Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”
Abstract
:1. Introduction
2. Etiology, Epidemiology, Diagnosis, and Treatment of Type 1 Diabetes
2.1. Etiology and Epidemiology of Type 1 Diabetes
2.2. Genetic Predisposition and Environment Triggers
2.3. T1D Diagnosis
2.4. T1D Treatment Options and Complications
2.5. T1D Cell Therapy
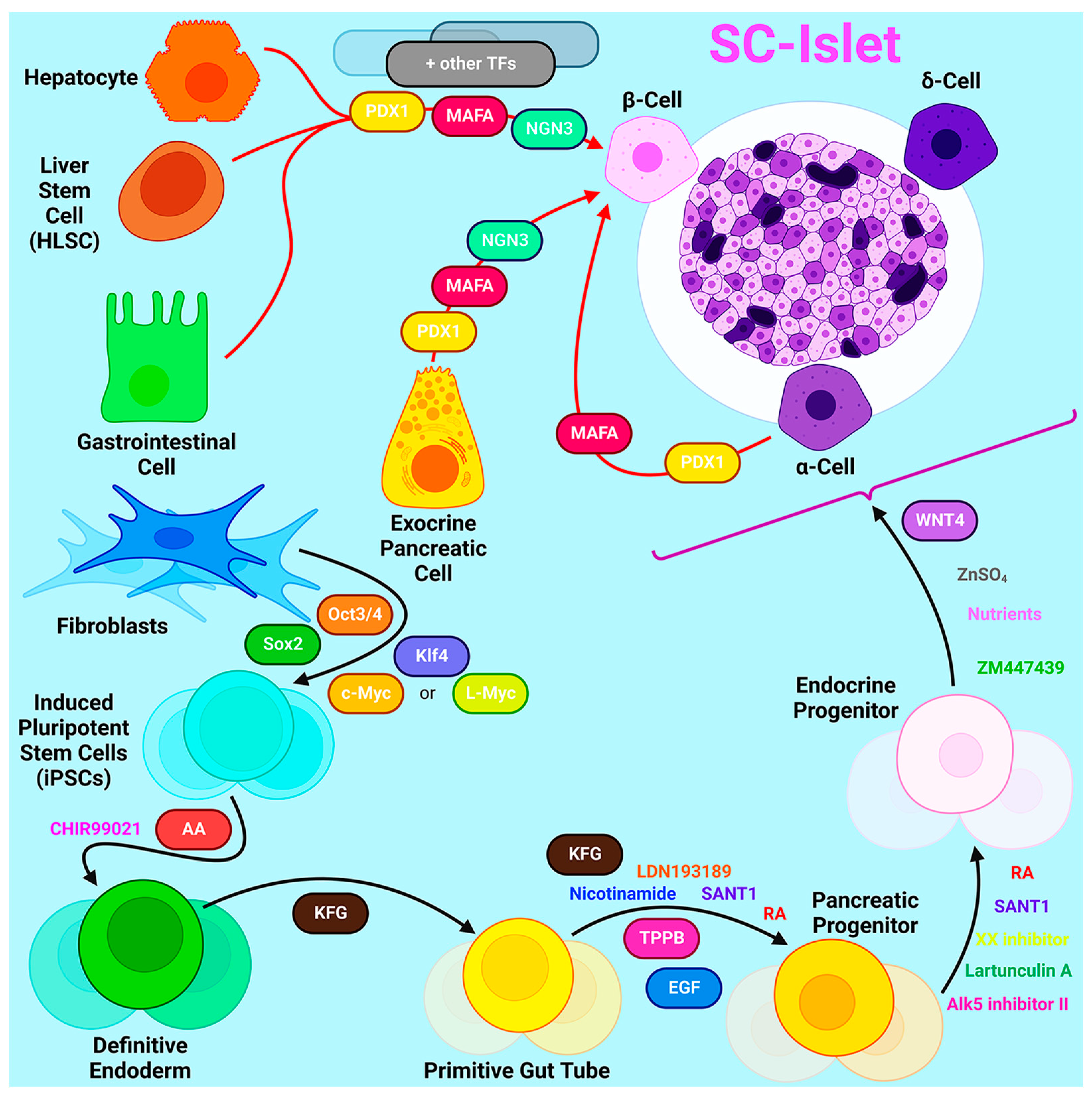
2.5.1. Sources for β-Cell Generation
2.5.2. HLA Haplotype Banks
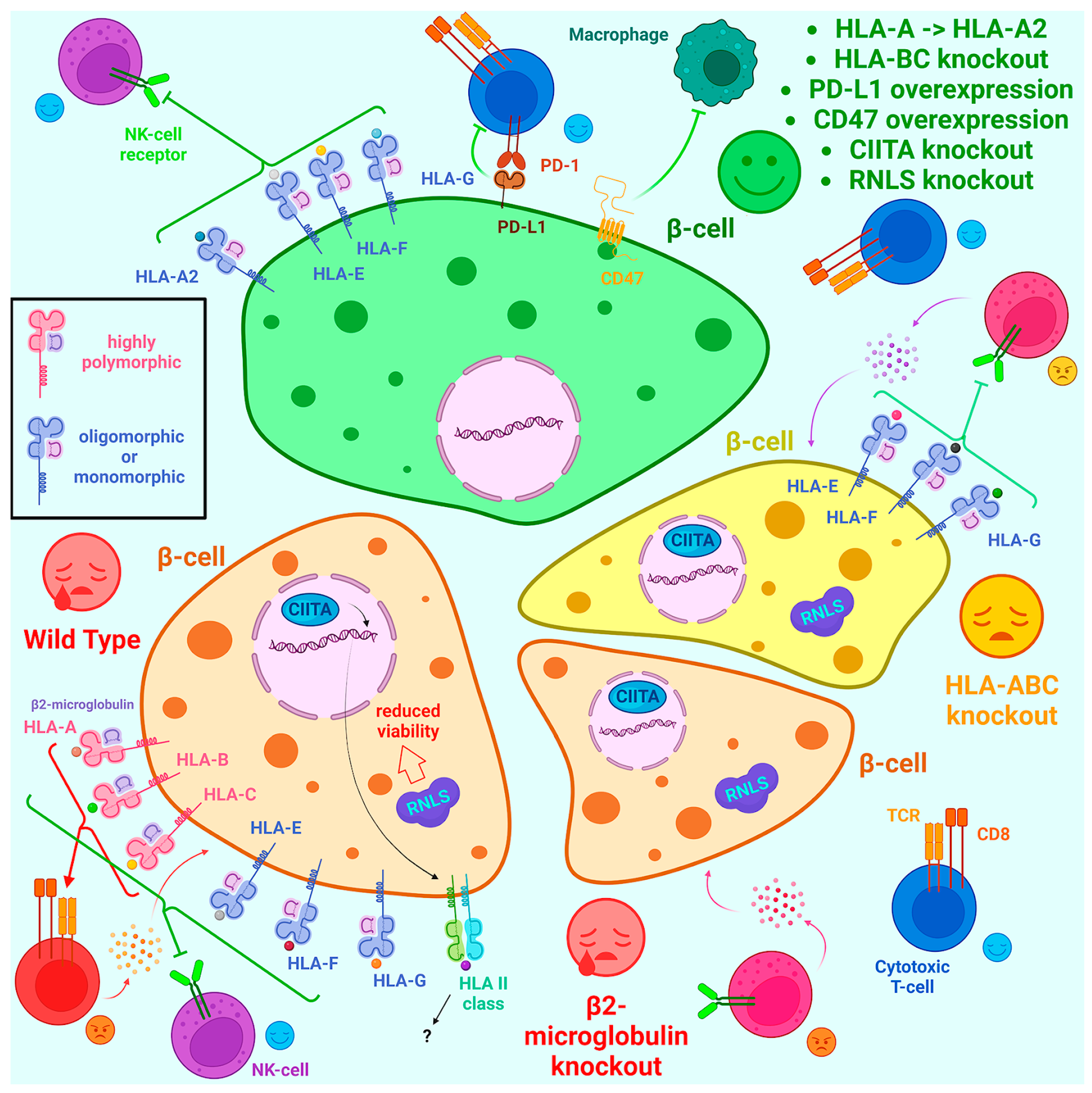
2.6. Engineering of Immunoprivileged iPSC Lines
2.6.1. Safety Concerns and Approaches to Overcome Them
2.6.2. Challenges in CRISPR/Cas9-Based Generation of Immunoprivileged iPSCs
- 50 kb away from known gene, to prevent genes located nearby from being affected;
- 300 kb away from known oncogene, to prevent insertional oncogenesis;
- 300 kb away from miRNAs, 150 kb away from lncRNAs and tRNAs, so as not to disrupt gene expression and cell cycle regulation;
- 300 kb away from telomeres and centromeres, so as not to disrupt cell division;
- 20 kb away from known enhancer region, so as not to interfere with enhancer–gene interaction.
2.6.3. The Potential of Advanced CRISPR/Cas9 Tools for Engineering Immunoprivileged iPSC Lines
CRISPR/Cas9 Artificial Transcription Factors
Prime Editors
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Norris, J.M.; Johnson, R.K.; Stene, L.C. Type 1 diabetes-early life origins and changing epidemiology. Lancet Diabetes Endocrinol. 2020, 8, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Gregory, G.A.; Robinson, T.I.G.; Linklater, S.E.; Wang, F.; Colagiuri, S.; de Beaufort, C.; Donaghue, K.C.; International Diabetes Federation Diabetes Atlas Type 1 Diabetes in Adults Special Interest Group; Magliano, D.J.; Maniam, J.; et al. Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: A modelling study. Lancet Diabetes Endocrinol. 2022, 10, 741–760. [Google Scholar] [CrossRef]
- Rodrigues Oliveira, S.M.; Rebocho, A.; Ahmadpour, E.; Nissapatorn, V.; de Lourdes Pereira, M. Type 1 diabetes mellitus: A review on advances and challenges in creating insulin producing devices. Micromachines 2023, 14, 151. [Google Scholar] [CrossRef]
- Nakatsuji, N.; Nakajima, F.; Tokunaga, K. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 2008, 26, 739–740. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Lei, A.; Tian, L.; Wang, X.; Correia, C.; Weiskittel, T.; Li, H.; Trounson, A.; Fu, Q.; Yao, K.; et al. Strategies for genetically engineering hypoimmunogenic universal pluripotent stem cells. iScience 2020, 23, 101162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.; Zhou, Z.; Fang, Y.; Ma, L.; Zhang, X.; Xiong, J.; Liu, L. Hypoimmunogenic human pluripotent stem cells are valid cell sources for cell therapeutics with normal self-renewal and multilineage differentiation capacity. Stem Cell Res. Ther. 2023, 14, 11. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- American Diabetes Association Professional Practice Committee. 6. Glycemic targets: Standards of medical care in diabetes-2022. Diabetes Care 2022, 45, S83–S96. [Google Scholar] [CrossRef]
- Li, Y.; Sun, F.; Yue, T.T.; Wang, F.X.; Yang, C.L.; Luo, J.H.; Rong, S.J.; Xiong, F.; Zhang, S.; Wang, C.Y. Revisiting the antigen-presenting function of beta cells in T1D pathogenesis. Front. Immunol. 2021, 12, 690783. [Google Scholar] [CrossRef]
- Insel, R.A.; Dunne, J.L.; Atkinson, M.A.; Chiang, J.L.; Dabelea, D.; Gottlieb, P.A.; Greenbaum, C.J.; Herold, K.C.; Krischer, J.P.; Lernmark, A.; et al. Staging presymptomatic type 1 diabetes: A scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015, 38, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, M.A. The pathogenesis and natural history of type 1 diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007641. [Google Scholar] [CrossRef]
- Mobasseri, M.; Shirmohammadi, M.; Amiri, T.; Vahed, N.; Hosseini Fard, H.; Ghojazadeh, M. Prevalence and incidence of type 1 diabetes in the world: A systematic review and meta-analysis. Health Promot. Perspect. 2020, 10, 98–115. [Google Scholar] [CrossRef]
- Ward, Z.J.; Yeh, J.M.; Reddy, C.L.; Gomber, A.; Ross, C.; Rittiphairoj, T.; Manne-Goehler, J.; Abdalla, A.T.; Abdullah, M.A.; Ahmed, A.; et al. Estimating the total incidence of type 1 diabetes in children and adolescents aged 0–19 years from 1990 to 2050: A global simulation-based analysis. Lancet Diabetes Endocrinol. 2022, 10, 848–858. [Google Scholar] [CrossRef]
- Ogle, G.D.; James, S.; Dabelea, D.; Pihoker, C.; Svennson, J.; Maniam, J.; Klatman, E.L.; Patterson, C.C. Global estimates of incidence of type 1 diabetes in children and adolescents: Results from the International Diabetes Federation Atlas, 10th edition. Diabetes Res. Clin. Pract. 2022, 183, 109083. [Google Scholar] [CrossRef] [PubMed]
- Roche, E.F.; McKenna, A.M.; O’Regan, M.; Ryder, K.J.; Fitzgerald, H.M.; Hoey, H. The incidence of type 1 diabetes in children under 15 years of age is rising again-a nationwide study. Eur. J. Pediatr. 2023, 182, 4615–4623. [Google Scholar] [CrossRef]
- Ruiz, P.L.D.; Chen, L.; Morton, J.I.; Salim, A.; Carstensen, B.; Gregg, E.W.; Pavkov, M.E.; Mata-Cases, M.; Mauricio, D.; Nichols, G.A.; et al. Mortality trends in type 1 diabetes: A multicountry analysis of six population-based cohorts. Diabetologia 2022, 65, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Giwa, A.M.; Ahmed, R.; Omidian, Z.; Majety, N.; Karakus, K.E.; Omer, S.M.; Donner, T.; Hamad, A.R.A. Current understandings of the pathogenesis of type 1 diabetes: Genetics to environment. World J. Diabetes 2020, 11, 13–25. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- IPD-IMGT/HLA. Available online: https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/ (accessed on 30 October 2023).
- Halpin, S.J.; McIvor, C.; Whyatt, G.; Adams, A.; Harvey, O.; McLean, L.; Walshaw, C.; Kemp, S.; Corrado, J.; Singh, R.; et al. Postdischarge symptoms and rehabilitation needs in survivors of COVID-19 infection: A cross-sectional evaluation. J. Med. Virol. 2021, 93, 1013–1022. [Google Scholar] [CrossRef]
- Primavera, M.; Giannini, C.; Chiarelli, F. Prediction and prevention of type 1 diabetes. Front. Endocrinol. 2020, 11, 248. [Google Scholar] [CrossRef]
- Oram, R.A.; Patel, K.; Hill, A.; Shields, B.; McDonald, T.J.; Jones, A.; Hattersley, A.T.; Weedon, M.N. A Type 1 Diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 2016, 39, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.A.; Oram, R.A.; Flanagan, S.E.; De Franco, E.; Colclough, K.; Shepherd, M.; Ellard, S.; Weedon, M.N.; Hattersley, A.T. Type 1 diabetes genetic risk score: A novel tool to discriminate monogenic and type 1 diabetes. Diabetes 2016, 65, 2094–2099. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.J.; Geyer, S.; Steck, A.K.; Sharp, S.; Wentworth, J.M.; Weedon, M.N.; Antinozzi, P.; Sosenko, J.; Atkinson, M.; Pugliese, A.; et al. A Type 1 Diabetes genetic risk score predicts progression of islet autoimmunity and development of type 1 diabetes in individuals at risk. Diabetes Care 2018, 41, 1887–1894. [Google Scholar] [CrossRef]
- Bonifacio, E.; Beyerlein, A.; Hippich, M.; Winkler, C.; Vehik, K.; Weedon, M.N.; Laimighofer, M.; Hattersley, A.T.; Krumsiek, J.; Frohnert, B.I.; et al. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children. PLoS Med. 2018, 15, e1002548. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, A.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- Maddaloni, E.; Bolli, G.B.; Frier, B.M.; Little, R.R.; Leslie, R.D.; Pozzilli, P.; Buzzetti, R. C-peptide determination in the diagnosis of type of diabetes and its management: A clinical perspective. Diabetes Obes. Metab. 2022, 24, 1912–1926. [Google Scholar] [CrossRef]
- Horber, S.; Orth, M.; Fritsche, A.; Peter, A. Comparability of C-peptide measurements—Current status and clinical relevance. Exp. Clin. Endocrinol. Diabetes 2023, 131, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, E. Anti-islet autoantibodies in type 1 diabetes. Int. J. Mol. Sci. 2023, 24, 10012. [Google Scholar] [CrossRef]
- Ziegler, A.G.; Rewers, M.; Simell, O.; Simell, T.; Lempainen, J.; Steck, A.; Winkler, C.; Ilonen, J.; Veijola, R.; Knip, M.; et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013, 309, 2473–2479. [Google Scholar] [CrossRef]
- Roep, B.O.; Thomaidou, S.; van Tienhoven, R.; Zaldumbide, A. Type 1 diabetes mellitus as a disease of the beta-cell (do not blame the immune system?). Nat. Rev. Endocrinol. 2021, 17, 150–161. [Google Scholar] [CrossRef]
- Blonde, L.; Umpierrez, G.E.; Reddy, S.S.; McGill, J.B.; Berga, S.L.; Bush, M.; Chandrasekaran, S.; DeFronzo, R.A.; Einhorn, D.; Galindo, R.J.; et al. American Association of Clinical Endocrinology Clinical Practice Guideline: Developing a diabetes mellitus comprehensive care plan-2022 update. Endocr. Pract. 2022, 28, 923–1049. [Google Scholar] [CrossRef]
- McQueen, R.B.; Geno Rasmussen, C.; Waugh, K.; Frohnert, B.I.; Steck, A.K.; Yu, L.; Baxter, J.; Rewers, M. Cost and cost-effectiveness of large-scale screening for type 1 diabetes in Colorado. Diabetes Care 2020, 43, 1496–1503. [Google Scholar] [CrossRef]
- Evans-Molina, C.; Oram, R.A. Teplizumab approval for type 1 diabetes in the USA. Lancet Diabetes Endocrinol. 2023, 11, 76–77. [Google Scholar] [CrossRef]
- Dayan, C.M.; Korah, M.; Tatovic, D.; Bundy, B.N.; Herold, K.C. Changing the landscape for type 1 diabetes: The first step to prevention. Lancet 2019, 394, 1286–1296. [Google Scholar] [CrossRef]
- With Type 1 Diabetes Delay Possible, Focus Now on Screening. Available online: https://www.medscape.com/viewarticle/984748?form=fpf (accessed on 23 October 2023).
- Nourelden, A.Z.; Elshanbary, A.A.; El-Sherif, L.; Benmelouka, A.Y.; Rohim, H.I.; Helmy, S.K.; Sayed, M.K.; Ismail, A.; Ali, A.S.; Ragab, K.M.; et al. Safety and efficacy of teplizumab for treatment of type one diabetes mellitus: A systematic review and meta-analysis. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 1895–1904. [Google Scholar] [CrossRef]
- Teplizumab-mzwv (Monograph). Available online: https://www.drugs.com/monograph/teplizumab-mzwv.html (accessed on 28 October 2023).
- Zhong, T.; Tang, R.; Gong, S.; Li, J.; Li, X.; Zhou, Z. The remission phase in type 1 diabetes: Changing epidemiology, definitions, and emerging immuno-metabolic mechanisms. Diabetes Metab. Res. Rev. 2020, 36, e3207. [Google Scholar] [CrossRef]
- Abdul-Rasoul, M.; Habib, H.; Al-Khouly, M. ‘The honeymoon phase’ in children with type 1 diabetes mellitus: Frequency, duration, and influential factors. Pediatr. Diabetes 2006, 7, 101–107. [Google Scholar] [CrossRef]
- Janez, A.; Guja, C.; Mitrakou, A.; Lalic, N.; Tankova, T.; Czupryniak, L.; Tabak, A.G.; Prazny, M.; Martinka, E.; Smircic-Duvnjak, L. Insulin therapy in adults with type 1 diabetes mellitus: A narrative review. Diabetes Ther. 2020, 11, 387–409. [Google Scholar] [CrossRef]
- Tauschmann, M.; Forlenza, G.; Hood, K.; Cardona-Hernandez, R.; Giani, E.; Hendrieckx, C.; DeSalvo, D.J.; Laffel, L.M.; Saboo, B.; Wheeler, B.J.; et al. ISPAD clinical practice consensus guidelines 2022: Diabetes technologies: Glucose monitoring. Pediatr Diabetes 2022, 23, 1390–1405. [Google Scholar] [CrossRef] [PubMed]
- Dermawan, D.; Purbayanto, M.A.K. An overview of advancements in closed-loop artificial pancreas system. Heliyon 2022, 8, e11648. [Google Scholar] [CrossRef] [PubMed]
- Lakshman, R.; Boughton, C.; Hovorka, R. The changing landscape of automated insulin delivery in the management of type 1 diabetes. Endocr. Connect. 2023, 12, e230132. [Google Scholar] [CrossRef] [PubMed]
- Whitticar, N.B.; Nunemaker, C.S. Reducing Glucokinase Activity to Enhance Insulin Secretion: A Counterintuitive Theory to Preserve Cellular Function and Glucose Homeostasis. Front. Endocrinol. 2020, 11, 378. [Google Scholar] [CrossRef]
- Kelkar, S.; Muley, S.; Ambardekar, P. Physiology of Insulin. In Towards Optimal Management of Diabetes in Surgery; Kelkar, S., Muley, S., Ambardekar, P., Eds.; Springer: Singapore, 2019; pp. 253–273. [Google Scholar]
- Campbell, J.E.; Newgard, C.B. Mechanisms controlling pancreatic islet cell function in insulin secretion. Nat. Rev. Mol. Cell Biol. 2021, 22, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Arcones, A.C.; Vila-Bedmar, R.; Mirasierra, M.; Cruces-Sande, M.; Vallejo, M.; Jones, B.; Tomas, A.; Mayor, F., Jr.; Murga, C. GRK2 regulates GLP-1R-mediated early phase insulin secretion in vivo. BMC Biol. 2021, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.I.G.; DeVries, J.H.; Hess-Fischl, A.; Hirsch, I.B.; Kirkman, M.S.; Klupa, T.; Ludwig, B.; Norgaard, K.; Pettus, J.; Renard, E.; et al. The Management of Type 1 Diabetes in Adults. A Consensus Report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care 2021, 44, 2589–2625. [Google Scholar] [CrossRef]
- Lin, Y.K.; Fisher, S.J.; Pop-Busui, R. Hypoglycemia unawareness and autonomic dysfunction in diabetes: Lessons learned and roles of diabetes technologies. J. Diabetes Investig. 2020, 11, 1388–1402. [Google Scholar] [CrossRef]
- Silver, B.; Ramaiya, K.; Andrew, S.B.; Fredrick, O.; Bajaj, S.; Kalra, S.; Charlotte, B.M.; Claudine, K.; Makhoba, A. EADSG Guidelines: Insulin Therapy in Diabetes. Diabetes Ther. 2018, 9, 449–492. [Google Scholar] [CrossRef]
- Sola-Gazagnes, A.; Pecquet, C.; Berre, S.; Achenbach, P.; Pierson, L.A.; Virmoux-Buisson, I.; M’Bemba, J.; Elgrably, F.; Moguelet, P.; Boitard, C.; et al. Insulin allergy: A diagnostic and therapeutic strategy based on a retrospective cohort and a case-control study. Diabetologia 2022, 65, 1278–1290. [Google Scholar] [CrossRef]
- Ansari-Moghaddam, A.; Setoodehzadeh, F.; Khammarnia, M.; Adineh, H.A. Economic cost of diabetes in the Eastern Mediterranean region countries: A meta-analysis. Diabetes Metab. Syndr. 2020, 14, 1101–1108. [Google Scholar] [CrossRef]
- Tommerdahl, K.L.; Shapiro, A.L.B.; Nehus, E.J.; Bjornstad, P. Early microvascular complications in type 1 and type 2 diabetes: Recent developments and updates. Pediatr. Nephrol. 2022, 37, 79–93. [Google Scholar] [CrossRef]
- Forlenza, G.P.; Ekhlaspour, L.; Breton, M.; Maahs, D.M.; Wadwa, R.P.; DeBoer, M.; Messer, L.H.; Town, M.; Pinnata, J.; Kruse, G.; et al. Successful At-Home Use of the Tandem Control-IQ Artificial Pancreas System in Young Children During a Randomized Controlled Trial. Diabetes Technol. Ther. 2019, 21, 159–169. [Google Scholar] [CrossRef]
- Kocova, M.; Milenkova, L. Old syndrome-new approach: Mauriac syndrome treated with continuous insulin delivery. SAGE Open Med. Case Rep. 2018, 6, 2050313X18785510. [Google Scholar] [CrossRef]
- Raman, S.; Dai, H.; DeLurgio, S.A.; Williams, D.D.; Lind, M.; Patton, S.R.; Spertus, J.A.; Kosiborod, M.; Clements, M.A. High hemoglobin A1c variability is associated with early risk of microalbuminuria in children with T1D. Pediatr. Diabetes 2016, 17, 398–406. [Google Scholar] [CrossRef]
- Scilletta, S.; Di Marco, M.; Miano, N.; Filippello, A.; Di Mauro, S.; Scamporrino, A.; Musmeci, M.; Coppolino, G.; Di Giacomo Barbagallo, F.; Bosco, G.; et al. Update on Diabetic Kidney Disease (DKD): Focus on Non-Albuminuric DKD and Cardiovascular Risk. Biomolecules 2023, 13, 752. [Google Scholar] [CrossRef]
- Selvarajah, D.; Kar, D.; Khunti, K.; Davies, M.J.; Scott, A.R.; Walker, J.; Tesfaye, S. Diabetic peripheral neuropathy: Advances in diagnosis and strategies for screening and early intervention. Lancet Diabetes Endocrinol. 2019, 7, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Qiao, G.H.; Wang, M.; Yu, L.; Sun, Y.; Shi, H.; Ma, T.L. Stem Cell-Based Therapy for Diabetic Foot Ulcers. Front. Cell Dev. Biol. 2022, 10, 812262. [Google Scholar] [CrossRef]
- Derry, S.; Bell, R.F.; Straube, S.; Wiffen, P.J.; Aldington, D.; Moore, R.A. Pregabalin for neuropathic pain in adults. Cochrane Database Syst. Rev. 2019, 2019, CD007076. [Google Scholar] [CrossRef]
- Gonzalez-Cortes, J.H.; Martinez-Pacheco, V.A.; Gonzalez-Cantu, J.E.; Bilgic, A.; de Ribot, F.M.; Sudhalkar, A.; Mohamed-Hamsho, J.; Kodjikian, L.; Mathis, T. Current Treatments and Innovations in Diabetic Retinopathy and Diabetic Macular Edema. Pharmaceutics 2022, 15, 122. [Google Scholar] [CrossRef]
- Shapiro, A.M.; Lakey, J.R.; Ryan, E.A.; Korbutt, G.S.; Toth, E.; Warnock, G.L.; Kneteman, N.M.; Rajotte, R.V. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 2000, 343, 230–238. [Google Scholar] [CrossRef]
- Shapiro, A.M.J.; Thompson, D.; Donner, T.W.; Bellin, M.D.; Hsueh, W.; Pettus, J.; Wilensky, J.; Daniels, M.; Wang, R.M.; Brandon, E.P.; et al. Insulin expression and C-peptide in type 1 diabetes subjects implanted with stem cell-derived pancreatic endoderm cells in an encapsulation device. Cell Rep. Med. 2021, 2, 100466. [Google Scholar] [CrossRef] [PubMed]
- Ramzy, A.; Thompson, D.M.; Ward-Hartstonge, K.A.; Ivison, S.; Cook, L.; Garcia, R.V.; Loyal, J.; Kim, P.T.W.; Warnock, G.L.; Levings, M.K.; et al. Implanted pluripotent stem-cell-derived pancreatic endoderm cells secrete glucose-responsive C-peptide in patients with type 1 diabetes. Cell Stem Cell 2021, 28, 2047–2061. [Google Scholar] [CrossRef] [PubMed]
- D’Amour, K.A.; Agulnick, A.D.; Eliazer, S.; Kelly, O.G.; Kroon, E.; Baetge, E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005, 23, 1534–1541. [Google Scholar] [CrossRef]
- Pagliuca, F.W.; Millman, J.R.; Gurtler, M.; Segel, M.; Van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of functional human pancreatic beta cells in vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Veres, A.; Faust, A.L.; Bushnell, H.L.; Engquist, E.N.; Kenty, J.H.; Harb, G.; Poh, Y.C.; Sintov, E.; Gurtler, M.; Pagliuca, F.W.; et al. Charting cellular identity during human in vitro beta-cell differentiation. Nature 2019, 569, 368–373. [Google Scholar] [CrossRef]
- Velazco-Cruz, L.; Song, J.; Maxwell, K.G.; Goedegebuure, M.M.; Augsornworawat, P.; Hogrebe, N.J.; Millman, J.R. Acquisition of Dynamic Function in Human Stem Cell-Derived beta Cells. Stem Cell Rep. 2019, 12, 351–365. [Google Scholar] [CrossRef]
- Sharon, N.; Vanderhooft, J.; Straubhaar, J.; Mueller, J.; Chawla, R.; Zhou, Q.; Engquist, E.N.; Trapnell, C.; Gifford, D.K.; Melton, D.A. Wnt Signaling Separates the Progenitor and Endocrine Compartments during Pancreas Development. Cell Rep. 2019, 27, 2281–2291. [Google Scholar] [CrossRef]
- Rosado-Olivieri, E.A.; Anderson, K.; Kenty, J.H.; Melton, D.A. YAP inhibition enhances the differentiation of functional stem cell-derived insulin-producing beta cells. Nat. Commun. 2019, 10, 1464. [Google Scholar] [CrossRef]
- Chen, S.; Du, K.; Zou, C. Current progress in stem cell therapy for type 1 diabetes mellitus. Stem Cell Res. Ther. 2020, 11, 275. [Google Scholar] [CrossRef]
- Helman, A.; Melton, D.A. A Stem Cell Approach to Cure Type 1 Diabetes. Cold Spring Harb. Perspect. Biol. 2021, 13, a035741. [Google Scholar] [CrossRef]
- Balboa, D.; Barsby, T.; Lithovius, V.; Saarimaki-Vire, J.; Omar-Hmeadi, M.; Dyachok, O.; Montaser, H.; Lund, P.E.; Yang, M.; Ibrahim, H.; et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 2022, 40, 1042–1055. [Google Scholar] [CrossRef] [PubMed]
- Augsornworawat, P.; Hogrebe, N.J.; Ishahak, M.; Schmidt, M.D.; Marquez, E.; Maestas, M.M.; Veronese-Paniagua, D.A.; Gale, S.E.; Miller, J.R.; Velazco-Cruz, L.; et al. Single-nucleus multi-omics of human stem cell-derived islets identifies deficiencies in lineage specification. Nat. Cell Biol. 2023, 25, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, G.; Nguyen-Ngoc, K.V.; Kim, D.; Miller, M.; Goss, G.; Kovsky, J.; Harrington, A.R.; Saunders, D.C.; Hopkirk, A.L.; et al. Understanding cell fate acquisition in stem-cell-derived pancreatic islets using single-cell multiome-inferred regulomes. Dev. Cell 2023, 58, 727–743.e711. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, C. Targeting beta-cell dedifferentiation and transdifferentiation: Opportunities and challenges. Endocr. Connect. 2021, 10, R213–R228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Bru-Tari, E.; Cobo-Vuilleumier, N.; Alonso-Magdalena, P.; Dos Santos, R.S.; Marroqui, L.; Nadal, A.; Gauthier, B.R.; Quesada, I. Pancreatic alpha-cell mass in the early-onset and advanced stage of a mouse model of experimental autoimmune diabetes. Sci. Rep. 2019, 9, 9515. [Google Scholar] [CrossRef] [PubMed]
- Furuyama, K.; Chera, S.; van Gurp, L.; Oropeza, D.; Ghila, L.; Damond, N.; Vethe, H.; Paulo, J.A.; Joosten, A.M.; Berney, T.; et al. Diabetes relief in mice by glucose-sensing insulin-secreting human alpha-cells. Nature 2019, 567, 43–48. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, T.; Lu, A.; Shiota, C.; Huard, M.; Whitney, K.E.; Huard, J. Specific reprogramming of alpha cells to insulin-producing cells by short glucagon promoter-driven Pdx1 and MafA. Mol. Ther. Methods Clin. Dev. 2023, 28, 355–365. [Google Scholar] [CrossRef]
- Banga, A.; Akinci, E.; Greder, L.V.; Dutton, J.R.; Slack, J.M. In Vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc. Natl. Acad. Sci. USA 2012, 109, 15336–15341. [Google Scholar] [CrossRef]
- Navarro-Tableros, V.; Gai, C.; Gomez, Y.; Giunti, S.; Pasquino, C.; Deregibus, M.C.; Tapparo, M.; Pitino, A.; Tetta, C.; Brizzi, M.F.; et al. Islet-Like Structures Generated In Vitro from Adult Human Liver Stem Cells Revert Hyperglycemia in Diabetic SCID Mice. Stem Cell Rev. Rep. 2019, 15, 93–111. [Google Scholar] [CrossRef]
- Klein, D.; Alvarez-Cubela, S.; Lanzoni, G.; Vargas, N.; Prabakar, K.R.; Boulina, M.; Ricordi, C.; Inverardi, L.; Pastori, R.L.; Dominguez-Bendala, J. BMP-7 Induces Adult Human Pancreatic Exocrine-to-Endocrine Conversion. Diabetes 2015, 64, 4123–4134. [Google Scholar] [CrossRef] [PubMed]
- Gourraud, P.A.; Gilson, L.; Girard, M.; Peschanski, M. The role of human leukocyte antigen matching in the development of multiethnic “haplobank” of induced pluripotent stem cell lines. Stem Cells 2012, 30, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.J.; Peacock, S.; Chaudhry, A.N.; Bradley, J.A.; Bolton, E.M. Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell 2012, 11, 147–152. [Google Scholar] [CrossRef]
- Abberton, K.M.; McDonald, T.L.; Diviney, M.; Holdsworth, R.; Leslie, S.; Delatycki, M.B.; Liu, L.; Klamer, G.; Johnson, P.; Elwood, N.J. Identification and Re-consent of Existing Cord Blood Donors for Creation of Induced Pluripotent Stem Cell Lines for Potential Clinical Applications. Stem Cells Transl. Med. 2022, 11, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Huh, J.Y.; Turner, D.M.; Lee, S.; Robinson, J.; Stein, J.E.; Shim, S.H.; Hong, C.P.; Kang, M.S.; Nakagawa, M.; et al. Repurposing the Cord Blood Bank for Haplobanking of HLA-Homozygous iPSCs and Their Usefulness to Multiple Populations. Stem Cells 2018, 36, 1552–1566. [Google Scholar] [CrossRef]
- Ichise, H.; Nagano, S.; Maeda, T.; Miyazaki, M.; Miyazaki, Y.; Kojima, H.; Yawata, N.; Yawata, M.; Tanaka, H.; Saji, H.; et al. NK Cell Alloreactivity against KIR-Ligand-Mismatched HLA-Haploidentical Tissue Derived from HLA Haplotype-Homozygous iPSCs. Stem Cell Rep. 2017, 9, 853–867. [Google Scholar] [CrossRef]
- Nowak, I.; Majorczyk, E.; Wisniewski, A.; Pawlik, A.; Magott-Procelewska, M.; Passowicz-Muszynska, E.; Malejczyk, J.; Ploski, R.; Giebel, S.; Barcz, E.; et al. Does the KIR2DS5 gene protect from some human diseases? PLoS ONE 2010, 5, e12381. [Google Scholar] [CrossRef]
- Wang, R.; Sun, Y.; Kuang, B.H.; Yan, X.; Lei, J.; Lin, Y.X.; Tian, J.; Li, Y.; Xie, X.; Chen, T.; et al. HLA-Bw4 in association with KIR3DL1 favors natural killer cell-mediated protection against severe COVID-19. Emerg. Microbes Infect. 2023, 12, 2185467. [Google Scholar] [CrossRef]
- Total Number of Donors and Cord Blood Units. Available online: https://statistics.wmda.info (accessed on 27 October 2023).
- Rim, Y.A.; Park, N.; Nam, Y.; Ham, D.S.; Kim, J.W.; Ha, H.Y.; Jung, J.W.; Jung, S.M.; Baek, I.C.; Kim, S.Y.; et al. Recent progress of national banking project on homozygous HLA-typed induced pluripotent stem cells in South Korea. J. Tissue Eng. Regen. Med. 2018, 12, e1531–e1536. [Google Scholar] [CrossRef]
- Umekage, M.; Sato, Y.; Takasu, N. Overview: An iPS cell stock at CiRA. Inflamm. Regen. 2019, 39, 17. [Google Scholar] [CrossRef]
- Yoshida, S.; Kato, T.M.; Sato, Y.; Umekage, M.; Ichisaka, T.; Tsukahara, M.; Takasu, N.; Yamanaka, S. A clinical-grade HLA haplobank of human induced pluripotent stem cells matching approximately 40% of the Japanese population. Med 2023, 4, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Callemeyn, J.; Lamarthee, B.; Koenig, A.; Koshy, P.; Thaunat, O.; Naesens, M. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022, 101, 692–710. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Jaramillo, E.; Haasnoot, G.W.; Kaider, A.; Schaefer, A.K.; Haberl, T.; Goekler, J.; Angleitner, P.; Moayedifar, R.; Zuckermann, A.; Fischer, G.F.; et al. Molecular-level HLA mismatch is associated with rejection and worsened graft survival in heart transplant recipients—A retrospective study. Transpl. Int. 2020, 33, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Alelign, T.; Ahmed, M.M.; Bobosha, K.; Tadesse, Y.; Howe, R.; Petros, B. Kidney Transplantation: The Challenge of Human Leukocyte Antigen and Its Therapeutic Strategies. J. Immunol. Res. 2018, 2018, 5986740. [Google Scholar] [CrossRef] [PubMed]
- Torikai, H.; Mi, T.; Gragert, L.; Maiers, M.; Najjar, A.; Ang, S.; Maiti, S.; Dai, J.; Switzer, K.C.; Huls, H.; et al. Genetic editing of HLA expression in hematopoietic stem cells to broaden their human application. Sci. Rep. 2016, 6, 21757. [Google Scholar] [CrossRef] [PubMed]
- Lanza, R.; Russell, D.W.; Nagy, A. Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 2019, 19, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef]
- Rehakova, D.; Souralova, T.; Koutna, I. Clinical-Grade Human Pluripotent Stem Cells for Cell Therapy: Characterization Strategy. Int. J. Mol. Sci. 2020, 21, 2435. [Google Scholar] [CrossRef]
- Frenet, E.M.; Scaradavou, A. Chapter 32—Human Leukocyte Antigens. In Transfusion Medicine and Hemostasis, 3rd ed.; Shaz, B.H., Hillyer, C.D., Reyes Gil, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 191–197. [Google Scholar]
- Tu, M.M.; Mahmoud, A.B.; Makrigiannis, A.P. Licensed and Unlicensed NK Cells: Differential Roles in Cancer and Viral Control. Front. Immunol. 2016, 7, 166. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Davis, B.W.; Ostrander, G.K. Transmissible Tumors: Breaking the Cancer Paradigm. Trends Genet. 2016, 32, 1–15. [Google Scholar] [CrossRef]
- Williams, T.M. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J. Mol. Diagn. 2001, 3, 98–104. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; Christmas, S.; Middleton, D.; Jones, A.R. Allele frequency net: A database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011, 39, D913–D919. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Suarez-Trujillo, F.; Juarez, I.; Rodriguez-Sainz, C.; Palacio-Gruber, J.; Vaquero-Yuste, C.; Molina-Alejandre, M.; Fernandez-Cruz, E.; Martin-Villa, J.M. Evolution and molecular interactions of major histocompatibility complex (MHC)-G, -E and -F genes. Cell Mol. Life Sci. 2022, 79, 464. [Google Scholar] [CrossRef] [PubMed]
- Dilthey, A.T. State-of-the-art genome inference in the human MHC. Int. J. Biochem. Cell Biol. 2021, 131, 105882. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, M.; Amiri, F.; Nourigorji, M.; Torabizadeh, F.; Dara, M.; Dianatpour, M. B2M gene knockout in HEK293T cells by non-viral delivery of CRISPR-Cas9 system for the generation of universal cells. Egypt. J. Med. Hum. Genet. 2022, 23, 62. [Google Scholar] [CrossRef]
- Lee, O.H.; Lee, S.; Park, M.; Moon, S.; Hwang, S.; Kim, B.; Kim, C.Y.; Lee, D.R.; Shim, S.H.; Park, K.H.; et al. Generation of a B2M homozygous knockout human somatic cell nuclear transfer-derived embryonic stem cell line using the CRISPR/Cas9 system. Stem Cell Res. 2022, 59, 102643. [Google Scholar] [CrossRef]
- Thongsin, N.; Suwanpitak, S.; Wattanapanitch, M. CRISPR-Cas9-mediated disruption of B2M and CIITA genes eliminates HLA class I and II expression in human induced pluripotent stem cells (MUSIi001-A-2). Stem Cell Res. 2023, 71, 103138. [Google Scholar] [CrossRef]
- Gornalusse, G.G.; Hirata, R.K.; Funk, S.E.; Riolobos, L.; Lopes, V.S.; Manske, G.; Prunkard, D.; Colunga, A.G.; Hanafi, L.A.; Clegg, D.O.; et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 2017, 35, 765–772. [Google Scholar] [CrossRef]
- Parent, A.V.; Faleo, G.; Chavez, J.; Saxton, M.; Berrios, D.I.; Kerper, N.R.; Tang, Q.; Hebrok, M. Selective deletion of human leukocyte antigens protects stem cell-derived islets from immune rejection. Cell Rep. 2021, 36, 109538. [Google Scholar] [CrossRef]
- Ji, T.T.; Niu, S.S.; Fang, M.H.; Xu, L.X.; Wang, X.; Zou, J.; Xu, F.; Zhang, M.; Niu, R.; Wu, J.; et al. Genome editing HLA alleles for a pilot immunocompatible hESC line in a Chinese hESC bank for cell therapies. Cell Prolif. 2023, 56, e13471. [Google Scholar] [CrossRef]
- Hu, X.; White, K.; Olroyd, A.G.; DeJesus, R.; Dominguez, A.A.; Dowdle, W.E.; Friera, A.M.; Young, C.; Wells, F.; Chu, E.Y.; et al. Hypoimmune induced pluripotent stem cells survive long term in fully immunocompetent, allogeneic rhesus macaques. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Jurewicz, M.M.; Stern, L.J. Class II MHC antigen processing in immune tolerance and inflammation. Immunogenetics 2019, 71, 171–187. [Google Scholar] [CrossRef]
- Quesada-Masachs, E.; Zilberman, S.; Rajendran, S.; Chu, T.; McArdle, S.; Kiosses, W.B.; Lee, J.M.; Yesildag, B.; Benkahla, M.A.; Pawlowska, A.; et al. Upregulation of HLA class II in pancreatic beta cells from organ donors with type 1 diabetes. Diabetologia 2022, 65, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Deuse, T.; Hu, X.; Gravina, A.; Wang, D.; Tediashvili, G.; De, C.; Thayer, W.O.; Wahl, A.; Garcia, J.V.; Reichenspurner, H.; et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 2019, 37, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, M.; Duan, S.; Franco, P.J.; Kenty, J.H.; Hedrick, P.; Xia, Y.; Allen, A.; Ferreira, L.M.R.; Strominger, J.L.; et al. Generation of hypoimmunogenic human pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2019, 116, 10441–10446. [Google Scholar] [CrossRef] [PubMed]
- Veres, A.; Gosis, B.S.; Ding, Q.; Collins, R.; Ragavendran, A.; Brand, H.; Erdin, S.; Cowan, C.A.; Talkowski, M.E.; Musunuru, K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 2014, 15, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Zhou, Y.; Kim, Y.J.; Zhu, Y.; Ma, F.; Yu, J.; Wang, Y.C.; Chen, X.; Li, Z.; Zeng, S.; et al. Development of allogeneic HSC-engineered iNKT cells for off-the-shelf cancer immunotherapy. Cell Rep. Med. 2021, 2, 100449. [Google Scholar] [CrossRef]
- Kitano, Y.; Nishimura, S.; Kato, T.M.; Ueda, A.; Takigawa, K.; Umekage, M.; Nomura, M.; Kawakami, A.; Ogawa, H.; Xu, H.; et al. Generation of hypoimmunogenic induced pluripotent stem cells by CRISPR-Cas9 system and detailed evaluation for clinical application. Mol. Ther. Methods Clin. Dev. 2022, 26, 15–25. [Google Scholar] [CrossRef]
- Xu, H.; Wang, B.; Ono, M.; Kagita, A.; Fujii, K.; Sasakawa, N.; Ueda, T.; Gee, P.; Nishikawa, M.; Nomura, M.; et al. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell 2019, 24, 566–578.e7. [Google Scholar] [CrossRef]
- Coronel, M.M.; Martin, K.E.; Hunckler, M.D.; Barber, G.; O’Neill, E.B.; Medina, J.D.; Opri, E.; McClain, C.A.; Batra, L.; Weaver, J.D.; et al. Immunotherapy via PD-L1-presenting biomaterials leads to long-term islet graft survival. Sci. Adv. 2020, 6, eaba5573. [Google Scholar] [CrossRef]
- Yoshihara, E.; O’Connor, C.; Gasser, E.; Wei, Z.; Oh, T.G.; Tseng, T.W.; Wang, D.; Cayabyab, F.; Dai, Y.; Yu, R.T.; et al. Immune-evasive human islet-like organoids ameliorate diabetes. Nature 2020, 586, 606–611. [Google Scholar] [CrossRef]
- Gerace, D.; Zhou, Q.; Kenty, J.H.; Veres, A.; Sintov, E.; Wang, X.; Boulanger, K.R.; Li, H.; Melton, D.A. Engineering human stem cell-derived islets to evade immune rejection and promote localized immune tolerance. Cell Rep. Med. 2023, 4, 100879. [Google Scholar] [CrossRef] [PubMed]
- Cai, E.P.; Ishikawa, Y.; Zhang, W.; Leite, N.C.; Li, J.; Hou, S.; Kiaf, B.; Hollister-Lock, J.; Yilmaz, N.K.; Schiffer, C.A.; et al. Genome-scale in vivo CRISPR screen identifies RNLS as a target for beta cell protection in type 1 diabetes. Nat. Metab. 2020, 2, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Tang, C.; Rao, M.S.; Weissman, I.L.; Wu, J.C. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 2013, 19, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Takizawa, N.; Narita, M.; Ichisaka, T.; Yamanaka, S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA 2010, 107, 14152–14157. [Google Scholar] [CrossRef]
- Chavez, J.C.; Bachmeier, C.; Kharfan-Dabaja, M.A. CAR T-cell therapy for B-cell lymphomas: Clinical trial results of available products. Ther. Adv. Hematol. 2019, 10, 2040620719841581. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Tang, Y.; Li, W.; Zeng, Q.; Chang, D. Efficiency of CAR-T Therapy for Treatment of Solid Tumor in Clinical Trials: A Meta-Analysis. Dis. Markers 2019, 2019, 3425291. [Google Scholar] [CrossRef] [PubMed]
- Sengsayadeth, S.; Savani, B.N.; Oluwole, O.; Dholaria, B. Overview of approved CAR-T therapies, ongoing clinical trials, and its impact on clinical practice. eJHaem 2022, 3, 6–10. [Google Scholar] [CrossRef]
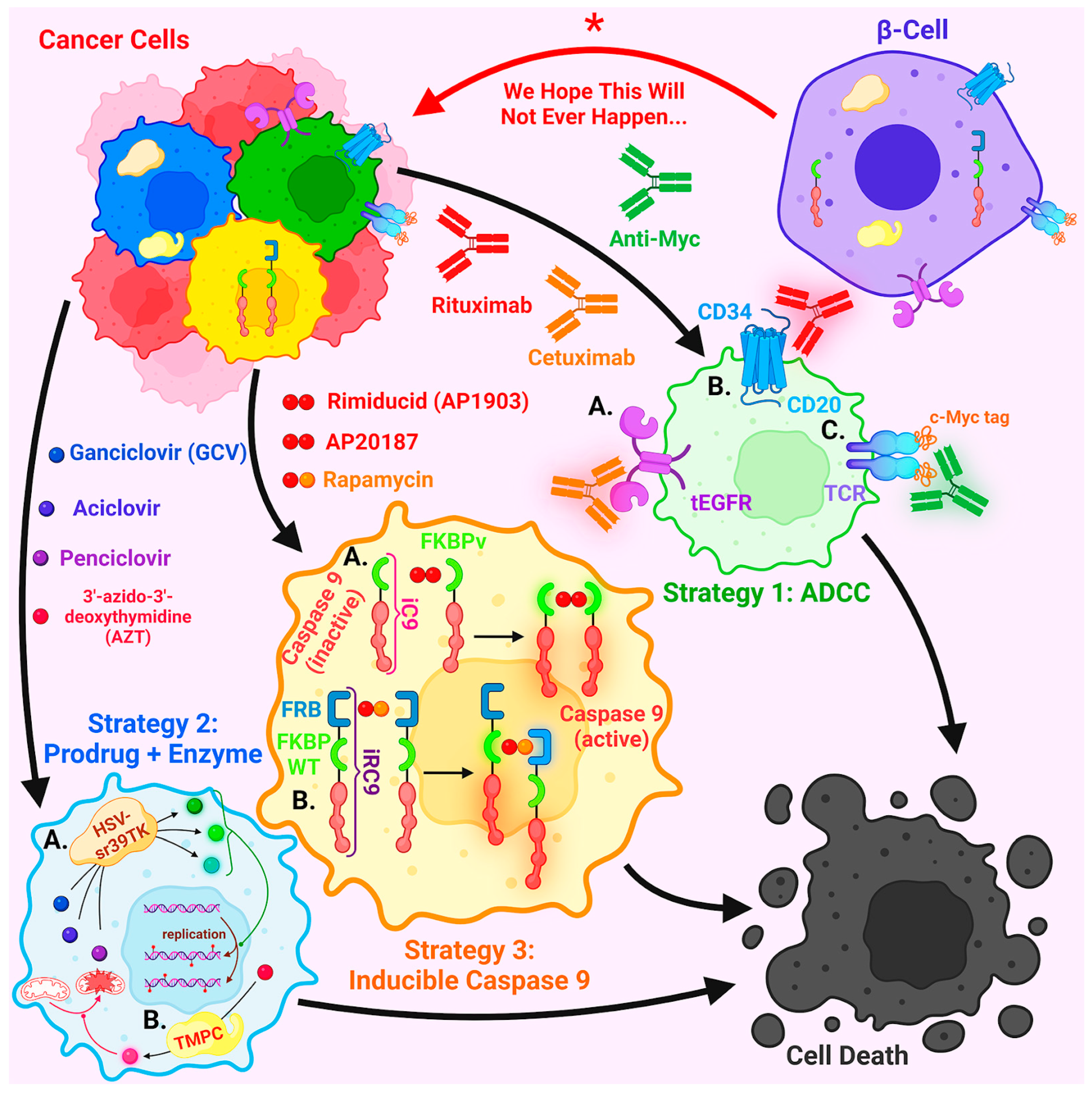
- Glowacki, P.; Rieske, P. Application and Design of Switches Used in CAR. Cells 2022, 11, 1910. [Google Scholar] [CrossRef]
- Wang, H.; Tang, L.; Kong, Y.; Liu, W.; Zhu, X.; You, Y. Strategies for Reducing Toxicity and Enhancing Efficacy of Chimeric Antigen Receptor T Cell Therapy in Hematological Malignancies. Int. J. Mol. Sci. 2023, 24, 9115. [Google Scholar] [CrossRef]
- Wang, Q.; He, F.; He, W.; Huang, Y.; Zeng, J.; Zi, F.; Zheng, J.; Fei, Y.; Xu, J.; Song, Y.; et al. A transgene-encoded truncated human epidermal growth factor receptor for depletion of anti- B-cell maturation antigen CAR-T cells. Cell Immunol. 2021, 363, 104342. [Google Scholar] [CrossRef] [PubMed]
- Paszkiewicz, P.J.; Frassle, S.P.; Srivastava, S.; Sommermeyer, D.; Hudecek, M.; Drexler, I.; Sadelain, M.; Liu, L.; Jensen, M.C.; Riddell, S.R.; et al. Targeted antibody-mediated depletion of murine CD19 CAR T cells permanently reverses B cell aplasia. J. Clin. Investig. 2016, 126, 4262–4272. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chang, W.C.; Wong, C.W.; Colcher, D.; Sherman, M.; Ostberg, J.R.; Forman, S.J.; Riddell, S.R.; Jensen, M.C. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 2011, 118, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Cheng, H.Y.; Nguyen, D.; Dettling, D.; Yeung, Y.A.; Sutton, J.; Hamze, M.; Valton, J.; Smith, J.; Djuretic, I.; et al. Allogeneic FLT3 CAR T Cells with an Off-Switch Exhibit Potent Activity against AML and Can Be Depleted to Expedite Bone Marrow Recovery. Mol. Ther. 2020, 28, 2237–2251. [Google Scholar] [CrossRef]
- Philip, B.; Kokalaki, E.; Mekkaoui, L.; Thomas, S.; Straathof, K.; Flutter, B.; Marin, V.; Marafioti, T.; Chakraverty, R.; Linch, D.; et al. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy. Blood 2014, 124, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Vogler, I.; Newrzela, S.; Hartmann, S.; Schneider, N.; von Laer, D.; Koehl, U.; Grez, M. An improved bicistronic CD20/tCD34 vector for efficient purification and in vivo depletion of gene-modified T cells for adoptive immunotherapy. Mol. Ther. 2010, 18, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Boldajipour, B.; Kuo, T.C.; Bentley, T.; Sutton, J.; Chen, A.; Geng, T.; Dong, H.; Galetto, R.; Valton, J.; et al. Preclinical Evaluation of Allogeneic CAR T Cells Targeting BCMA for the Treatment of Multiple Myeloma. Mol. Ther. 2019, 27, 1126–1138. [Google Scholar] [CrossRef]
- Kieback, E.; Charo, J.; Sommermeyer, D.; Blankenstein, T.; Uckert, W. A safeguard eliminates T cell receptor gene-modified autoreactive T cells after adoptive transfer. Proc. Natl. Acad. Sci. USA 2008, 105, 623–628. [Google Scholar] [CrossRef]
- Tran, V.L.; Novell, A.; Tournier, N.; Gerstenmayer, M.; Schweitzer-Chaput, A.; Mateos, C.; Jego, B.; Bouleau, A.; Nozach, H.; Winkeler, A.; et al. Impact of blood-brain barrier permeabilization induced by ultrasound associated to microbubbles on the brain delivery and kinetics of cetuximab: An immunoPET study using (89)Zr-cetuximab. J. Control. Release 2020, 328, 304–312. [Google Scholar] [CrossRef]
- Bonnan, M.; Ferrari, S.; Bertandeau, E.; Demasles, S.; Krim, E.; Miquel, M.; Barroso, B. Intrathecal rituximab therapy in multiple sclerosis: Review of evidence supporting the need for future trials. Curr. Drug Targets 2014, 15, 1205–1214. [Google Scholar] [CrossRef]
- Kao, R.L.; Truscott, L.C.; Chiou, T.T.; Tsai, W.; Wu, A.M.; De Oliveira, S.N. A Cetuximab-Mediated Suicide System in Chimeric Antigen Receptor-Modified Hematopoietic Stem Cells for Cancer Therapy. Hum. Gene Ther. 2019, 30, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Caratelli, S.; Arriga, R.; Sconocchia, T.; Ottaviani, A.; Lanzilli, G.; Pastore, D.; Cenciarelli, C.; Venditti, A.; Del Principe, M.I.; Lauro, D.; et al. In vitro elimination of epidermal growth factor receptor-overexpressing cancer cells by CD32A-chimeric receptor T cells in combination with cetuximab or panitumumab. Int. J. Cancer 2020, 146, 236–247. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Q.; Liu, H.; Lin, S.; Chen, H.; Ding, R.; Gu, Y.; Chao, C.C.; Dong, X. The Targeting Effect of Cetuximab Combined with PD-L1 Blockade against EGFR-Expressing Tumors in a Tailored CD16-CAR T-Cell Reporter System. Cancer Investig. 2021, 39, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.N.; Johnson, B.D.; Schneider, D.; Zhu, F.; Szabo, A.; Keever-Taylor, C.A.; Krueger, W.; Worden, A.A.; Kadan, M.J.; Yim, S.; et al. Bispecific anti-CD20, anti-CD19 CAR T cells for relapsed B cell malignancies: A phase 1 dose escalation and expansion trial. Nat. Med. 2020, 26, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Q.; Shen, F.; Xu, X.Y.; Zhang, H.; Yang, X.F.; Liu, W.G. Gene therapy with HSV1-sr39TK/GCV exhibits a stronger therapeutic efficacy than HSV1-TK/GCV in rat C6 glioma cells. Sci. World J. 2013, 2013, 951343. [Google Scholar] [CrossRef]
- Dilip, D.; Gregory, R.D.E. Suicide Gene Therapy by Herpes Simplex Virus-1 Thymidine Kinase (HSV-TK). In Targets in Gene Therapy; Yongping, Y., Ed.; IntechOpen: Rijeka, Croatia, 2011; p. 4. [Google Scholar]
- Hsu, C.; Abad, J.D.; Morgan, R.A. Characterization of human T lymphocytes engineered to express interleukin-15 and herpes simplex virus-thymidine kinase. J. Surg. Res. 2013, 184, 282–289. [Google Scholar] [CrossRef]
- Zhan, H.; Gilmour, K.; Chan, L.; Farzaneh, F.; McNicol, A.M.; Xu, J.H.; Adams, S.; Fehse, B.; Veys, P.; Thrasher, A.; et al. Production and first-in-man use of T cells engineered to express a HSVTK-CD34 sort-suicide gene. PLoS ONE 2013, 8, e77106. [Google Scholar] [CrossRef]
- Casucci, M.; Falcone, L.; Camisa, B.; Norelli, M.; Porcellini, S.; Stornaiuolo, A.; Ciceri, F.; Traversari, C.; Bordignon, C.; Bonini, C.; et al. Extracellular NGFR Spacers Allow Efficient Tracking and Enrichment of Fully Functional CAR-T Cells Co-Expressing a Suicide Gene. Front. Immunol. 2018, 9, 507. [Google Scholar] [CrossRef]
- Greco, R.; Oliveira, G.; Stanghellini, M.T.; Vago, L.; Bondanza, A.; Peccatori, J.; Cieri, N.; Marktel, S.; Mastaglio, S.; Bordignon, C.; et al. Improving the safety of cell therapy with the TK-suicide gene. Front. Pharmacol. 2015, 6, 95. [Google Scholar] [CrossRef]
- Traversari, C.; Marktel, S.; Magnani, Z.; Mangia, P.; Russo, V.; Ciceri, F.; Bonini, C.; Bordignon, C. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies. Blood 2007, 109, 4708–4715. [Google Scholar] [CrossRef]
- Klopp, A.; Schreiber, S.; Kosinska, A.D.; Pule, M.; Protzer, U.; Wisskirchen, K. Depletion of T cells via Inducible Caspase 9 Increases Safety of Adoptive T-Cell Therapy Against Chronic Hepatitis B. Front. Immunol. 2021, 12, 734246. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Neschadim, A.; Konrad, M.; Fowler, D.H.; Lavie, A.; Medin, J.A. Engineered human tmpk/AZT as a novel enzyme/prodrug axis for suicide gene therapy. Mol. Ther. 2007, 15, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Di Stasi, A.; Tey, S.K.; Dotti, G.; Fujita, Y.; Kennedy-Nasser, A.; Martinez, C.; Straathof, K.; Liu, E.; Durett, A.G.; Grilley, B.; et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011, 365, 1673–1683. [Google Scholar] [CrossRef]
- Amatya, C.; Pegues, M.A.; Lam, N.; Vanasse, D.; Geldres, C.; Choi, S.; Hewitt, S.M.; Feldman, S.A.; Kochenderfer, J.N. Development of CAR T Cells Expressing a Suicide Gene Plus a Chimeric Antigen Receptor Targeting Signaling Lymphocytic-Activation Molecule F7. Mol. Ther. 2021, 29, 702–717. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, K.; Al-Obaidi, M.; Di Stasi, A. Generation of Suicide Gene-Modified Chimeric Antigen Receptor-Redirected T-Cells for Cancer Immunotherapy. Methods Mol. Biol. 2019, 1895, 57–73. [Google Scholar] [CrossRef]
- Minagawa, K.; Jamil, M.O.; Al-Obaidi, M.; Pereboeva, L.; Salzman, D.; Erba, H.P.; Lamb, L.S.; Bhatia, R.; Mineishi, S.; Di Stasi, A. In Vitro Pre-Clinical Validation of Suicide Gene Modified Anti-CD33 Redirected Chimeric Antigen Receptor T-Cells for Acute Myeloid Leukemia. PLoS ONE 2016, 11, e0166891. [Google Scholar] [CrossRef]
- Guercio, M.; Manni, S.; Boffa, I.; Caruso, S.; Di Cecca, S.; Sinibaldi, M.; Abbaszadeh, Z.; Camera, A.; Ciccone, R.; Polito, V.A.; et al. Inclusion of the Inducible Caspase 9 Suicide Gene in CAR Construct Increases Safety of CAR.CD19 T Cell Therapy in B-Cell Malignancies. Front. Immunol. 2021, 12, 755639. [Google Scholar] [CrossRef]
- Duong, M.T.; Collinson-Pautz, M.R.; Morschl, E.; Lu, A.; Szymanski, S.P.; Zhang, M.; Brandt, M.E.; Chang, W.C.; Sharp, K.L.; Toler, S.M.; et al. Two-Dimensional Regulation of CAR-T Cell Therapy with Orthogonal Switches. Mol. Ther. Oncolytics 2019, 12, 124–137. [Google Scholar] [CrossRef]
- Diaconu, I.; Ballard, B.; Zhang, M.; Chen, Y.; West, J.; Dotti, G.; Savoldo, B. Inducible Caspase-9 Selectively Modulates the Toxicities of CD19-Specific Chimeric Antigen Receptor-Modified T Cells. Mol. Ther. 2017, 25, 580–592. [Google Scholar] [CrossRef]
- Warda, W.; Da Rocha, M.N.; Trad, R.; Haderbache, R.; Salma, Y.; Bouquet, L.; Roussel, X.; Nicod, C.; Deschamps, M.; Ferrand, C. Overcoming target epitope masking resistance that can occur on low-antigen-expresser AML blasts after IL-1RAP chimeric antigen receptor T cell therapy using the inducible caspase 9 suicide gene safety switch. Cancer Gene Ther. 2021, 28, 1365–1375. [Google Scholar] [CrossRef]
- Kim, A.; Lee, K.G.; Kwon, Y.; Lee, K.I.; Yang, H.M.; Habib, O.; Kim, J.; Kim, S.T.; Kim, S.J.; Kim, J.S.; et al. Off-the-Shelf, Immune-Compatible Human Embryonic Stem Cells Generated Via CRISPR-Mediated Genome Editing. Stem Cell Rev. Rep. 2021, 17, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Li, W.; Liu, Y.; Chen, Z.; Hui, Y.; Hao, P.; Xu, X.; Zhang, S.; Feng, H.; Zhang, B.; et al. Generation of hypoimmunogenic human pluripotent stem cells via expression of membrane-bound and secreted beta2m-HLA-G fusion proteins. Stem Cells 2020, 38, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Choi, J.; Park, N.; Kang, J.; Kim, M.; Kim, Y.; Ju, J.H. Development of immunocompatible pluripotent stem cells via CRISPR-based human leukocyte antigen engineering. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Ferreira, L.M.; Collins, R.; Meissner, T.B.; Boutwell, C.L.; Friesen, M.; Vrbanac, V.; Garrison, B.S.; Stortchevoi, A.; Bryder, D.; et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 2014, 15, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Flahou, C.; Yoshikawa, N.; Stirblyte, I.; Hayashi, Y.; Sawaguchi, A.; Akasaka, M.; Nakamura, S.; Higashi, N.; Xu, H.; et al. iPSC-Derived Platelets Depleted of HLA Class I Are Inert to Anti-HLA Class I and Natural Killer Cell Immunity. Stem Cell Rep. 2020, 14, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Iriguchi, S.; Waseda, M.; Ueda, N.; Ueda, T.; Xu, H.; Minagawa, A.; Ishikawa, A.; Yano, H.; Ishi, T.; et al. Generation of hypoimmunogenic T cells from genetically engineered allogeneic human induced pluripotent stem cells. Nat Biomed Eng 2021, 5, 429–440. [Google Scholar] [CrossRef]
- Bogomiakova, M.E.; Sekretova, E.K.; Anufrieva, K.S.; Khabarova, P.O.; Kazakova, A.N.; Bobrovsky, P.A.; Grigoryeva, T.V.; Eremeev, A.V.; Lebedeva, O.S.; Bogomazova, A.N.; et al. iPSC-derived cells lack immune tolerance to autologous NK-cells due to imbalance in ligands for activating and inhibitory NK-cell receptors. Stem Cell Res. Ther. 2023, 14, 77. [Google Scholar] [CrossRef]
- Mattapally, S.; Pawlik, K.M.; Fast, V.G.; Zumaquero, E.; Lund, F.E.; Randall, T.D.; Townes, T.M.; Zhang, J. Human Leukocyte Antigen Class I and II Knockout Human Induced Pluripotent Stem Cell-Derived Cells: Universal Donor for Cell Therapy. J. Am. Heart Assoc. 2018, 7, e010239. [Google Scholar] [CrossRef]
- Andrade da Silva, L.H.; Heuer, R.A.; Roque, C.B.; McGuire, T.L.; Hosoya, T.; Kimura, H.; Tamura, K.; Matsuoka, A.J. Enhanced survival of hypoimmunogenic otic progenitors following intracochlear xenotransplantation: Repercussions for stem cell therapy in hearing loss models. Stem Cell Res. Ther. 2023, 14, 83. [Google Scholar] [CrossRef]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef]
- Pacesa, M.; Lin, C.H.; Clery, A.; Saha, A.; Arantes, P.R.; Bargsten, K.; Irby, M.J.; Allain, F.H.; Palermo, G.; Cameron, P.; et al. Structural basis for Cas9 off-target activity. Cell 2022, 185, 4067–4081. [Google Scholar] [CrossRef]
- AlJanahi, A.A.; Lazzarotto, C.R.; Chen, S.; Shin, T.H.; Cordes, S.; Fan, X.; Jabara, I.; Zhou, Y.; Young, D.J.; Lee, B.C.; et al. Prediction and validation of hematopoietic stem and progenitor cell off-target editing in transplanted rhesus macaques. Mol. Ther. 2022, 30, 209–222. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef] [PubMed]
- Sherkatghanad, Z.; Abdar, M.; Charlier, J.; Makarenkov, V. Using traditional machine learning and deep learning methods for on- and off-target prediction in CRISPR/Cas9: A review. Brief. Bioinform. 2023, 24, bbad131. [Google Scholar] [CrossRef]
- Malinin, N.L.; Lee, G.; Lazzarotto, C.R.; Li, Y.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Iafrate, A.J.; Le, L.P.; et al. Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq. Nat. Protoc. 2021, 16, 5592–5615. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.Q.; Liu, P.; Smith, J.L.; Mintzer, E.; Maitland, S.; Dong, X.; Yang, Q.; Lee, J.; Haynes, C.M.; Zhu, L.J.; et al. Genome-wide detection of CRISPR editing in vivo using GUIDE-tag. Nat. Commun. 2022, 13, 437. [Google Scholar] [CrossRef] [PubMed]
- Dario, G.; Quan, Z.; Jennifer Hyoje-Ryu, K.; Elad, S.; Xi, W.; Kyle, R.B.; Hongfei, L.; Douglas, A.M. Secreted cytokines provide local immune tolerance for human stem cell-derived islets. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Lamothe, R.C.; Storlie, M.D.; Espinosa, D.A.; Rudlaff, R.; Browne, P.; Liu, J.; Rivas, A.; Devoto, A.; Oki, J.; Khoubyari, A.; et al. Novel CRISPR-Associated Gene-Editing Systems Discovered in Metagenomic Samples Enable Efficient and Specific Genome Engineering. CRISPR J. 2023, 6, 243–260. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.P.K.; Liu, M.S.; Hibshman, G.N.; Dangerfield, T.L.; Jung, K.; McCool, R.S.; Johnson, K.A.; Taylor, D.W. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 2022, 603, 343–347. [Google Scholar] [CrossRef]
- Vos, P.D.; Rossetti, G.; Mantegna, J.L.; Siira, S.J.; Gandadireja, A.P.; Bruce, M.; Raven, S.A.; Khersonsky, O.; Fleishman, S.J.; Filipovska, A.; et al. Computationally designed hyperactive Cas9 enzymes. Nat. Commun. 2022, 13, 3023. [Google Scholar] [CrossRef] [PubMed]
- Cerchione, D.; Loveluck, K.; Tillotson, E.L.; Harbinski, F.; DaSilva, J.; Kelley, C.P.; Keston-Smith, E.; Fernandez, C.A.; Myer, V.E.; Jayaram, H.; et al. SMOOT libraries and phage-induced directed evolution of Cas9 to engineer reduced off-target activity. PLoS ONE 2020, 15, e0231716. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Gao, L.; Li, D.; Gardner, Z.; Strecker, J.; Lash, B.; Zhang, F. Highly Parallel Profiling of Cas9 Variant Specificity. Mol. Cell 2020, 78, 794–800.e798. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, K.I.; Sutrisnoh, N.B.; Srinivasan, H.; Zhang, J.; Li, J.; Zhang, F.; Lalith, C.R.J.; Xing, H.; Shanmugam, R.; et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol. 2018, 19, 62. [Google Scholar] [CrossRef]
- Shor, O.; Rabinowitz, R.; Offen, D.; Benninger, F. Computational normal mode analysis accurately replicates the activity and specificity profiles of CRISPR-Cas9 and high-fidelity variants. Comput. Struct. Biotechnol. J. 2022, 20, 2013–2019. [Google Scholar] [CrossRef]
- Fontes, A.; Lakshmipathy, U. Advances in genetic modification of pluripotent stem cells. Biotechnol. Adv. 2013, 31, 994–1001. [Google Scholar] [CrossRef]
- Rapti, K.; Stillitano, F.; Karakikes, I.; Nonnenmacher, M.; Weber, T.; Hulot, J.S.; Hajjar, R.J. Effectiveness of gene delivery systems for pluripotent and differentiated cells. Mol. Ther. Methods Clin. Dev. 2015, 2, 14067. [Google Scholar] [CrossRef]
- Choi, G.C.G.; Zhou, P.; Yuen, C.T.L.; Chan, B.K.C.; Xu, F.; Bao, S.; Chu, H.Y.; Thean, D.; Tan, K.; Wong, K.H.; et al. Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9. Nat. Methods 2019, 16, 722–730. [Google Scholar] [CrossRef]
- Spasskaya, D.S.; Davletshin, A.I.; Bachurin, S.S.; Tutyaeva, V.V.; Garbuz, D.G.; Karpov, D.S. Improving the on-target activity of high-fidelity Cas9 editors by combining rational design and random mutagenesis. Appl. Microbiol. Biotechnol. 2023, 107, 2385–2401. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzoli, E.; Bianchi, A.; Umbach, A.; Amistadi, S.; Brusson, M.; Frati, G.; Ciciani, M.; Badowska, K.A.; Arosio, D.; Miccio, A.; et al. An optimized SpCas9 high-fidelity variant for direct protein delivery. Mol. Ther. 2023, 31, 2257–2265. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, N.; Okafor, I.; Choi, S.; Min, S.; Lee, J.; Bae, S.M.; Choi, K.; Choi, J.; Harihar, V.; et al. Sniper2L is a high-fidelity Cas9 variant with high activity. Nat. Chem. Biol. 2023, 19, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef]
- Ranzani, M.; Cesana, D.; Bartholomae, C.C.; Sanvito, F.; Pala, M.; Benedicenti, F.; Gallina, P.; Sergi, L.S.; Merella, S.; Bulfone, A.; et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat. Methods 2013, 10, 155–161. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Hauer, J.; Lim, A.; Picard, C.; Wang, G.P.; Berry, C.C.; Martinache, C.; Rieux-Laucat, F.; Latour, S.; Belohradsky, B.H.; et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2010, 363, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, H.B.; Cooray, S.; Gilmour, K.C.; Parsley, K.L.; Adams, S.; Howe, S.J.; Al Ghonaium, A.; Bayford, J.; Brown, L.; Davies, E.G.; et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2011, 3, 97ra79. [Google Scholar] [CrossRef]
- Grez, M.; Reichenbach, J.; Schwable, J.; Seger, R.; Dinauer, M.C.; Thrasher, A.J. Gene therapy of chronic granulomatous disease: The engraftment dilemma. Mol. Ther. 2011, 19, 28–35. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.P.; Wulffraat, N.; Leboulch, P.; Lim, A.; Osborne, C.S.; Pawliuk, R.; Morillon, E.; et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef]
- Howe, S.J.; Mansour, M.R.; Schwarzwaelder, K.; Bartholomae, C.; Hubank, M.; Kempski, H.; Brugman, M.H.; Pike-Overzet, K.; Chatters, S.J.; de Ridder, D.; et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Investig. 2008, 118, 3143–3150. [Google Scholar] [CrossRef]
- Mitchell, R.S.; Beitzel, B.F.; Schroder, A.R.; Shinn, P.; Chen, H.; Berry, C.C.; Ecker, J.R.; Bushman, F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004, 2, E234. [Google Scholar] [CrossRef]
- Dalwadi, D.A.; Calabria, A.; Tiyaboonchai, A.; Posey, J.; Naugler, W.E.; Montini, E.; Grompe, M. AAV integration in human hepatocytes. Mol. Ther. 2021, 29, 2898–2909. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.A.; Dominguez, D.A.; Chapman, L.M.; Gertz, E.M.; Budhu, A.; Forgues, M.; Chaisaingmongkol, J.; Rabibhadana, S.; Pupacdi, B.; Wu, X.; et al. Integration of adeno-associated virus (AAV) into the genomes of most Thai and Mongolian liver cancer patients does not induce oncogenesis. BMC Genom. 2021, 22, 814. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Salvetti, A. Integration of AAV vectors and insertional mutagenesis. Med. Sci. 2016, 32, 167–174. [Google Scholar] [CrossRef]
- Nault, J.C.; Datta, S.; Imbeaud, S.; Franconi, A.; Mallet, M.; Couchy, G.; Letouze, E.; Pilati, C.; Verret, B.; Blanc, J.F.; et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015, 47, 1187–1193. [Google Scholar] [CrossRef]
- Bayard, Q.; Meunier, L.; Peneau, C.; Renault, V.; Shinde, J.; Nault, J.C.; Mami, I.; Couchy, G.; Amaddeo, G.; Tubacher, E.; et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nat. Commun. 2018, 9, 5235. [Google Scholar] [CrossRef] [PubMed]
- Tatsuno, K.; Midorikawa, Y.; Takayama, T.; Yamamoto, S.; Nagae, G.; Moriyama, M.; Nakagawa, H.; Koike, K.; Moriya, K.; Aburatani, H. Impact of AAV2 and Hepatitis B Virus Integration Into Genome on Development of Hepatocellular Carcinoma in Patients with Prior Hepatitis B Virus Infection. Clin. Cancer Res. 2019, 25, 6217–6227. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019, 10, 4439. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, C.P.; Qiu, H.Y.; Zhang, H.X.; Chen, Q.B.; Zhang, Y.M.; Lei, X.L.; Zhang, C.X.; Yin, H.; Zhang, Y. Enhancement of the viability of T cells electroporated with DNA via osmotic dampening of the DNA-sensing cGAS-STING pathway. Nat. Biomed. Eng. 2023. [Google Scholar] [CrossRef]
- Cornu, T.I.; Mussolino, C.; Cathomen, T. Refining strategies to translate genome editing to the clinic. Nat. Med. 2017, 23, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Nichols, W.W.; Ledwith, B.J.; Manam, S.V.; Troilo, P.J. Potential DNA vaccine integration into host cell genome. Ann. N. Y. Acad. Sci. 1995, 772, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Ledwith, B.J.; Manam, S.; Troilo, P.J.; Barnum, A.B.; Pauley, C.J.; Griffiths, T.G., 2nd; Harper, L.B.; Beare, C.M.; Bagdon, W.J.; Nichols, W.W. Plasmid DNA vaccines: Investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 2000, 43, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Stadler, J.; Lemmens, R.; Nyhammar, T. Plasmid DNA purification. J. Gene Med. 2004, 6 (Suppl. S1), S54–S66. [Google Scholar] [CrossRef]
- Geng, K.; Merino, L.G.; Wedemann, L.; Martens, A.; Sobota, M.; Sanchez, Y.P.; Sondergaard, J.N.; White, R.J.; Kutter, C. Target-enriched nanopore sequencing and de novo assembly reveals co-occurrences of complex on-target genomic rearrangements induced by CRISPR-Cas9 in human cells. Genome Res. 2022, 32, 1876–1891. [Google Scholar] [CrossRef] [PubMed]
- Roth, T.L.; Puig-Saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, P.J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018, 559, 405–409. [Google Scholar] [CrossRef]
- Cromer, M.K.; Vaidyanathan, S.; Ryan, D.E.; Curry, B.; Lucas, A.B.; Camarena, J.; Kaushik, M.; Hay, S.R.; Martin, R.M.; Steinfeld, I.; et al. Global Transcriptional Response to CRISPR/Cas9-AAV6-Based Genome Editing in CD34(+) Hematopoietic Stem and Progenitor Cells. Mol. Ther. 2018, 26, 2431–2442. [Google Scholar] [CrossRef]
- Dever, D.P.; Bak, R.O.; Reinisch, A.; Camarena, J.; Washington, G.; Nicolas, C.E.; Pavel-Dinu, M.; Saxena, N.; Wilkens, A.B.; Mantri, S.; et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 2016, 539, 384–389. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Lattanzi, A.; Meneghini, V.; Pavani, G.; Amor, F.; Ramadier, S.; Felix, T.; Antoniani, C.; Masson, C.; Alibeu, O.; Lee, C.; et al. Optimization of CRISPR/Cas9 Delivery to Human Hematopoietic Stem and Progenitor Cells for Therapeutic Genomic Rearrangements. Mol. Ther. 2019, 27, 137–150. [Google Scholar] [CrossRef]
- Gravina, A.; Tediashvili, G.; Rajalingam, R.; Quandt, Z.; Deisenroth, C.; Schrepfer, S.; Deuse, T. Protection of cell therapeutics from antibody-mediated killing by CD64 overexpression. Nat. Biotechnol. 2023, 41, 717–727. [Google Scholar] [CrossRef]
- Ordovas, L.; Boon, R.; Pistoni, M.; Chen, Y.; Wolfs, E.; Guo, W.; Sambathkumar, R.; Bobis-Wozowicz, S.; Helsen, N.; Vanhove, J.; et al. Efficient Recombinase-Mediated Cassette Exchange in hPSCs to Study the Hepatocyte Lineage Reveals AAVS1 Locus-Mediated Transgene Inhibition. Stem Cell Rep. 2015, 5, 918–931. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, C.; Cerbini, T.; San, H.; Lin, Y.; Chen, G.; Rao, M.S.; Zou, J. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl. Med. 2014, 3, 821–835. [Google Scholar] [CrossRef]
- Shi, Z.D.; Tchao, J.; Wu, L.; Carman, A.J. Precision installation of a highly efficient suicide gene safety switch in human induced pluripotent stem cells. Stem Cells Transl. Med. 2020, 9, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Li, R.; Haga, H.; Kawabata, K. Transgene integration into the human AAVS1 locus enhances myosin II-dependent contractile force by reducing expression of myosin binding subunit 85. Biochem. Biophys. Res. Commun. 2015, 465, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xiong, Y.; Zhao, C.; Xie, S.; Li, C.; Li, X.; Liu, X.; Li, K.; Zhao, S.; Ruan, J. Identification of Glyceraldehyde-3-Phosphate Dehydrogenase Gene as an Alternative Safe Harbor Locus in Pig Genome. Genes 2019, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Paxton, W.A.; Choe, S.; Ceradini, D.; Martin, S.R.; Horuk, R.; MacDonald, M.E.; Stuhlmann, H.; Koup, R.A.; Landau, N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996, 86, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Sadelain, M.; Papapetrou, E.P.; Bushman, F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 2011, 12, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, G.; Soriano, P. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes. Dev. 1991, 5, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Aznauryan, E.; Yermanos, A.; Kinzina, E.; Devaux, A.; Kapetanovic, E.; Milanova, D.; Church, G.M.; Reddy, S.T. Discovery and validation of human genomic safe harbor sites for gene and cell therapies. Cell Rep. Methods 2022, 2, 100154. [Google Scholar] [CrossRef] [PubMed]
- Odak, A.; Yuan, H.; Feucht, J.; Cantu, V.A.; Mansilla-Soto, J.; Kogel, F.; Eyquem, J.; Everett, J.; Bushman, F.D.; Leslie, C.S.; et al. Novel extragenic genomic safe harbors for precise therapeutic T-cell engineering. Blood 2023, 141, 2698–2712. [Google Scholar] [CrossRef]
- Filion, T.M.; Qiao, M.; Ghule, P.N.; Mandeville, M.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Altieri, D.C.; Stein, G.S. Survival responses of human embryonic stem cells to DNA damage. J. Cell. Physiol. 2009, 220, 586–592. [Google Scholar] [CrossRef]
- Ihry, R.J.; Worringer, K.A.; Salick, M.R.; Frias, E.; Ho, D.; Theriault, K.; Kommineni, S.; Chen, J.; Sondey, M.; Ye, C.; et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018, 24, 939–946. [Google Scholar] [CrossRef]
- Marion, R.M.; Strati, K.; Li, H.; Murga, M.; Blanco, R.; Ortega, S.; Fernandez-Capetillo, O.; Serrano, M.; Blasco, M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009, 460, 1149–1153. [Google Scholar] [CrossRef]
- Bendixen, L.; Jensen, T.I.; Bak, R.O. CRISPR-Cas-mediated transcriptional modulation: The therapeutic promises of CRISPRa and CRISPRi. Mol. Ther. 2023, 31, 1920–1937. [Google Scholar] [CrossRef]
- Tse, H.M.; Kozlovskaya, V.; Kharlampieva, E.; Hunter, C.S. Minireview: Directed Differentiation and Encapsulation of Islet beta-Cells-Recent Advances and Future Considerations. Mol. Endocrinol. 2015, 29, 1388–1399. [Google Scholar] [CrossRef]
- Gheibi, S.; Singh, T.; da Cunha, J.; Fex, M.; Mulder, H. Insulin/Glucose-Responsive Cells Derived from Induced Pluripotent Stem Cells: Disease Modeling and Treatment of Diabetes. Cells 2020, 9, 2465. [Google Scholar] [CrossRef]
- Tran, R.; Moraes, C.; Hoesli, C.A. Controlled clustering enhances PDX1 and NKX6.1 expression in pancreatic endoderm cells derived from pluripotent stem cells. Sci. Rep. 2020, 10, 1190. [Google Scholar] [CrossRef] [PubMed]
- Alzhanuly, B.; Mukhatayev, Z.Y.; Botbayev, D.M.; Ashirbekov, Y.; Katkenov, N.D.; Dzhaynakbaev, N.T.; Sharipov, K.O. Modulation of Insulin Gene Expression with CRISPR/Cas9-based Transcription Factors. Open Access Maced. J. Med. Sci. 2021, 9, 876–881. [Google Scholar] [CrossRef]
- Gimenez Carla, A.; Curti, L.; Hyon Sung, H.; Grosembacher, L.; Ross Pablo, J.; Pereyra-Bonnet, F. Activation of pancreatic β-cell genes by multiplex epigenetic CRISPR-editing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lee, M.H.; Thomas, J.L.; Lin, C.Y.; Li, Y.E.; Lin, H.Y. Nanoparticle-mediated CRISPR/dCas9a activation of multiple transcription factors to engineer insulin-producing cells. J. Mater. Chem. B 2023, 11, 1866–1870. [Google Scholar] [CrossRef]
- Lee, M.H.; Thomas, J.L.; Lin, C.Y.; Li, Y.E.; Lin, H.Y. Activation of Insulin Gene Expression via Transfection of a CRISPR/dCas9a System Using Magnetic Peptide-Imprinted Nanoparticles. Pharmaceutics 2023, 15, 1311. [Google Scholar] [CrossRef] [PubMed]
- Omole, A.E.; Fakoya, A.O.J. Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ 2018, 6, e4370. [Google Scholar] [CrossRef]
- Boyle, E.A.; Becker, W.R.; Bai, H.B.; Chen, J.S.; Doudna, J.A.; Greenleaf, W.J. Quantification of Cas9 binding and cleavage across diverse guide sequences maps landscapes of target engagement. Sci. Adv. 2021, 7, eabe5496. [Google Scholar] [CrossRef]
- Lee, J.; Lim, K.; Kim, A.; Mok, Y.G.; Chung, E.; Cho, S.I.; Lee, J.M.; Kim, J.S. Prime editing with genuine Cas9 nickases minimizes unwanted indels. Nat. Commun. 2023, 14, 1786. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023, 24, 161–177. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef]
- Sun, R.; Cui, Y.; Liu, Z.; Guo, J.; Zhang, X.; Zhu, P.; Sha, J.; Yang, X.; Yuan, Y. A prime editor efficiently repaired human induced pluripotent stem cells with AR gene mutation (c.2710G>A; p. V904M). Stem Cell Res. 2023, 69, 103102. [Google Scholar] [CrossRef] [PubMed]
- Chemello, F.; Chai, A.C.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Atmanli, A.; Mireault, A.A.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021, 7, eabg4910. [Google Scholar] [CrossRef] [PubMed]
- Eggenschwiler, R.; Gschwendtberger, T.; Felski, C.; Jahn, C.; Langer, F.; Sterneckert, J.; Hermann, A.; Luhmann, J.; Steinemann, D.; Haase, A.; et al. A selectable all-in-one CRISPR prime editing piggyBac transposon allows for highly efficient gene editing in human cell lines. Sci. Rep. 2021, 11, 22154. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, Z.; Tao, W.; Liu, Y.; Li, X.; Wang, X.; Harati, J.; Wang, P.Y.; Huang, X.; Lin, C.P. Broadening prime editing toolkits using RNA-Pol-II-driven engineered pegRNA. Mol. Ther. 2022, 30, 2923–2932. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, N.S.; Bak, R.O. Enrichment strategies to enhance genome editing. J. Biomed. Sci. 2023, 30, 51. [Google Scholar] [CrossRef]
| Fasting Glucose Plasma (Fasting Is Defined as No Caloric Intake for at Least 8 h) | Random Plasma Glucose or 2-h Plasma Glucose during OGTT | Glycated Hemoglobin |
|---|---|---|
| ≥7.0 mmol/L (126 mg/dL) | ≥11.1 mmol/L (200 mg/dL). | ≥6.5% (48 mmol/mol) |
| Clinical Trial NCT | Company | Product | Cells | Administration |
|---|---|---|---|---|
| NCT03163511 | Viacyte | VC-02 | Pancreatic endoderm cells (PEC-01 cells) | Subcutaneous in a protective device |
| NCT03513939 | Sernova | Cell Pouch | Therapeutic cells including islets | Abdominal musculature |
| NCT04786262 | Vertex | VX-880 | Allogeneic fully differentiated insulin-producing islets | Infused into the hepatic portal vein |
| NCT05565248 | CRISPR Therapeutics AG in collaboration with Viacyte | VCTX211 | Allogeneic pancreatic endoderm cells (PEC211) genetically modified using CRISPR/Cas9 | In a surgically implanted durable, removable, perforated device |
| NCT05791201 | Vertex | VX-264 | Allogeneic fully differentiated insulin-producing islets | In a surgically implanted channel array protective device |
| Type of Nuclease | Type of Delivery | Modification | Reference |
|---|---|---|---|
| spCas9n | Transfection plasmids | knockout B2M gene | [111] |
| spCas9 | Nucleofection RNP | knockout CIITA gene | [113] |
| knockout HLA-B, and CIITA genes | [125] | ||
| knockout HLA-B, HLA-C, and CIITA genes | [124] | ||
| knockout B2M, and CIITA genes | [123] | ||
| knockout HLA-A, HLA-B, and HLA-DR | [168] | ||
| Nucleofection plasmids | knockout HLA-B, HLA-C, and CIITA genes | [116] | |
| knockout B2M, knockin HLA-G in B2M locus | [169] | ||
| knockout HLA-B, HLA-C, and CIITA genes | [121] | ||
| knockin PD-L1, HLA-G, CD47 in AAVS1 locus | |||
| knockout CIITA gene | [122] | ||
| knockout HLA-B gene | [170] | ||
| knockout B2M gene | [171,172] | ||
| knockout HLA-B, HLA-C, and CIITA genes | [116] | ||
| knockin iC9 in AAVS1 | |||
| knockout B2M, CIITA, and PVR genes | [173] | ||
| Lipofection plasmids | knockout B2M gene | [112,174] | |
| knockout B2M, and CIITA genes | [120] | ||
| Lipofection gRNA in cells with knockin inducible Cas9 in AAVS1 locus | knockout HLA-A, HLA-B, HLA-C, and CIITA genes | [115] | |
| Lentiviral particles | knockout B2M, and CIITA genes | [175] | |
| Hi-Fi spCas9 | Nucleofection RNP | knockout HLA-A, HLA-B, and HLA-C | [176] |
| knockout B2M gene | [128] | ||
| Nucleofection plasmids | knockin HLA-E, PD-L1, IL-2 mutein, IL-10, and TGFB1 in GAPDH locus | [128] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karpov, D.S.; Sosnovtseva, A.O.; Pylina, S.V.; Bastrich, A.N.; Petrova, D.A.; Kovalev, M.A.; Shuvalova, A.I.; Eremkina, A.K.; Mokrysheva, N.G. Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”. Int. J. Mol. Sci. 2023, 24, 17320. https://doi.org/10.3390/ijms242417320
Karpov DS, Sosnovtseva AO, Pylina SV, Bastrich AN, Petrova DA, Kovalev MA, Shuvalova AI, Eremkina AK, Mokrysheva NG. Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”. International Journal of Molecular Sciences. 2023; 24(24):17320. https://doi.org/10.3390/ijms242417320
Chicago/Turabian StyleKarpov, Dmitry S., Anastasiia O. Sosnovtseva, Svetlana V. Pylina, Asya N. Bastrich, Darya A. Petrova, Maxim A. Kovalev, Anastasija I. Shuvalova, Anna K. Eremkina, and Natalia G. Mokrysheva. 2023. "Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”" International Journal of Molecular Sciences 24, no. 24: 17320. https://doi.org/10.3390/ijms242417320
APA StyleKarpov, D. S., Sosnovtseva, A. O., Pylina, S. V., Bastrich, A. N., Petrova, D. A., Kovalev, M. A., Shuvalova, A. I., Eremkina, A. K., & Mokrysheva, N. G. (2023). Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”. International Journal of Molecular Sciences, 24(24), 17320. https://doi.org/10.3390/ijms242417320










