C. Bringing the gentle properties of daylight photodynamic therapy indoors: A systematic review of efficacy and safety. Photodiagnosis Photodyn. Ther. 2022, 39, 102858. [Google Scholar] [CrossRef]
- Mazur, A.; Koziorowska, K.; Dynarowicz, K.; Aebisher, D.; Bartusik-Aebisher, D. Photodynamic Therapy for Treatment of Disease in Children—A Review of the Literature. Children 2022, 9, 695. [Google Scholar] [CrossRef]
- Morita, A.; Tateishi, C.; Ikumi, K.; Hayashi, D.; Nakada, A.; Nishihara, H.; Torii, K.; Nishida, E.; Tsuruta, D. Comparison of the Efficacy and Safety of Bexarotene and Photo(Chemo)Therapy Combination Therapy and Bexarotene Monotherapy for Cutaneous T-Cell Lymphoma. Dermatol. Ther. 2022, 12, 615–629. [Google Scholar] [CrossRef]
- Hu, S.; Dong, C.; Wang, J.; Liu, K.; Zhou, Q.; Xiang, J.; Zhou, Z.; Liu, F.; Shen, Y. Assemblies of indocyanine green and chemotherapeutic drug to cure established tumors by synergistic chemo-photo therapy. J. Control. Release 2020, 324, 250–259. [Google Scholar] [CrossRef]
- Ghosh, S.; Gul, A.R.; Xu, P.; Lee, S.Y.; Rafique, R.; Kim, Y.H.; Park, T.J. Target delivery of photo-triggered nanocarrier for externally activated chemo-photodynamic therapy of prostate cancer. Mater. Today Chem. 2022, 23, 100688. [Google Scholar] [CrossRef]
- Zamani, M.; Aghajanzadeh, M.; Jashnani, S.; Darvishzad, S.; Khoramabadi, H.; Shirin Shahangian, S.; Shirini, F. Combination of chemo and photo dynamic therapy using pH triggered bio-coated spinels for treatment of breast cancer. J. Mol. Liq. 2022, 358, 119211. [Google Scholar] [CrossRef]
- Ma, C.-H.; Ma, H.-H.; Deng, X.-B.; Yu, R.; Song, K.-W.; Wei, K.-K.; Wang, C.-J.; Li, H.-X.; Chen, H. Photodynamic Therapy in Combination with Chemotherapy, Targeted, and Immunotherapy As a Successful Therapeutic Approach for Advanced Gastric Adenocarcinoma: A Case Report and Literature Review. Photobiomodulation Photomed. Laser Surg. 2022, 40, 308–314. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Guo, X.; Wang, L.; Zeng, J.; Qiu, H.; Tan, Y.; Chen, D.; Zhao, H.; Gu, Y. Enhanced antimicrobial activity through the combination of antimicrobial photodynamic therapy and low-frequency ultrasonic irradiation. Adv. Drug Deliv. Rev. 2022, 183, 114168. [Google Scholar] [CrossRef]
- Ferrisse, T.M.; Dias, L.M.; de Oliveira, A.B.; Jordão, C.C.; de Oliveira Mima, E.G.; Pavarina, A.C. Efficacy of curcumin-mediated antibacterial photodynamic therapy for oral antisepsis: A systematic review and network meta-analysis of randomized clinical trials. Photodiagnosis Photodyn. Ther. 2022, 39, 102876. [Google Scholar] [CrossRef]
- Sales, L.S.; Miranda, M.L.; de Oliveira, A.B.; Ferrisse, T.M.; Fontana, C.R.; Milward, M.; Brighenti, F.L. Effect of the technique of photodynamic therapy against the main microorganisms responsible for periodontitis: A systematic review of in-vitro studies. Arch. Oral Biol. 2022, 138, 105425. [Google Scholar] [CrossRef]
- Maisch, T. Resistance in antimicrobial photodynamic inactivation of bacteria. Photochem. Photobiol. Sci. 2015, 14, 1518–1526. [Google Scholar] [CrossRef]
- Hamblin, M.R. Antimicrobial photodynamic inactivation: A bright new technique to kill resistant microbes. Curr. Opin. Microbiol. 2016, 33, 67–73. [Google Scholar] [CrossRef]
- Qiu, D.; Lian, C.; Mao, J.; Fagnoni, M.; Protti, S. Dyedauxiliary Groups, an Emerging Approach in Organic Chemistry. The Case of Arylazo Sulfones. J. Org. Chem. 2020, 85, 12813–12822. [Google Scholar] [CrossRef]
- Crespi, S.; Fagnoni, M. Generation of Alkyl Radicals: From the Tyranny of Tin to the Photon Democracy. Chem. Rev. 2020, 120, 9790–9833. [Google Scholar] [CrossRef]
- Coyle, R.; Fahey, K.; Aldabbagh, F. Barton esters for initiator-free radical cyclisation with heteroaromatic substitution. Org. Biomol. Chem. 2013, 11, 1672–1682. [Google Scholar] [CrossRef] [PubMed]
- Coyle, R.; McArdle, P.; Aldabbagh, F. Tandem reactions via barton esters with intermolecular addition and vinyl radical substitution onto indole. J. Org. Chem. 2014, 79, 5903–5907. [Google Scholar] [CrossRef]
- Saraiva, M.F.; Couri, M.R.C.; Le Hyaric, M.; de Almeida, M.V. The Barton ester free-radical reaction: A brief review of applications. Tetrahedron 2009, 65, 3563–3572. [Google Scholar] [CrossRef]
- Protti, S.; Ravelli, D.; Fagnoni, M. Designing Radical Chemistry by Visible-Light Promoted Homolysis. Trends Chem. 2022, 4, 305–317. [Google Scholar] [CrossRef]
- Blom, P.; Xiang, A.X.; Kao, D.; Theodorakis, E.A. Design, synthesis, and evaluation of N-aroyloxy-2-thiopyridones as DNA photocleaving reagents. Bioorg. Med. Chem. 1999, 7, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Theodorakis, E.A.; Xiang, X.; Blom, P. Photochemical cleavage of duplex DNA by N -benzoyloxy-2-thiopyridone linked to 9-aminoacridine. Chem. Commun. 1997, 5, 1463–1464. [Google Scholar] [CrossRef]
- Theodorakis, E.A.; Wilcoxen, K.M. N-Aroyloxy-2-thiopyridones as efficient oxygen-radical generators: Novel time-controlled DNA photocleaving reagents. Chem. Commun. 1996, 16, 1927–1928. [Google Scholar] [CrossRef]
- Theodorakis, E.A.; Xiang, X.; Lee, M.; Gibson, T. On the Mechanism of Photo-induced Nucleic Acid Cleavage Using N-Aroyloxy-2-thiopyridones. Tetrahedron Lett. 1998, 39, 3383–3386. [Google Scholar] [CrossRef]
- Qian, X.; Yao, W.; Chen, G.; Huang, X.; Mao, P. N-Aroyloxynaphthalimides as novel highly efficient DNA photocleavers: Substituent effects. Tetrahedron Lett. 2001, 42, 6175–6178. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, X.; Qian, X.; Yao, W. N-Aroyloxylthioxo-naphthalimides as DNA photocleavers of aroyloxyl oxygen radicals: Synthesis, evaluation, and substituents’ effect. Bioorg. Med. Chem. 2004, 12, 2335–2341. [Google Scholar] [CrossRef]
- Alonso, R.; Campos, P.J.; Rodríguez, M.A.; Sampedro, D. Photocyclization of iminyl radicals: Theoretical study and photochemical aspects. J. Org. Chem. 2008, 73, 2234–2239. [Google Scholar] [CrossRef]
- Lalevée, J.; Allonas, X.; Fouassier, J.P.; Tachi, H.; Izumitani, A.; Shirai, M.; Tsunooka, M. Investigation of the photochemical properties of an important class of photobase generators: The O-acyloximes. J. Photochem. Photobiol. A Chem. 2002, 151, 27–37. [Google Scholar] [CrossRef]
- Alonso, R.; Caballero, A.; Campos, P.J.; Rodríguez, M.A. Photochemistry of acyloximes: Synthesis of heterocycles and natural products. Tetrahedron 2010, 66, 8828–8831. [Google Scholar] [CrossRef]
- Walton, J.C. Functionalised oximes: Emergent precursors for carbon-, nitrogen- and oxygen-centred radicals. Molecules 2016, 21, 63. [Google Scholar] [CrossRef]
- Pang, Y.; Fan, S.; Wang, Q.; Oprych, D.; Feilen, A.; Reiner, K.; Keil, D.; Slominsky, Y.L.; Popov, S.; Zou, Y.; et al. NIR-Sensitized Activated Photoreaction between Cyanines and Oxime Esters: Free-Radical Photopolymerization. Angew. Chem.—Int. Ed. 2020, 59, 11440–11447. [Google Scholar] [CrossRef]
- Karamtzioti, P.; Papastergiou, A.; Stefanakis, J.G.; Koumbis, A.E.; Anastasiou, I.; Koffa, M.; Fylaktakidou, K.C. O-Benzoyl pyridine aldoxime and amidoxime derivatives: Novel efficient DNA photo-cleavage agents. Medchemcomm 2015, 6, 719–726. [Google Scholar] [CrossRef]
- Hwu, J.R.; Tsay, S.C.; Hong, S.C.; Hsu, M.H.; Liu, C.F.; Chou, S.S.P. Relationship between structure of conjugated oxime esters and their ability to cleave DNA. Bioconjug. Chem. 2013, 24, 1778–1783. [Google Scholar] [CrossRef]
- Bindu, P.J.; Mahadevan, K.M.; Satyanarayan, N.D.; Ravikumar Naik, T.R. Synthesis and DNA cleavage studies of novel quinoline oxime esters. Bioorg. Med. Chem. Lett. 2012, 22, 898–900. [Google Scholar] [CrossRef]
- Chowdhury, N.; Dutta, S.; Dasgupta, S.; Singh, N.D.P.; Baidya, M.; Ghosh, S.K. Synthesis, photophysical, photochemical, DNA cleavage/binding and cytotoxic properties of pyrene oxime ester conjugates. Photochem. Photobiol. Sci. 2012, 11, 1239–1250. [Google Scholar] [CrossRef]
- Pasolli, M.; Dafnopoulos, K.; Andreou, N.P.; Gritzapis, P.S.; Koffa, M.; Koumbis, A.E.; Psomas, G.; Fylaktakidou, K.C. Pyridine and p-nitrophenyl oxime esters with possible photochemotherapeutic activity: Synthesis, DNA photocleavage and DNA binding studies. Molecules 2016, 21, 864. [Google Scholar] [CrossRef]
- Andreou, N.P.; Dafnopoulos, K.; Tortopidis, C.; Koumbis, A.E.; Koffa, M.; Psomas, G.; Fylaktakidou, K.C. Alkyl and aryl sulfonyl p-pyridine ethanone oximes are efficient DNA photo-cleavage agents. J. Photochem. Photobiol. B Biol. 2016, 158, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Papastergiou, A.; Perontsis, S.; Gritzapis, P.; Koumbis, A.E.; Koffa, M.; Psomas, G.; Fylaktakidou, K.C. Evaluation of O-alkyl and aryl sulfonyl aromatic and heteroaromatic amidoximes as novel potent DNA photo-cleavers. Photochem. Photobiol. Sci. 2016, 15, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Gritzapis, P.S.; Varras, P.C.; Andreou, N.P.; Katsani, K.R.; Dafnopoulos, K.; Psomas, G.; Peitsinis, Z.V.; Koumbis, A.E.; Fylaktakidou, K.C. P-Pyridinyl oxime carbamates: Synthesis, DNA binding, DNA photocleaving activity and theoretical photodegradation studies. Beilstein J. Org. Chem. 2020, 16, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Yue, G.; Mao, J.; Liu, D.; Ding, Y.; Liu, Z.; Qiu, D.; Zhao, X.; Lu, K.; Fagnoni, M.; et al. Visible-Light-Driven Synthesis of Arylstannanes from Arylazo Sulfones. Org. Lett. 2019, 21, 5187–5191. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Lian, C.; Mao, J.; Ding, Y.; Liu, Z.; Wei, L.; Fagnoni, M.; Protti, S. Visible Light-Driven, Photocatalyst-Free Arbuzov-Like Reaction via Arylazo Sulfones. Adv. Synth. Catal. 2019, 361, 5239–5244. [Google Scholar] [CrossRef]
- Liu, J.; Tian, M.; Li, Y.; Shan, X.; Li, A.; Lu, K.; Fagnoni, M.; Protti, S.; Zhao, X. Metal-Free Synthesis of Unsymmetrical Aryl Selenides and Tellurides via Visible Light-Driven Activation of Arylazo Sulfones. Eur. J. Org. Chem. 2020, 2020, 7358–7367. [Google Scholar] [CrossRef]
- Di Terlizzi, L.; Scaringi, S.; Raviola, C.; Pedrazzani, R.; Bandini, M.; Fagnoni, M.; Protti, S. Visible Light-Driven, Gold(I)-Catalyzed Preparation of Symmetrical (Hetero)biaryls by Homocoupling of Arylazo Sulfones. J. Org. Chem. 2022, 87, 4863–4872. [Google Scholar] [CrossRef]
- Blank, L.; Fagnoni, M.; Protti, S.; Rueping, M. Visible-Light Promoted Formation of C-B and C-S Bonds under metal and photocatalyst-free conditions. Synthesis 2019, 51, 1243–1252. [Google Scholar] [CrossRef]
- Bui, T.T.; Tran, V.H.; Kim, H.K. Visible-Light-Mediated Synthesis of Sulfonyl Fluorides from Arylazo Sulfones. Adv. Synth. Catal. 2022, 364, 341–347. [Google Scholar] [CrossRef]
- Chawla, R.; Jaiswal, S.; Dutta, P.K.; Yadav, L.D.S. A photocatalyst-free visible-light-mediated solvent-switchable route to stilbenes/vinyl sulfones from β-nitrostyrenes and arylazo sulfones. Org. Biomol. Chem. 2021, 19, 6487–6492. [Google Scholar] [CrossRef]
- Li, A.; Li, Y.; Liu, J.; Chen, J.; Lu, K.; Qiu, D.; Fagnoni, M.; Protti, S.; Zhao, X. Metal-Free Trifluoromethylthiolation of Arylazo Sulfones. J. Org. Chem. 2021, 86, 1292–1299. [Google Scholar] [CrossRef]
- Chawla, R.; Jaiswal, S.; Dutta, P.K.; Yadav, L.D.S. Photocatalyst-free visible light driven synthesis of (E)-vinyl sulfones from cinnamic acids and arylazo sulfones. Tetrahedron Lett. 2020, 61, 151898. [Google Scholar] [CrossRef]
- Nitti, A.; Martinelli, A.; Batteux, F.; Protti, S.; Fagnoni, M.; Pasini, D. Blue light driven free-radical polymerization using arylazo sulfones as initiators. Polym. Chem. 2021, 12, 5747–5751. [Google Scholar] [CrossRef]
- Lombardi, L.; Kovtun, A.; Mantovani, S.; Bertuzzi, G.; Favaretto, L.; Bettini, C.; Palermo, V.; Melucci, M.; Bandini, M. Visible-Light Assisted Covalent Surface Functionalization of Reduced Graphene Oxide Nanosheets with Arylazo Sulfones. Chem.—Eur. J. 2022, 28, e202200333. [Google Scholar] [CrossRef]
- Médard, J.; Decorse, P.; Mangeney, C.; Pinson, J.; Fagnoni, M.; Protti, S. Simultaneous Photografting of Two Organic Groups on a Gold Surface by using Arylazo Sulfones as Single Precursors. Langmuir 2020, 36, 2786–2793. [Google Scholar] [CrossRef]
- Panagopoulos, A.; Balalas, T.; Mitrakas, A.; Vrazas, V.; Katsani, K.R.; Koumbis, A.E.; Koukourakis, M.I.; Litinas, K.E.; Fylaktakidou, K.C. 6-Nitro-Quinazolin−4(3H)−one Exhibits Photodynamic Effects and Photodegrades Human Melanoma Cell Lines. A Study on the Photoreactivity of Simple Quinazolin−4(3H)−ones. Photochem. Photobiol. 2021, 97, 826–836. [Google Scholar] [CrossRef]
- Hansda, S.; Ghosh, G.; Ghosh, R. 9-phenyl acridine photosensitizes A375 cells to UVA radiation. Heliyon 2020, 6, e04733. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic Aromatic Hydrocarbons Physically Intercalate into Duplex Regions of Denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Dimitrakopoulou, A.; Dendrinou-Samara, C.; Pantazaki, A.A.; Alexiou, M.; Nordlander, E.; Kessissoglou, D.P. Synthesis, structure and interactions with DNA of novel tetranuclear, [Mn4(II/II/II/IV)] mixed valence complexes. J. Inorg. Biochem. 2008, 102, 618–628. [Google Scholar] [CrossRef]
- Luis García-Giménez, J.; González-Álvarez, M.; Liu-González, M.; Macías, B.; Borrás, J.; Alzuet, G. Toward the development of metal-based synthetic nucleases: DNA binding and oxidative DNA cleavage of a mixed copper(II) complex with N-(9H-purin-6-yl)benzenesulfonamide and 1,10-phenantroline. Antitumor activity in human Caco-2 cells and Jurkat T lymphocy. J. Inorg. Biochem. 2009, 103, 923–934. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.; Chen, C.; Deng, H.; Lu, T.; Ji, L. Interaction of macrocyclic copper(II) complexes with calf thymus DNA: Effects of the side chains of the ligands on the DNA-binding behaviors. Dalt. Trans. 2003, 1, 114–119. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Wilson, W.D.; Ratmeyer, L.; Zhao, M.; Strekowski, L.; Boykin, D. The Search for Structure-Specific Nucleic Acid-Interactive Drugs: Effects of Compound Structure on RNA versus DNA Interaction Strength. Biochemistry 1993, 32, 4098–4104. [Google Scholar] [CrossRef] [PubMed]
- Heller, D.P.; Greenstock, C.L. Fluorescence lifetime analysis of DNA intercalated ethidium bromide and quenching by free dye. Biophys. Chem. 1994, 50, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Pang, E.; Zhao, S.; Wang, B.; Niu, G.; Song, X.; Lan, M. Strategies to construct efficient singlet oxygen-generating photosensitizers. Coord. Chem. Rev. 2022, 472, 214780. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Malacarne, M.; Protti, S.; Fagnoni, M. A Visible-Light-Driven, Metal-free Route to Aromatic Amides via Radical Arylation of Isonitriles. Adv. Synth. Catal. 2017, 359, 3826–3830. [Google Scholar] [CrossRef]
- Di Terlizzi, L.; Cola, I.; Raviola, C.; Fagnoni, M.; Protti, S. A Dyedauxiliary Group Strategy for the a-Functionalization of Ketones and Esters. ACS Org. Inorg. Au 2021, 1, 68–71. [Google Scholar] [CrossRef]
- Nan, G.; Ren, F.; Luo, M. Suzuki–Miyaura cross-coupling reaction of 1-aryltriazenes with arylboronic acids catalyzed by a recyclable polymer-supported N-heterocyclic carbene–palladium complex catalyst. Beilstein J. Org. Chem. 2010, 28, 70. [Google Scholar] [CrossRef]
- Disli, A.; Yildirir, Y. New Synthesis Of Phenylthioglycolic Acids Via Related Tri-azene Compounds. Org. Prep. Proc. Int. 1998, 30, 349–352. [Google Scholar] [CrossRef]
- Novi, M.; Petrillo, G.; Dell’Erba, C. An SRN1 approach to some aromatic nitriles diazosulfides. Tetrahedron Lett. 1987, 28, 1345–1348. [Google Scholar] [CrossRef]
- Zhou, X.; Luo, J.; Liu, J.; Peng, S.; Deng, G.-J. Pd-Catalyzed desulfitative Heck coupling with dioxygen as the terminal oxidant. Org. Lett. 2011, 13, 1432–1435. [Google Scholar]
- Frisch, M.J.; Trucks, G.; Schlegel, H.; Scuseria, G.E.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09; Revision B.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Drew, H.R.; Dickerson, R.E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 1981, 151, 535–556. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System; Version 1.2r3pre; Schrödinger LLC: New York, NY, USA, 2022. [Google Scholar]

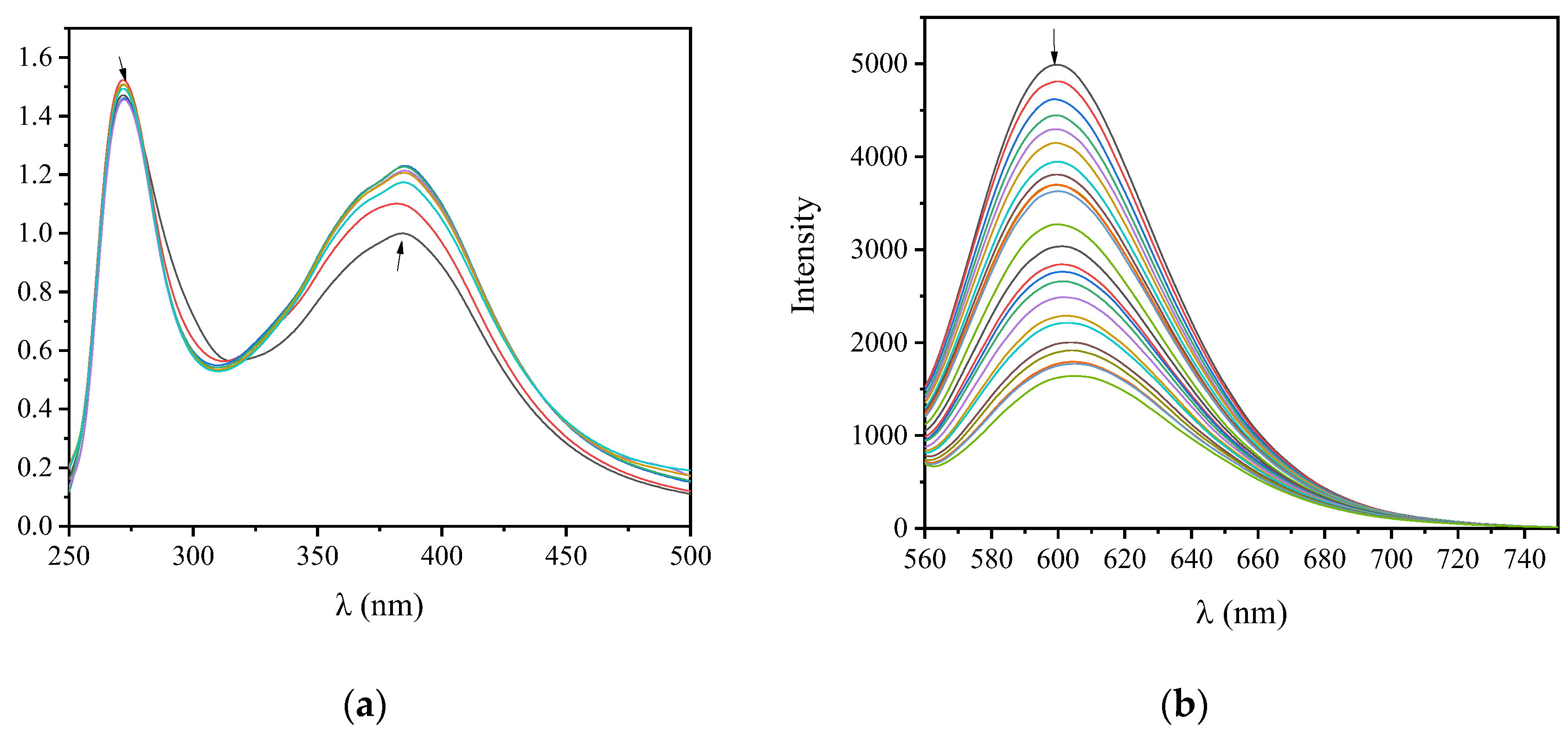



| Groups | No of Compound | Band (nm) (ΔA/A0 (%) 1, Δλ (nm) 2) | Kb (M−1) |
|---|---|---|---|
| Group A | 1 | 272 (+2,+1); 385 (+17, 0) | 3.73(±0.24) × 105 |
| 2 | 300 (+2, +1) | 1.12(±0.15) × 105 | |
| 3 | 347 (−4, +1) | 5.04(±0.16) × 105 | |
| 4 | 387 (−68, +11); 489 (+54, +6) | 6.08(±0.15) × 105 | |
| Group B | 5 | 288 (+6, −2); 359 (sh) 3 (−43, +20) | 9.31(±0.18) × 103 |
| 6 | 269 (+8, +2) | 2.87(±0.10) × 107 | |
| 7 | 289 (+2, +3) | 4.59(±0.10) × 105 | |
| 16 | 274 (+6, +3) | 1.05(±0.08) × 106 | |
| Group C | 8 | 301 (+1, +1) | 2.67(±0.27) × 105 |
| 9 | 298 (+2, +0) | 3.81(±0.35) × 105 | |
| 10 | 290 (+8, −1) | 1.31(±0.08) × 106 | |
| 11 | 308 (+4, +1) | 6.41(±0.32) × 105 | |
| 12 | 312 (+0.5, +2) | 9.13(±0.15) × 105 | |
| 13 | 330 (+8, −1) | 6.02(±0.44) × 105 | |
| Group D | 14 | 279 (+12, 0) | 7.42(±0.10) × 105 |
| 15 | 280 (+23, +8) | 1.02(±0.04) × 105 | |
| 17 | 322 (−3, +0) | 3.82(±0.30) × 105 |
| Groups | Compound | (∆I/Io, %) | KSV (M−1) | kq, M−1 s−1 |
|---|---|---|---|---|
| Group A | 1 | 65.2 | 2.40(±0.03) × 104 | 1.04(±0.01) × 1012 |
| 2 | 57.1 | 3.51(±0.06) × 104 | 1.53(±0.03) × 1012 | |
| 3 | 41.8 | 4.41(±0.08) × 104 | 1.92(±0.03) × 1012 | |
| 4 | 42.4 | 8.51(±0.26) × 104 | 3.70(±0.11) × 1012 | |
| Group B | 5 | 51.6 | 3.58(±0.06) × 104 | 1.56(±0.02) × 1012 |
| 6 | 54.2 | 3.41(±0.07) × 104 | 1.48(±0.03) × 1012 | |
| 7 | 54.8 | 1.31(±0.03) × 104 | 5.70(±0.14) × 1011 | |
| 16 | 49.5 | 3.31(±0.05) × 104 | 1.44(±0.02) × 1012 | |
| Group C | 8 | 58.0 | 8.40(±0.13) × 104 | 3.65(±0.05) × 1012 |
| 9 | 67.5 | 5.88(±0.13) × 104 | 2.56(±0.06) × 1012 | |
| 10 | 59 | 4.65(±0.12) × 104 | 2.02(±0.05) × 1012 | |
| 11 | 58.7 | 4.20(±0.08) × 104 | 1.83(±0.04) × 1012 | |
| 12 | 56.5 | 3.90(±0.11) × 104 | 1.70(±0.05) × 1012 | |
| 13 | 53.3 | 3.69(±0.07) × 104 | 1.60(±0.03) × 1012 | |
| Group D | 14 | 61.2 | 4.37(±0.11) × 104 | 1.90(±0.05) × 1012 |
| 15 | 52.9 | 3.61(±0.08) × 104 | 1.57(±0.03) × 1012 | |
| 17 | 50.7 | 3.77(±0.05) × 104 | 1.64(±0.02) × 1012 |
| Groups | Compound | Energy (Kcal/mol) | Interactions (PyMol) Polar Contacts |
|---|---|---|---|
| Group A | 1 | −7.8 | DG16, DG10 |
| 2 | −6.2 | DA17 | |
| 3 | −6.8 | DG16, DA17 | |
| 4 | −7.1 | DG16, DA17 | |
| Group B | 5 | −7.2 | DA17, DG10 |
| 6 | −7.3 | DG10, DG12, DG14 | |
| 7 | −7.3 | DG10, DG14, DG16 | |
| 16 | −8.9 | DA16 | |
| Group C | 8 | −6.6 | DA17, DG10 |
| 9 | −6.5 | DG16, DA17 | |
| 10 | −6.7 | DG10, DA17 | |
| 11 | −6.0 | DG10, DA17 | |
| 12 | −6.5 | DG16, DA17 | |
| 13 | −6.6 | DG10, DA17 | |
| Group D | 14 | −7.3 | DA5 |
| 15 | −7.3 | No Polar Contacts | |
| 17 | −7.1 | No Polar Contacts |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikra, C.; Mitrakas, A.; Ghizzani, V.; Katsani, K.R.; Koffa, M.; Koukourakis, M.; Psomas, G.; Protti, S.; Fagnoni, M.; Fylaktakidou, K.C. Effect of Arylazo Sulfones on DNA: Binding, Cleavage, Photocleavage, Molecular Docking Studies and Interaction with A375 Melanoma and Non-Cancer Cells. Int. J. Mol. Sci. 2023, 24, 1834. https://doi.org/10.3390/ijms24031834
Mikra C, Mitrakas A, Ghizzani V, Katsani KR, Koffa M, Koukourakis M, Psomas G, Protti S, Fagnoni M, Fylaktakidou KC. Effect of Arylazo Sulfones on DNA: Binding, Cleavage, Photocleavage, Molecular Docking Studies and Interaction with A375 Melanoma and Non-Cancer Cells. International Journal of Molecular Sciences. 2023; 24(3):1834. https://doi.org/10.3390/ijms24031834
Chicago/Turabian StyleMikra, Chrysoula, Achilleas Mitrakas, Virginia Ghizzani, Katerina R. Katsani, Maria Koffa, Michael Koukourakis, George Psomas, Stefano Protti, Maurizio Fagnoni, and Konstantina C. Fylaktakidou. 2023. "Effect of Arylazo Sulfones on DNA: Binding, Cleavage, Photocleavage, Molecular Docking Studies and Interaction with A375 Melanoma and Non-Cancer Cells" International Journal of Molecular Sciences 24, no. 3: 1834. https://doi.org/10.3390/ijms24031834
APA StyleMikra, C., Mitrakas, A., Ghizzani, V., Katsani, K. R., Koffa, M., Koukourakis, M., Psomas, G., Protti, S., Fagnoni, M., & Fylaktakidou, K. C. (2023). Effect of Arylazo Sulfones on DNA: Binding, Cleavage, Photocleavage, Molecular Docking Studies and Interaction with A375 Melanoma and Non-Cancer Cells. International Journal of Molecular Sciences, 24(3), 1834. https://doi.org/10.3390/ijms24031834