Intron Editing Reveals SNORD-Dependent Maturation of the Small Nucleolar RNA Host Gene GAS5 in Human Cells
Abstract
:1. Introduction
2. Results
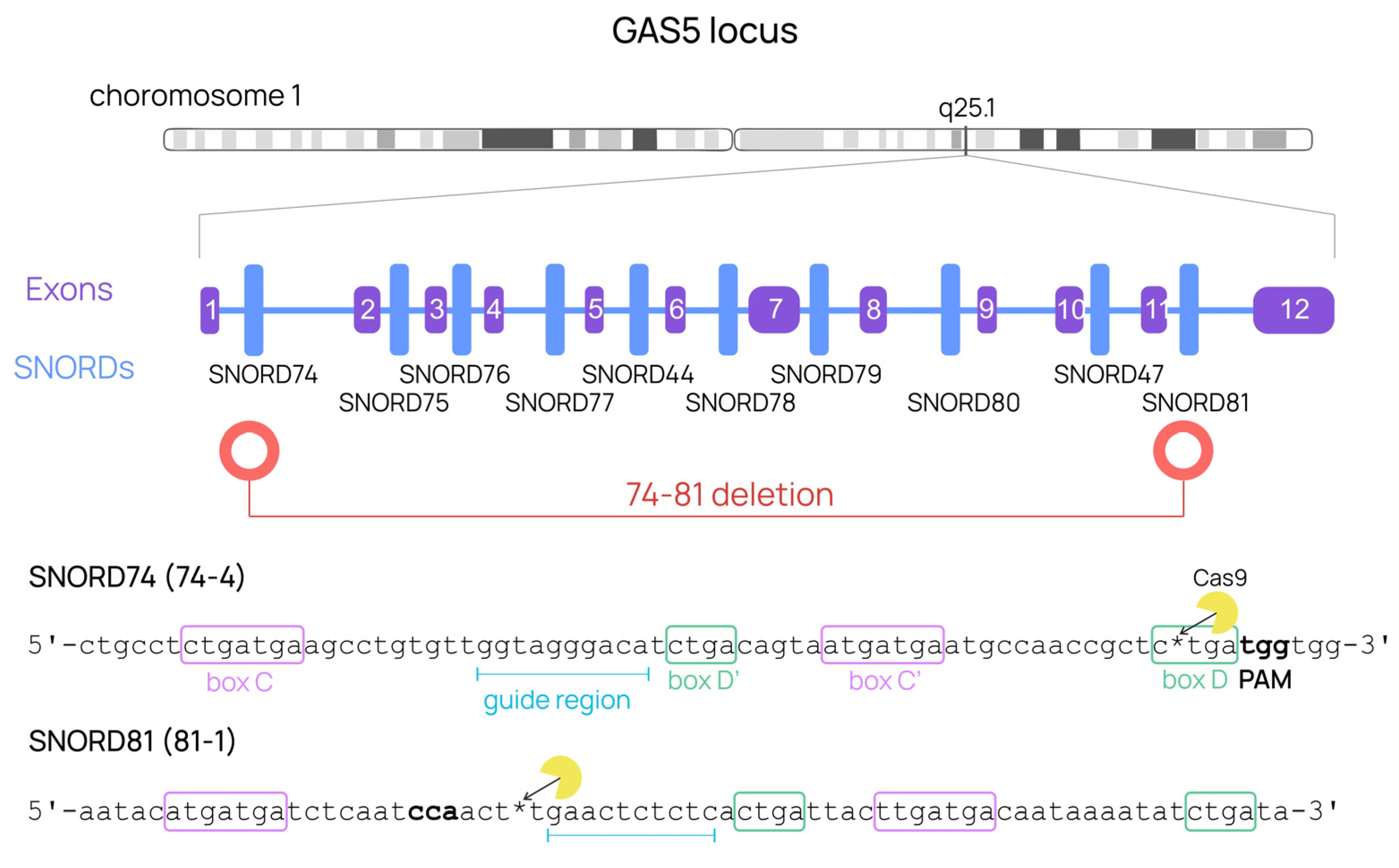
2.1. Selection and Design of Single Guide RNAs Targeted at snoRNAs
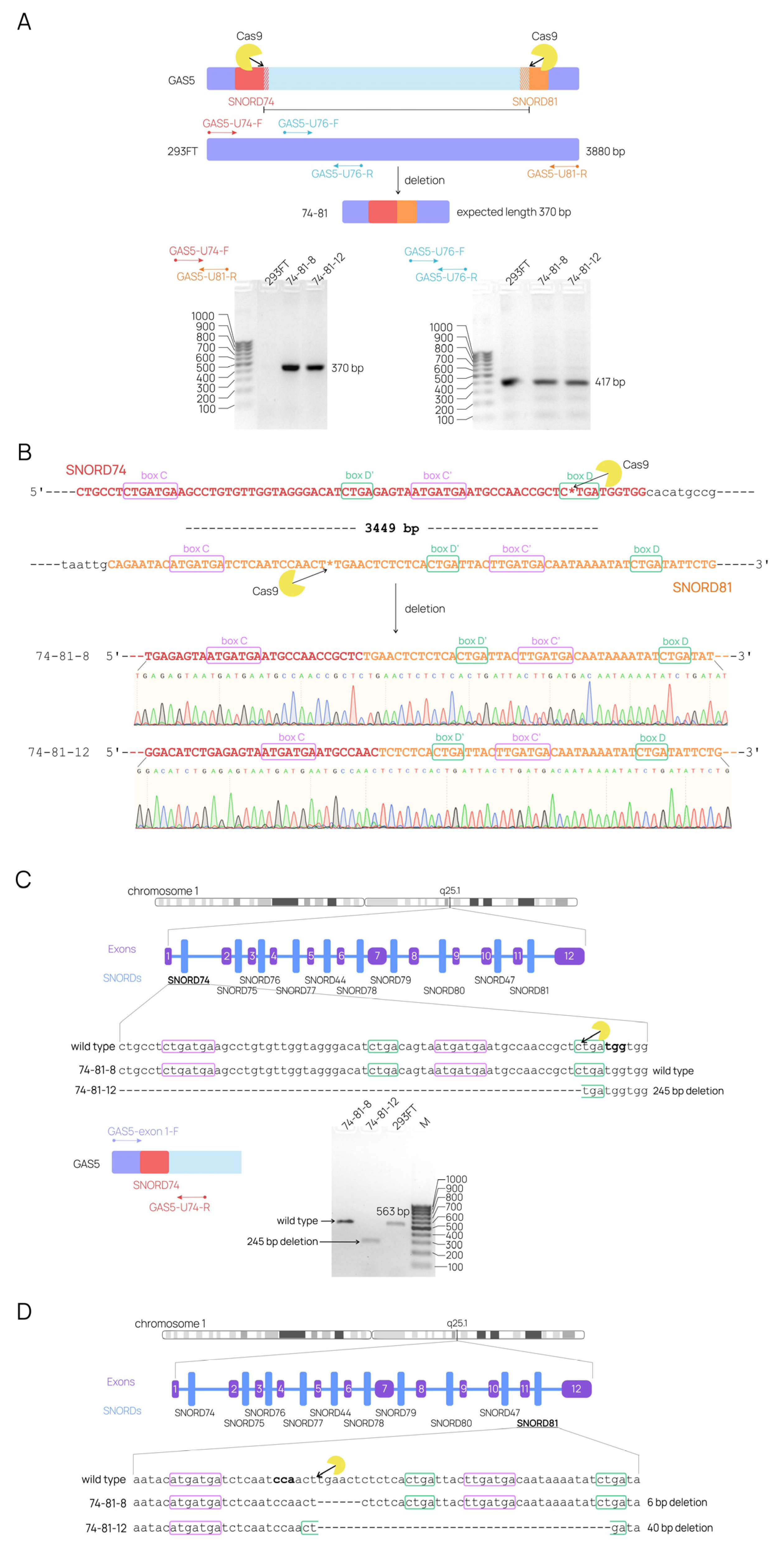
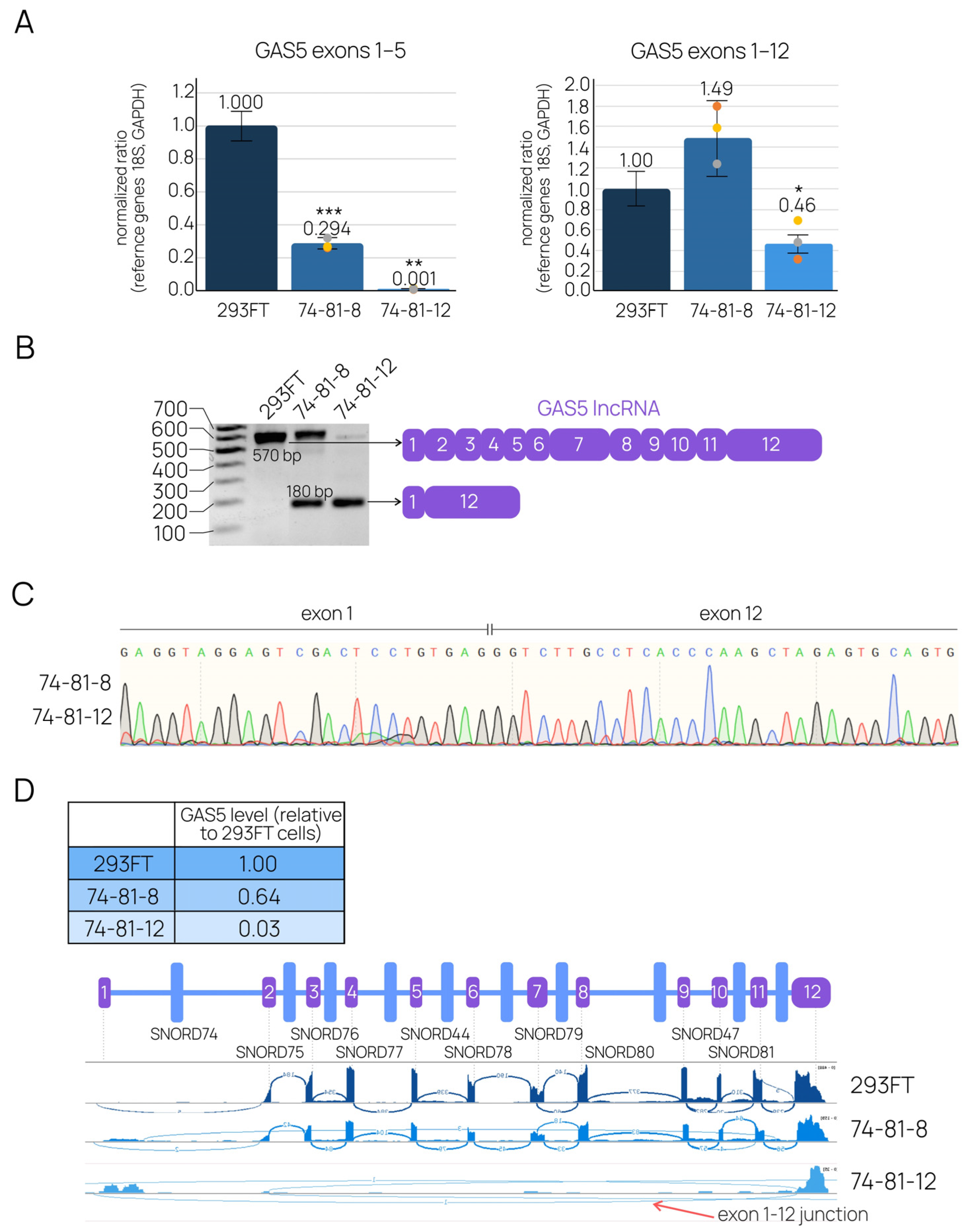
2.2. CRISPR-Mediated Large Deletions in the GAS5 Gene Are Present in Only One Allele and Result in the Downregulation of the Target lncRNA
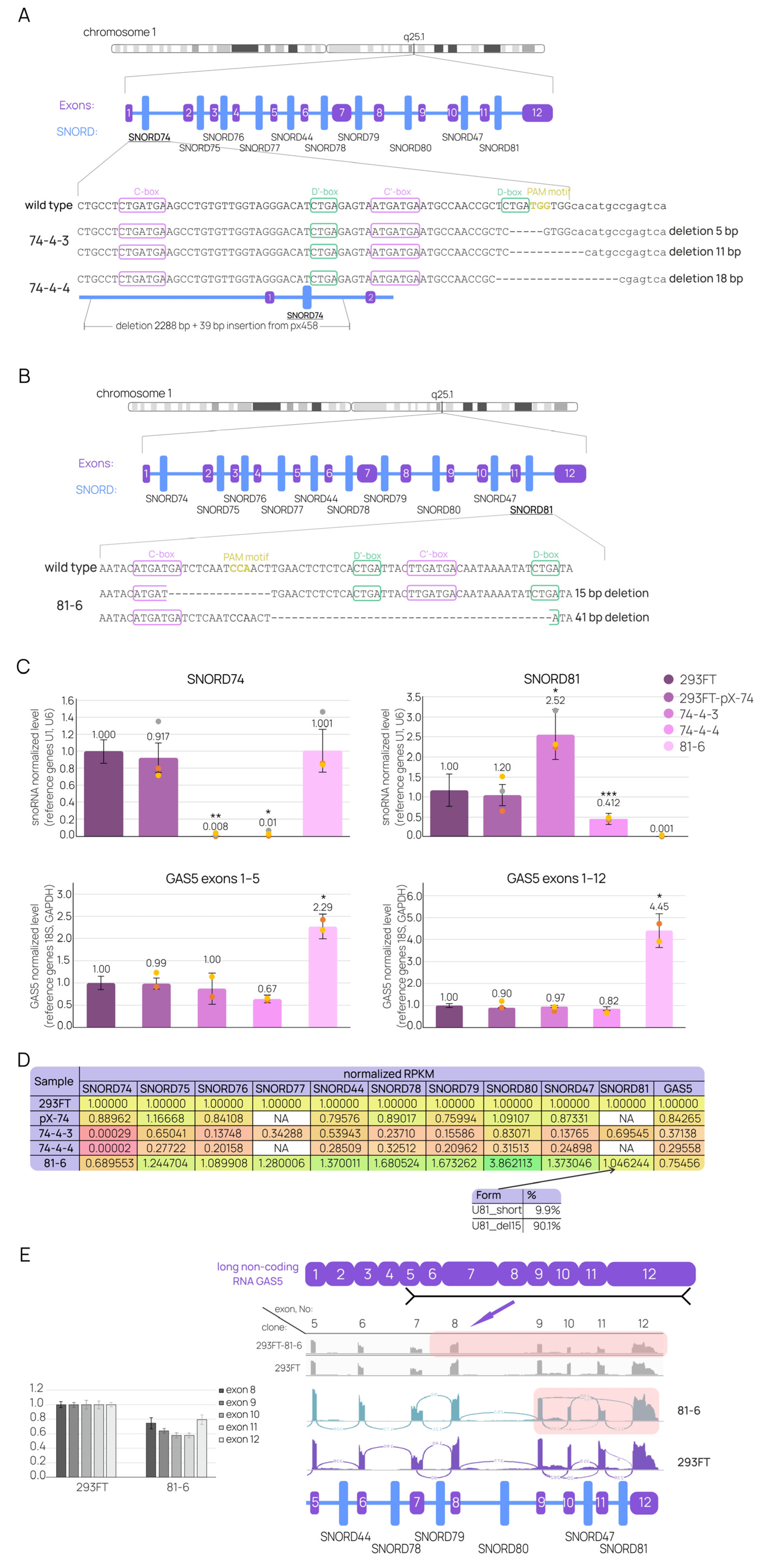
2.3. Individual SNORD Mutations Affect GAS5 and snoRNAs Maturation
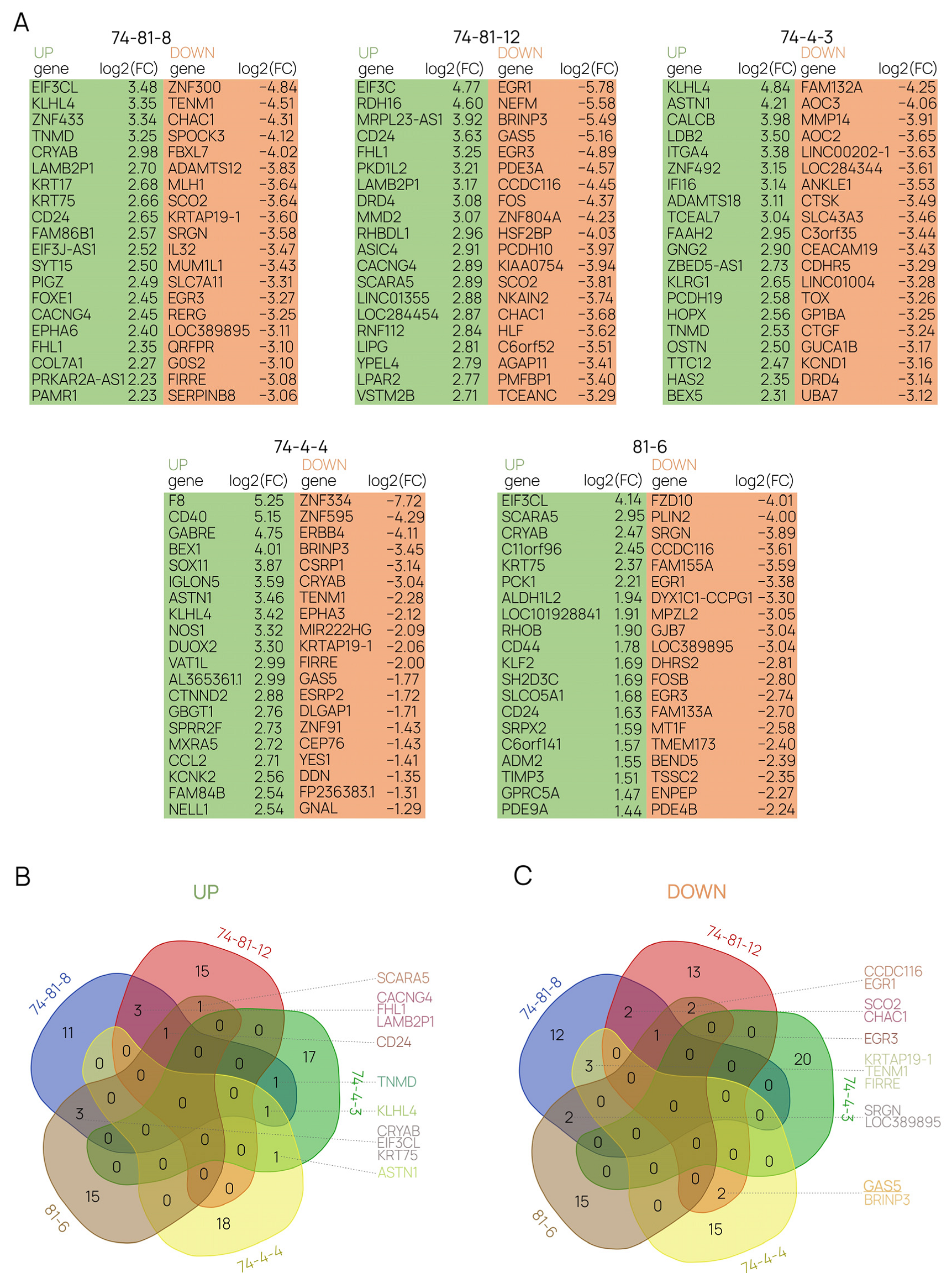
2.4. Differential Gene-Expression Analysis Sheds Light on the Molecular Mechanisms behind the Functioning of GAS5 lncRNA in Human Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Generation of CRISPR/Cas9 Constructs
4.3. Cell Culture and Transfection
4.4. Individual Clone Selection and the Identification of Mutations
4.5. Isolation of Total Cell RNA
4.6. Library Preparation and Sequencing
4.7. RNA-Seq and Differential Expression Analysis
4.8. Real-Time RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneider, C.; King, R.M.; Philipson, L. Genes Specifically Expressed at Growth Arrest of Mammalian Cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.; Williams, G. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef] [PubMed]
- Goustin, A.; Thepsuwan, P.; Kosir, M.; Lipovich, L. The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating LncRNA Widely Expressed in Cancers. Noncoding RNA 2019, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Steitz, J.A. Classification of Gas5 as a Multi-Small-Nucleolar-RNA (SnoRNA) Host Gene and a Member of the 5′-Terminal Oligopyrimidine Gene Family Reveals Common Features of SnoRNA Host Genes. Mol. Cell Biol. 1998, 18, 6897–6909. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a Non-Protein-Coding RNA, Controls Apoptosis and Is Downregulated in Breast Cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Decreased Expression of Long Non-Coding RNA GAS5 Promotes Cell Proliferation, Migration and Invasion, and Indicates a Poor Prognosis in Ovarian Cancer. Oncol. Rep. 2016, 36, 3241–3250. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cai, Y.; Wang, C.; Huang, M.; Chen, J. LncRNA GAS5 Regulates the Proliferation, Migration, Invasion and Apoptosis of Brain Glioma Cells through Targeting GSTM3 Expression. The Effect of LncRNA GAS5 on Glioma Cells. J. Neuro-Oncol. 2019, 143, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal 2010, 3, ra8. [Google Scholar] [CrossRef]
- Hudson, W.H.; Pickard, M.R.; de Vera, I.M.S.; Kuiper, E.G.; Mourtada-Maarabouni, M.; Conn, G.L.; Kojetin, D.J.; Williams, G.T.; Ortlund, E.A. Conserved Sequence-Specific LincRNA–Steroid Receptor Interactions Drive Transcriptional Repression and Direct Cell Fate. Nat. Commun. 2014, 5, 5395. [Google Scholar] [CrossRef]
- Zong, Y.; Zhang, Y.; Sun, X.; Xu, T.; Cheng, X.; Qin, Y. MiR-221/222 Promote Tumor Growth and Suppress Apoptosis by Targeting LncRNA GAS5 in Breast Cancer. Biosci. Rep. 2019, 39, BSR20181859. [Google Scholar] [CrossRef]
- Wang, K.; Li, J.; Xiong, G.; He, G.; Guan, X.; Yang, K.; Bai, Y. Negative Regulation of LncRNA GAS5 by MiR-196a Inhibits Esophageal Squamous Cell Carcinoma Growth. Biochem. Biophys. Res. Commun. 2018, 495, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative Regulation of LncRNA GAS5 by MiR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Shi, Y.; Parag, S.; Patel, R.; Lui, A.; Murr, M.; Cai, J.; Patel, N.A. Stabilization of LncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes. Cell Chem. Biol. 2019, 26, 319–330.e6. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lv, Y.; Shao, C.; Chen, C.; Zhang, T.; Wei, Y.; Fan, H.; Lv, T.; Liu, H.; Song, Y. Tumor-Derived Exosomal LncRNA GAS5 as a Biomarker for Early-Stage Non-Small-Cell Lung Cancer Diagnosis. J. Cell Physiol. 2019, 234, 20721–20727. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal LncRNAs as New Players in Cell-to-Cell Communication. Transl. Cancer Res. 2018, 7, S243–S252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Wang, B.; Wang, X.H.; Xu, C.W. Effect of LncRNA GAS5 on the Apoptosis of Neurons via the Notch1 Signaling Pathway in Rats with Cerebral Infarction. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10083–10091. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, X.; Bao, Y.; Yu, H.; Jia, D.; Ma, C. Down-Regulation of GAS5 Ameliorates Myocardial Ischaemia/Reperfusion Injury via the MiR-335/ROCK1/AKT/GSK-3β Axis. J. Cell Mol. Med. 2019, 23, 8420–8431. [Google Scholar] [CrossRef]
- Miao, X.; Liang, A. Knockdown of Long Noncoding RNA GAS5 Attenuates H2O2-Induced Damage in Retinal Ganglion Cells through Upregulating MiR-124: Potential Role in Traumatic Brain Injury. J. Cell Biochem. 2019, 120, 2313–2322. [Google Scholar] [CrossRef]
- Wang, Y.-N.-Z.; Shan, K.; Yao, M.-D.; Yao, J.; Wang, J.-J.; Li, X.; Liu, B.; Zhang, Y.-Y.; Ji, Y.; Jiang, Q.; et al. Long Noncoding RNA-GAS5. Hypertension 2016, 68, 736–748. [Google Scholar] [CrossRef]
- Fang, Y.; Hu, J.F.; Wang, Z.H.; Zhang, S.G.; Zhang, R.F.; Sun, L.M.; Cui, H.W.; Yang, F. GAS5 Promotes Podocyte Injury in Sepsis by Inhibiting PTEN Expression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8423–8430. [Google Scholar] [CrossRef]
- Han, X.; Xu, J.; Chen, Z.; Li, P.; Zhao, L.; Tao, J.; Shen, Y.; Zhu, S.; Yu, B.; Zhu, J.; et al. Gas5 Inhibition Promotes the Axon Regeneration in the Adult Mammalian Nervous System. Exp. Neurol. 2022, 356, 114157. [Google Scholar] [CrossRef] [PubMed]
- Jorjani, H.; Kehr, S.; Jedlinski, D.J.; Gumienny, R.; Hertel, J.; Stadler, P.F.; Zavolan, M.; Gruber, A.R. An Updated Human SnoRNAome. Nucleic Acids Res. 2016, 44, 5068–5082. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Dickmanns, A.; Luhrmann, R. Conserved Stem II of the Box C/D Motif Is Essential for Nucleolar Localization and Is Required, Along with the 15.5K Protein, for the Hierarchical Assembly of the Box C/D SnoRNP. Mol. Cell Biol. 2002, 22, 8342–8352. [Google Scholar] [CrossRef] [PubMed]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and Trafficking of Box C/D and H/ACA SnoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional Diversity of Small Nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef] [PubMed]
- Baldini, L.; Charpentier, B.; Labialle, S. Emerging Data on the Diversity of Molecular Mechanisms Involving C/D SnoRNAs. Noncoding RNA 2021, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yang, J.; van Nues, R.; Watzinger, P.; Kötter, P.; Lafontaine, D.L.J.; Granneman, S.; Entian, K.-D. Specialized Box C/D SnoRNPs Act as Antisense Guides to Target RNA Base Acetylation. PLoS Genet. 2017, 13, e1006804. [Google Scholar] [CrossRef]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual Function of C/D Box Small Nucleolar RNAs in RRNA Modification and Alternative Pre-MRNA Splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M.; Yamada, K.; Endo, A.; Barton, G.J.; Lamond, A.I. Human Box C/D SnoRNA Processing Conservation across Multiple Cell Types. Nucleic Acids Res. 2012, 40, 3676–3688. [Google Scholar] [CrossRef]
- Stepanov, G.A.; Semenov, D.V.; Kuligina, E.V.; Koval, O.A.; Rabinov, I.V.; Kit, Y.Y.; Richter, V.A. Analogues of Artificial Human Box C/D Small Nucleolar RNA As Regulators of Alternative Splicing of a Pre-MRNA Target. Acta Nat. 2012, 4, 32–41. [Google Scholar] [CrossRef]
- Kishore, S. The SnoRNA HBII-52 Regulates Alternative Splicing of the Serotonin Receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Ardal, B.K.; Hollensen, A.K.; Damgaard, C.K.; Jensen, T.H. Box C/D SnoRNP Autoregulation by a Cis-Acting SnoRNA in the NOP56 Pre-MRNA. Mol. Cell 2018, 72, 99–111.e5. [Google Scholar] [CrossRef] [PubMed]
- Brandis, K.A.; Gale, S.; Jinn, S.; Langmade, S.J.; Dudley-Rucker, N.; Jiang, H.; Sidhu, R.; Ren, A.; Goldberg, A.; Schaffer, J.E.; et al. Box C/D Small Nucleolar RNA (SnoRNA) U60 Regulates Intracellular Cholesterol Trafficking. J. Biol. Chem. 2013, 288, 35703–35713. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, D.; Fafard-Couture, É; Scott, M.S. Small Nucleolar RNAs: Continuing Identification of Novel Members and Increasing Diversity of Their Molecular Mechanisms of Action. Biochem. Soc. Trans. 2020, 48, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Youssef, O.A.; Safran, S.A.; Nakamura, T.; Nix, D.A.; Hotamisligil, G.S.; Bass, B.L. Potential Role for SnoRNAs in PKR Activation during Metabolic Stress. Proc. Natl. Acad. Sci. USA 2015, 112, 5023–5028. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Harris, A.N.; Holley, C.L.; Mahadevan, J.; Pyles, K.D.; Lavagnino, Z.; Scherrer, D.E.; Fujiwara, H.; Sidhu, R.; Zhang, J.; et al. Rpl13a Small Nucleolar RNAs Regulate Systemic Glucose Metabolism. J. Clin. Investig. 2016, 126, 4616–4625. [Google Scholar] [CrossRef]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small Nucleolar RNAs U32a, U33, and U35a Are Critical Mediators of Metabolic Stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic Accumulation of Small Nucleolar RNAs (SnoRNAs) Is Dynamically Regulated by NADPH Oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef]
- Rogelj, B.; Hartmann, C.E.A.; Yeo, C.H.; Hunt, S.P.; Giese, K.P. Contextual Fear Conditioning Regulates the Expression of Brain-Specific Small Nucleolar RNAs in Hippocampus. Eur. J. Neurosci. 2003, 18, 3089–3096. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Wang, M.; Li, X.; Gong, H.; Tang, H.; Chen, L.; Wan, L.; Liu, Q. Activity Dependent LoNA Regulates Translation by Coordinating RRNA Transcription and Methylation. Nat. Commun. 2018, 9, 1726. [Google Scholar] [CrossRef]
- Vitali, P.; Kiss, T. Cooperative 2′-O-Methylation of the Wobble Cytidine of Human Elongator TRNA Met (CAT) by a Nucleolar and a Cajal Body-Specific Box C/D RNP. Genes. Dev. 2019, 33, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, J.; Guo, Y.; Huang, W.; Huang, S.; Ming, S.; Wu, X.; Zhang, R.; Ding, J.; Zhao, W.; et al. A SnoRNA Modulates MRNA 3′ End Processing and Regulates the Expression of a Subset of MRNAs. Nucleic Acids Res. 2017, 45, 8647–8660. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Huang, C.; Huang, S.; Yao, C. SnoRNAs Associate with MRNA 3′ Processing Complex: New Wine in Old Bottles. RNA Biol. 2018, 15, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Surface, J.; Shen, M.; de la Grange, P.; Stamm, S. SNORD116 and SNORD115 Change Expression of Multiple Genes and Modify Each Other’s Activity. Gene 2015, 572, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Bortolin-Cavaillé, M.L.; Cavaillé, J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) Gene Clusters at the Imprinted Prader-Willi Locus Generate Canonical Box C/D SnoRNAs. Nucleic Acids Res. 2012, 40, 6800–6807. [Google Scholar] [CrossRef] [PubMed]
- Cavaillé, J. Box C/D Small Nucleolar RNA Genes and the Prader-Willi Syndrome: A Complex Interplay. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs Derived from SnoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human Box C/D SnoRNAs with MiRNA like Functions: Expanding the Range of Regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-Sequencing of Human Argonaute-Associated Small RNAs Provides Insight into MiRNA Sorting and Reveals Argonaute Association with RNA Fragments of Diverse Origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef]
- Ono, M.; Yamada, K.; Bensaddek, D.; Afzal, V.; Biddlestone, J.; Ortmann, B.; Mudie, S.; Boivin, V.; Scott, M.S.; Rocha, S.; et al. Enhanced SnoMEN Vectors Facilitate Establishment of GFP-HIF-1α Protein Replacement Human Cell Lines. PLoS ONE 2016, 11, e0154759. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Yamada, K.; Avolio, F.; Scott, M.S.; Van Koningsbruggen, S.; Barton, G.J.; Lamond, A.I. Analysis of Human Small Nucleolar RNAs (SnoRNA) and the Development of SnoRNA Modulator of Gene Expression Vectors. Mol. Biol. Cell 2010, 21, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long Noncoding RNAs with SnoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chen, K.; Song, B.; Ma, J.; Wu, X.; Xu, Q.; Wei, Z.; Su, J.; Liu, G.; Rong, R.; et al. M6A-Atlas: A Comprehensive Knowledgebase for Unraveling the N6-Methyladenosine (M6A) Epitranscriptome. Nucleic Acids Res. 2021, 49, D134–D143. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.; Wei, Z.; Zhang, S.; Hua, G.; Zhang, S.-W.; Zhang, L.; Gao, S.-J.; Meng, J.; Chen, X.; et al. MeT-DB V2.0: Elucidating Context-Specific Functions of N6-Methyl-Adenosine Methyltranscriptome. Nucleic Acids Res. 2018, 46, D281–D287. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Leu, J.-S.; Ma, L.; Xie, K.; Huang, S. Methylation of RNA N6-Methyladenosine in Modulation of Cytokine Responses and Tumorigenesis. Cytokine 2018, 118, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6 -Methyladenosine-Dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Zhu, Z.; Huo, F.; Zhang, J.; Shan, H.; Pei, D. Crosstalk between M6A Modification and Alternative Splicing during Cancer Progression. Clin. Transl. Med. 2023, 13, e1460. [Google Scholar] [CrossRef]
- Louloupi, A.; Ntini, E.; Conrad, T.; Ørom, U.A.V. Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of M6A in Splicing Efficiency. Cell Rep. 2018, 23, 3429–3437. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 SnRNA m 6 A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [PubMed]
- Lafaille, F.G.; Harschnitz, O.; Lee, Y.S.; Zhang, P.; Hasek, M.L.; Kerner, G.; Itan, Y.; Ewaleifoh, O.; Rapaport, F.; Carlile, T.M.; et al. Human SNORA31 Variations Impair Cortical Neuron-Intrinsic Immunity to HSV-1 and Underlie Herpes Simplex Encephalitis. Nat. Med. 2019, 25, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.; Liu, Y.; Rohde, C.; Cui, C.; Fijalkowska, D.; Gerloff, D.; Walter, C.; Krijgsveld, J.; Dugas, M.; Edemir, B.; et al. Site-Specific Methylation of 18S Ribosomal RNA by SNORD42A Is Required for Acute Myeloid Leukemia Cell Proliferation. Blood 2020, 135, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Siprashvili, Z.; Webster, D.E.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The Noncoding RNAs SNORD50A and SNORD50B Bind K-Ras and Are Recurrently Deleted in Human Cancer. Nat. Genet. 2016, 48, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Toden, S.; Weng, W.; Shigeyasu, K.; Miyoshi, J.; Turner, J.; Nagasaka, T.; Ma, Y.; Takayama, T.; Fujiwara, T.; et al. SNORA21–An Oncogenic Small Nucleolar RNA, with a Prognostic Biomarker Potential in Human Colorectal Cancer. EBioMedicine 2017, 22, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Filippova, J.A.; Matveeva, A.M.; Zhuravlev, E.S.; Balakhonova, E.A.; Prokhorova, D.V.; Malanin, S.J.; Shah Mahmud, R.; Grigoryeva, T.V.; Anufrieva, K.S.; Semenov, D.V.; et al. Are Small Nucleolar RNAs “CRISPRable”? A Report on Box C/D Small Nucleolar RNA Editing in Human Cells. Front. Pharmacol. 2019, 10, 1246. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, Y.; Han, J.; Su, H. GAS5 Knockdown Ameliorates Apoptosis and Inflammatory Response by Modulating MiR-26b-5p/Smad1 Axis in Cerebral Ischaemia/Reperfusion Injury. Behav. Brain Res. 2020, 379, 112370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, X.; Yang, M.; Shangguan, J.; Yin, Y. GAS5 Knockdown Suppresses Inflammation and Oxidative Stress Induced by Oxidized Low-Density Lipoprotein in Macrophages by Sponging MiR-135a. Mol. Cell Biochem. 2021, 476, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chen, J.; Ou, B.; Liu, C.; Zou, Y.; Chen, Q. GAS5 Knockdown Reduces the Chemo-Sensitivity of Non-Small Cell Lung Cancer (NSCLC) Cell to Cisplatin (DDP) through Regulating MiR-21/PTEN Axis. Biomed. Pharmacother. 2017, 93, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Salehen, N.; Norazit, A.; Rahman, A.A.; Murad, N.A.A.; Rahman, N.M.A.N.A.; Ibrahim, K. Tumor Suppressive Effects of GAS5 in Cancer Cells. Noncoding RNA 2022, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, W.; Jiang, J.; Bao, E.; Xu, D.; Zeng, Y.; Tao, L.; Qiu, J. Downregulation of GAS5 Promotes Bladder Cancer Cell Proliferation, Partly by Regulating CDK6. PLoS ONE 2013, 8, e73991. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Kong, R.; Chen, F.; Song, Y. A Critical Role for the Long Non-coding RNA GAS5 in Proliferation and Apoptosis in Non-small-cell Lung Cancer. Mol. Carcinog. 2015, 54, E1–E12. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Wu, T.; Gao, X.; He, Z.; Nong, W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front. Oncol. 2022, 12, 846497. [Google Scholar] [CrossRef]
- Sun, D.; Yu, Z.; Fang, X.; Liu, M.; Pu, Y.; Shao, Q.; Wang, D.; Zhao, X.; Huang, A.; Xiang, Z.; et al. LncRNA GAS5 Inhibits Microglial M2 Polarization and Exacerbates Demyelination. EMBO Rep. 2017, 18, 1801–1816. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Y.; Yu, M. LncRNA GAS5 Facilitates Nasopharyngeal Carcinoma Progression through Epigenetically Silencing PTEN via EZH2. RSC Adv. 2019, 9, 31691–31698. [Google Scholar] [CrossRef]
- Wang, M.; Guo, C.; Wang, L.; Luo, G.; Huang, C.; Li, Y.; Liu, D.; Zeng, F.; Jiang, G.; Xiao, X. Long Noncoding RNA GAS5 Promotes Bladder Cancer Cells Apoptosis through Inhibiting EZH2 Transcription. Cell Death Dis. 2018, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zhao, J.; Zhang, Z.-P.; Wu, M.; Li, J.; Xiao, G.-L.; Liu, B.; Liao, Y.-X.; Liu, J.-P. Long Non-Coding RNA GAS5, by up-Regulating PRC2 and Targeting the Promoter Methylation of MiR-424, Suppresses Multiple Malignant Phenotypes of Glioma. J. Neuro-Oncol. 2020, 148, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Grzechnik, P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Coker, H.; Wei, G.; Brockdorff, N. M6A Modification of Non-Coding RNA and the Control of Mammalian Gene Expression. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 310–318. [Google Scholar] [CrossRef]
- Parker, M.T.; Soanes, B.K.; Kusakina, J.; Larrieu, A.; Knop, K.; Joy, N.; Breidenbach, F.; Sherwood, A.V.; Barton, G.J.; Fica, S.M.; et al. M6A Modification of U6 SnRNA Modulates Usage of Two Major Classes of Pre-MRNA 5′ Splice Site. Elife 2022, 11, e78808. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ashraf, S.; Wang, J.; Lilley, D.M. Control of Box C/D SnoRNP Assembly by N6-Methylation of Adenine. EMBO Rep. 2017, 18, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy Quantitative Assessment of Genome Editing by Sequence Trace Decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 August 2020).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Katz, Y.; Wang, E.T.; Silterra, J.; Schwartz, S.; Wong, B.; Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P.; Airoldi, E.M.; Burge, C.B. Quantitative Visualization of Alternative Exon Expression from RNA-Seq Data. Bioinformatics 2015, 31, 2400–2402. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress Update–From Bulk to Single-Cell Expression Data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveeva, A.; Vinogradov, D.; Zhuravlev, E.; Semenov, D.; Vlassov, V.; Stepanov, G. Intron Editing Reveals SNORD-Dependent Maturation of the Small Nucleolar RNA Host Gene GAS5 in Human Cells. Int. J. Mol. Sci. 2023, 24, 17621. https://doi.org/10.3390/ijms242417621
Matveeva A, Vinogradov D, Zhuravlev E, Semenov D, Vlassov V, Stepanov G. Intron Editing Reveals SNORD-Dependent Maturation of the Small Nucleolar RNA Host Gene GAS5 in Human Cells. International Journal of Molecular Sciences. 2023; 24(24):17621. https://doi.org/10.3390/ijms242417621
Chicago/Turabian StyleMatveeva, Anastasiya, Dmitry Vinogradov, Evgenii Zhuravlev, Dmitriy Semenov, Valentin Vlassov, and Grigory Stepanov. 2023. "Intron Editing Reveals SNORD-Dependent Maturation of the Small Nucleolar RNA Host Gene GAS5 in Human Cells" International Journal of Molecular Sciences 24, no. 24: 17621. https://doi.org/10.3390/ijms242417621
APA StyleMatveeva, A., Vinogradov, D., Zhuravlev, E., Semenov, D., Vlassov, V., & Stepanov, G. (2023). Intron Editing Reveals SNORD-Dependent Maturation of the Small Nucleolar RNA Host Gene GAS5 in Human Cells. International Journal of Molecular Sciences, 24(24), 17621. https://doi.org/10.3390/ijms242417621





