High-Throughput Transcriptomics Differentiates Toxic versus Non-Toxic Chemical Exposures Using a Rat Liver Model
Abstract
1. Introduction
2. Results and Discussion
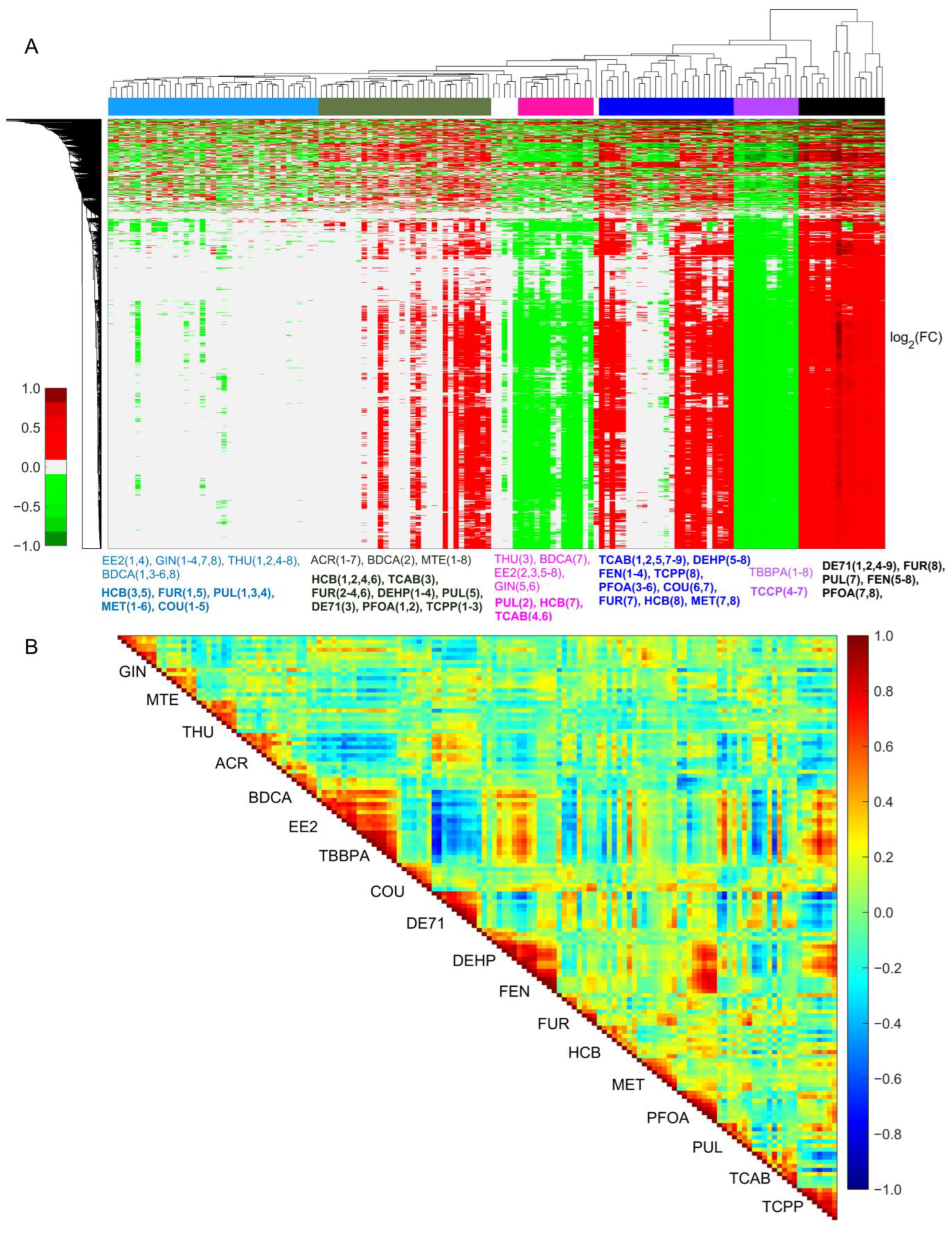
2.1. Gene Expression Patterns in Dose–Response Studies Predict Toxicity Outcomes
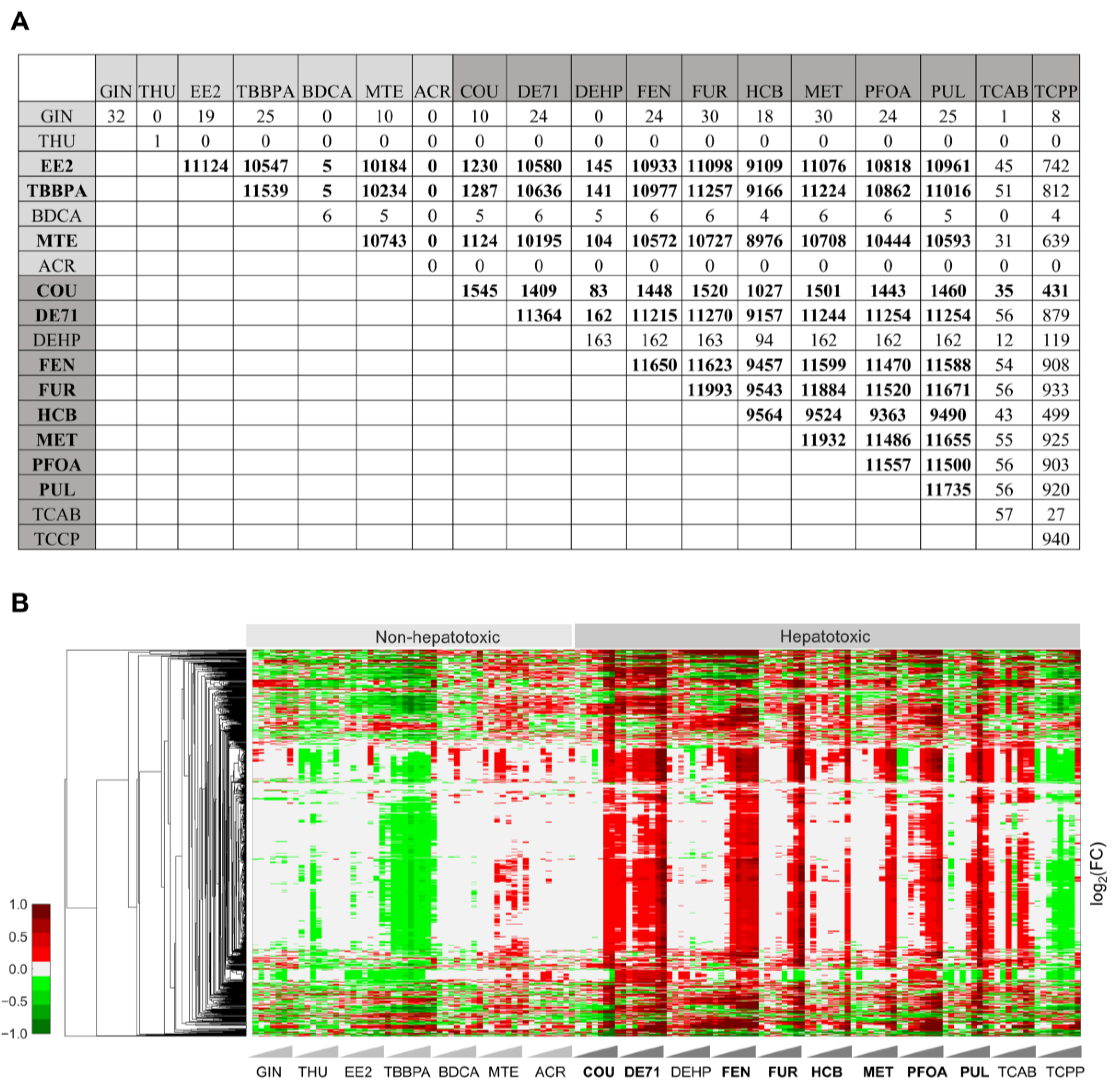
2.2. Significantly Changed Common Genes Differentiate Non-Hepatotoxic Chemicals from Hepatotoxic Chemicals
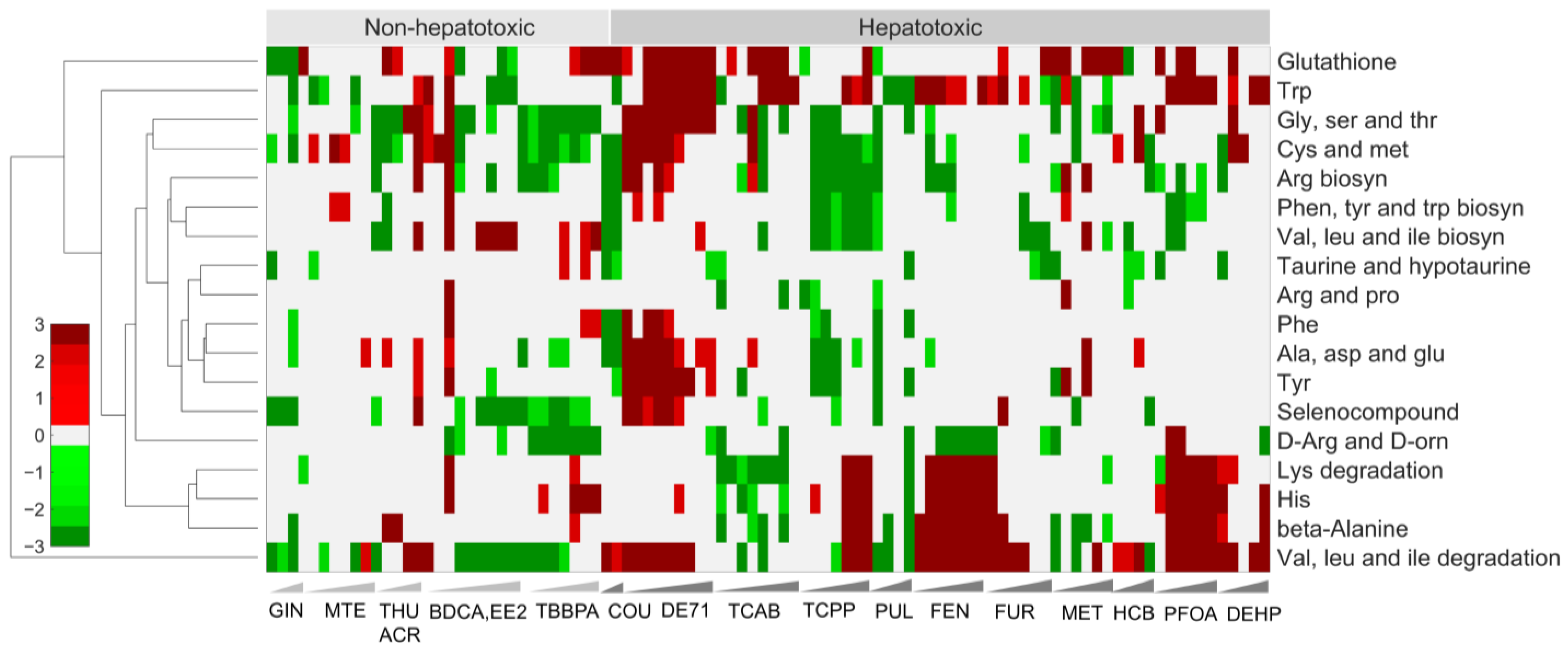
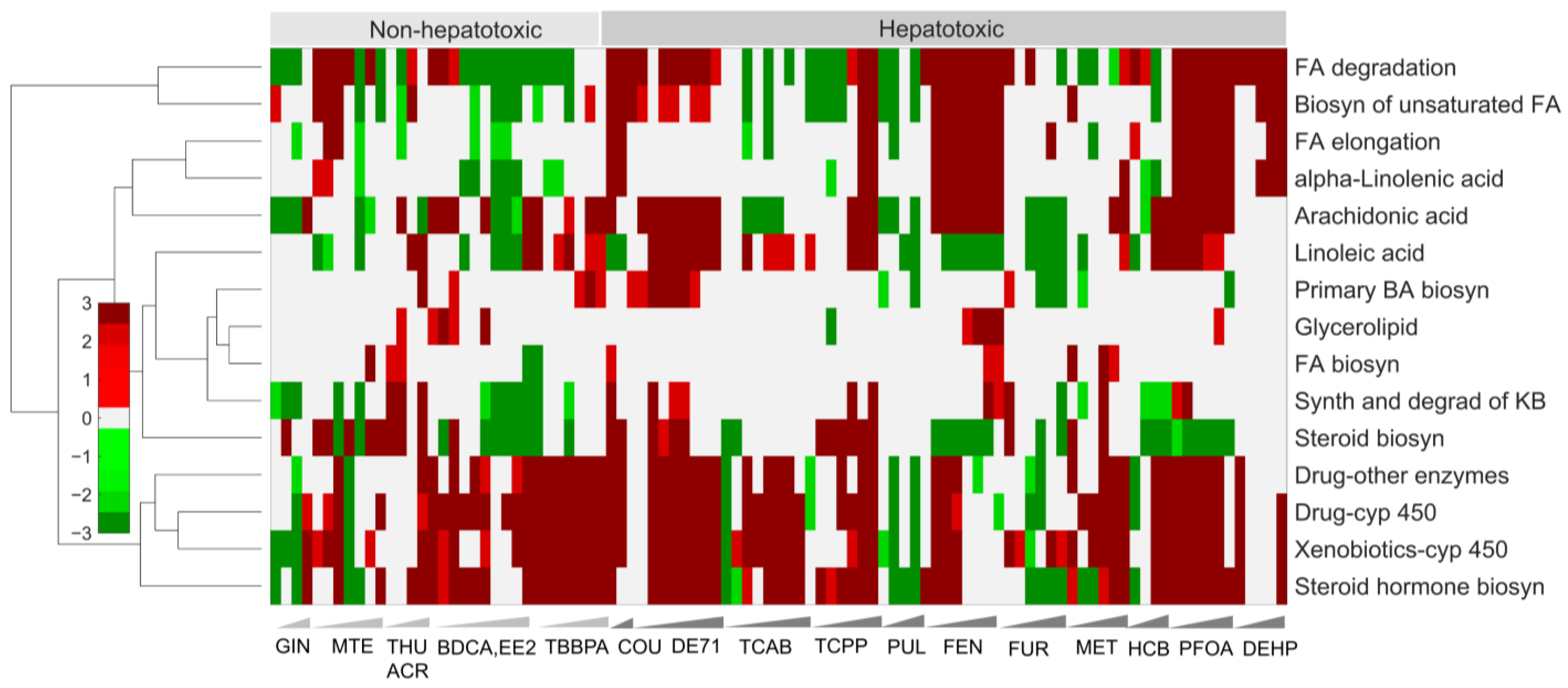
2.3. Alterations of Pathways in Organismal, Environmental, and Metabolism Processes Are Potential Indicators of Toxicity
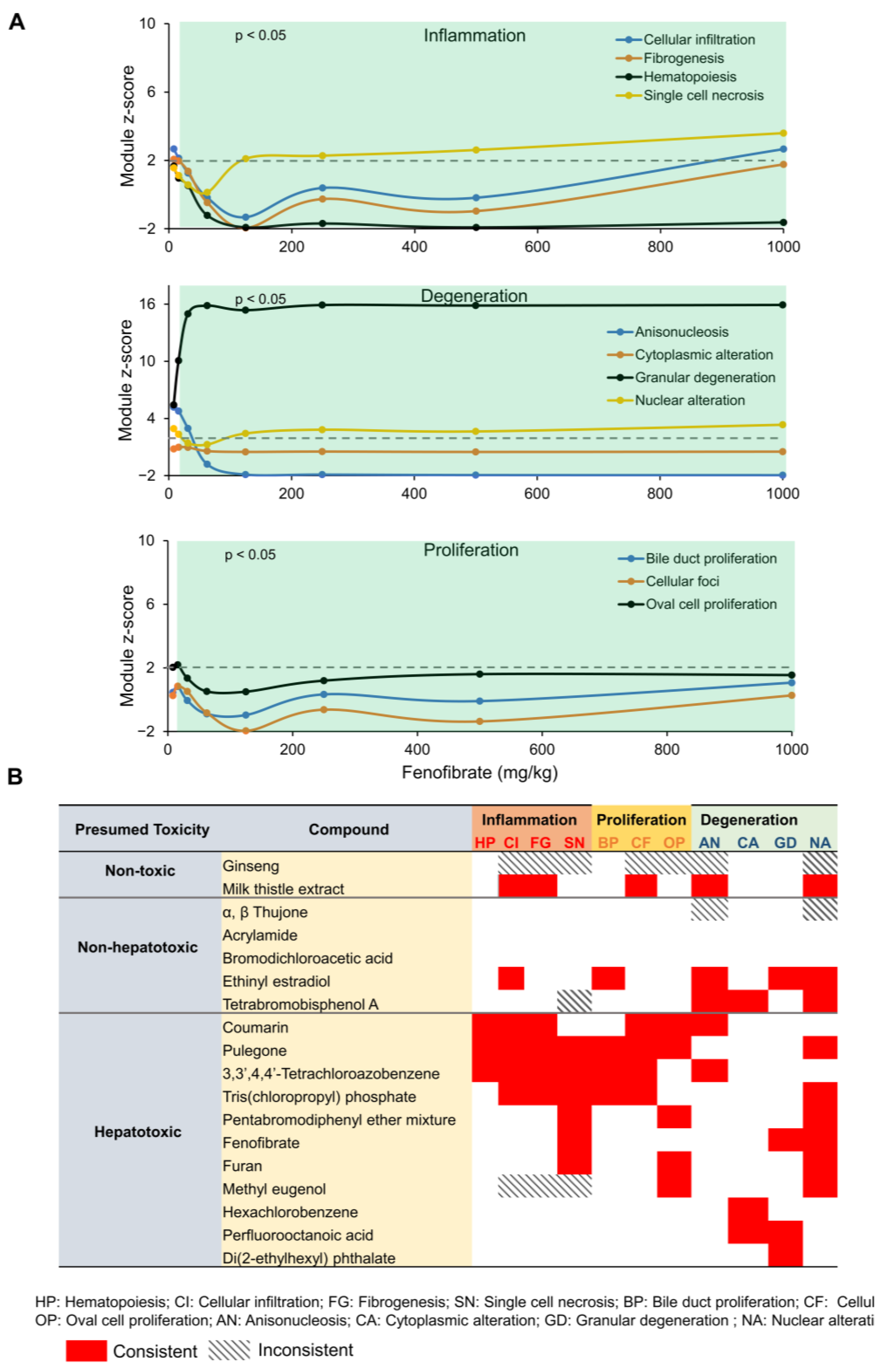
2.4. Toxicant-Induced Changes in Gene Expression Predict Liver Histopathological Phenotypes
3. Materials and Methods
3.1. Animals and Chemical Selection
3.2. Liver RNA Extraction and HTT Using the Rat S1500+
3.3. Gene Expression, KEGG Pathway, and Liver Injury Module Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waters, M.D.; Fostel, J.M. Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 2004, 5, 936–948. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; Auerbach, S.S.; Saddler, T.O.; Rice, J.R.; Dunlap, P.E.; Sipes, N.S.; DeVito, M.J.; Shah, R.R.; Bushel, P.R.; Merrick, B.A.; et al. The Power of Resolution: Contextualized Understanding of Biological Responses to Liver Injury Chemicals Using High-throughput Transcriptomics and Benchmark Concentration Modeling. Toxicol. Sci. 2019, 169, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Aardema, M.J.; MacGregor, J.T. Toxicology and genetic toxicology in the new era of “toxicogenomics”: Impact of “-omics” technologies. Mutat. Res. 2002, 499, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Kullak-Ublick, G.A.; Andrade, R.J.; Merz, M.; End, P.; Benesic, A.; Gerbes, A.L.; Aithal, G.P. Drug-induced liver injury: Recent advances in diagnosis and risk assessment. Gut 2017, 66, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Stine, J.G.; Chalasani, N.P. Drug Hepatotoxicity: Environmental Factors. Clin. Liver Dis. 2017, 21, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Kiyosawa, N.; Ando, Y.; Manabe, S.; Yamoto, T. Toxicogenomic biomarkers for liver toxicity. J. Toxicol. Pathol. 2009, 22, 35–52. [Google Scholar] [CrossRef]
- Lauschke, V.M. Toxicogenomics of drug induced liver injury—From mechanistic understanding to early prediction. Drug Metab. Rev. 2021, 53, 245–252. [Google Scholar] [CrossRef]
- Pannala, V.R.; Estes, S.K.; Rahim, M.; Trenary, I.; O’Brien, T.P.; Shiota, C.; Printz, R.L.; Reifman, J.; Shiota, M.; Young, J.D.; et al. Toxicant-Induced Metabolic Alterations in Lipid and Amino Acid Pathways Are Predictive of Acute Liver Toxicity in Rats. Int. J. Mol. Sci. 2020, 21, 8250. [Google Scholar] [CrossRef]
- Schyman, P.; Printz, R.L.; Pannala, V.R.; AbdulHameed, M.D.M.; Estes, S.K.; Shiota, C.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Genomics and metabolomics of early-stage thioacetamide-induced liver injury: An interspecies study between guinea pig and rat. Toxicol. Appl. Pharmacol. 2021, 430, 115713. [Google Scholar] [CrossRef]
- Judson, R.S.; Mortensen, H.M.; Shah, I.; Knudsen, T.B.; Elloumi, F. Using pathway modules as targets for assay development in xenobiotic screening. Mol. Biosyst. 2012, 8, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Mav, D.; Shah, R.R.; Howard, B.E.; Auerbach, S.S.; Bushel, P.R.; Collins, J.B.; Gerhold, D.L.; Judson, R.S.; Karmaus, A.L.; Maull, E.A.; et al. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS ONE 2018, 13, e0191105. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, W.M.; Auerbach, S.S.; Parham, F.; Stout, M.D.; Waidyanatha, S.; Mutlu, E.; Collins, B.; Paules, R.S.; Merrick, B.A.; Ferguson, S.; et al. Evaluation of 5-day In Vivo Rat Liver and Kidney with High-throughput Transcriptomics for Estimating Benchmark Doses of Apical Outcomes. Toxicol. Sci. 2020, 176, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Bushel, P.R.; Paules, R.S.; Auerbach, S.S. A Comparison of the TempO-Seq S1500+ Platform to RNA-Seq and Microarray Using Rat Liver Mode of Action Samples. Front. Genet. 2018, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Everett, L.J.; Mav, D.; Phadke, D.P.; Balik-Meisner, M.R.; Shah, R.R. Impact of Aligner, Normalization Method, and Sequencing Depth on TempO-seq Accuracy. Bioinform. Biol. Insights 2022, 16, 11779322221095216. [Google Scholar] [CrossRef]
- Schyman, P.; Xu, Z.; Desai, V.; Wallqvist, A. TOXPANEL: A Gene-Set Analysis Tool to Assess Liver and Kidney Injuries. Front. Pharmacol. 2021, 12, 601511. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar]
- Lo, E.K.K.; Felicianna; Xu, J.H.; Zhan, Q.; Zeng, Z.; El-Nezami, H. The Emerging Role of Branched-Chain Amino Acids in Liver Diseases. Biomedicines 2022, 10, 1444. [Google Scholar] [CrossRef]
- Pastore, A.; Alisi, A.; di Giovamberardino, G.; Crudele, A.; Ceccarelli, S.; Panera, N.; Dionisi-Vici, C.; Nobili, V. Plasma levels of homocysteine and cysteine increased in pediatric NAFLD and strongly correlated with severity of liver damage. Int. J. Mol. Sci. 2014, 15, 21202–21214. [Google Scholar] [CrossRef]
- Pannala, V.R.; Vinnakota, K.C.; Rawls, K.D.; Estes, S.K.; O’Brien, T.P.; Printz, R.L.; Papin, J.A.; Reifman, J.; Shiota, M.; Young, J.D.; et al. Mechanistic identification of biofluid metabolite changes as markers of acetaminophen-induced liver toxicity in rats. Toxicol. Appl. Pharmacol. 2019, 372, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Vairetti, M.; Di Pasqua, L.G.; Cagna, M.; Richelmi, P.; Ferrigno, A.; Berardo, C. Changes in Glutathione Content in Liver Diseases: An Update. Antioxidants 2021, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.; Leray, V.; Diez, M.; Serisier, S.; Le Bloc’h, J.; Siliart, B.; Dumon, H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008, 92, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Pei, K.; Gui, T.; Kan, D.; Feng, H.; Jin, Y.; Yang, Y.; Zhang, Q.; Du, Z.; Gai, Z.; Wu, J.; et al. An overview of lipid metabolism and nonalcoholic fatty liver disease. BioMed Res. Int. 2020, 2020, 4020249. [Google Scholar] [CrossRef]
- Tawa, G.J.; AbdulHameed, M.D.; Yu, X.; Kumar, K.; Ippolito, D.L.; Lewis, J.A.; Stallings, J.D.; Wallqvist, A. Characterization of chemically induced liver injuries using gene co-expression modules. PLoS ONE 2014, 9, e107230. [Google Scholar] [CrossRef]
- Uehara, T.; Ono, A.; Maruyama, T.; Kato, I.; Yamada, H.; Ohno, Y.; Urushidani, T. The Japanese toxicogenomics project: Application of toxicogenomics. Mol. Nutr. Food Res. 2010, 54, 218–227. [Google Scholar] [CrossRef]
- Zubrzycki, A.; Wronska, A.; Kotulak-Chrzaszcz, A.; Wierzbicki, P.M.; Kmiec, Z. Fenofibrate impairs liver function and structure more pronounced in old than young rats. Arch. Gerontol. Geriatr. 2020, 91, 104244. [Google Scholar] [CrossRef]
- Tamayo, C.; Diamond, S. Review of clinical trials evaluating safety and efficacy of milk thistle (Silybum marianum [L.] Gaertn.). Integr. Cancer Ther. 2007, 6, 146–157. [Google Scholar] [CrossRef]
- Hackett, E.S.; Twedt, D.C.; Gustafson, D.L. Milk thistle and its derivative compounds: A review of opportunities for treatment of liver disease. J. Vet. Intern. Med. 2013, 27, 10–16. [Google Scholar] [CrossRef]
- Pandey, G.; Pandey, S.P.; Sharma, M. Experimental hepatotoxicity produced by ethinyl estradiol. Toxicol. Int. 2011, 18, 160–162. [Google Scholar] [CrossRef][Green Version]
- Yao, L.; Wang, Y.; Shi, J.; Liu, Y.; Guo, H.; Yang, X.; Liu, Y.; Ma, J.; Li, D.; Wang, Z.; et al. Toxicity of tetrabromobisphenol A and its derivative in the mouse liver following oral exposure at environmentally relevant levels. Environ. Sci. Technol. 2021, 55, 8191–8202. [Google Scholar] [CrossRef]
- Tanaka, Y.; Fujii, W.; Hori, H.; Kitagawa, Y.; Ozaki, K. Relationship between coumarin-induced hepatocellular toxicity and mitochondrial function in rats. Food Chem. Toxicol. 2016, 90, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Silva, C.M.; Faustino-Rocha, A.I.; Gil da Costa, R.M.; Medeiros, R.; Pires, M.J.; Gaivao, I.; Gama, A.; Neuparth, M.J.; Barbosa, J.V.; Peixoto, F.; et al. Pulegone and eugenol oral supplementation in laboratory animals: Results from acute and chronic studies. Biomedicines 2022, 10, 2595. [Google Scholar] [CrossRef]
- van Birgelen, A.P.; Hebert, C.D.; Wenk, M.L.; Grimes, L.K.; Chapin, R.E.; Mahler, J.; Travlos, G.S.; Bucher, J.R. Toxicity of 3,3’,4,4’-tetrachloroazobenzene in rats and mice. Toxicol. Appl. Pharmacol. 1999, 156, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Dunnick, J.K.; Pandiri, A.R.; Merrick, B.A.; Kissling, G.E.; Cunny, H.; Mutlu, E.; Waidyanatha, S.; Sills, R.; Hong, H.L.; Ton, T.V.; et al. Carcinogenic activity of pentabrominated diphenyl ether mixture (DE-71) in rats and mice. Toxicol. Rep. 2018, 5, 615–624. [Google Scholar] [CrossRef]
- Giribaldi, L.; Chiappini, F.; Pontillo, C.; Randi, A.S.; Kleiman de Pisarev, D.L.; Alvarez, L. Hexachlorobenzene induces deregulation of cellular growth in rat liver. Toxicology 2011, 289, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Thorup, I.; Wurtzen, G.; Carstensen, J.; Olsen, P. Short term toxicity study in rats dosed with pulegone and menthol. Toxicol. Lett. 1983, 19, 207–210. [Google Scholar] [CrossRef] [PubMed]
- NTP Technical Report on the Toxicity Studies of 3,3’,4,4’-Tetrachloroazobenzene (CAS No. 14047-09-7) Administered by Gavage to F344/N rats and B6C3F1 mice. Toxic Rep. Ser. 1998, 65, 1-F6.
- Moser, G.J.; Foley, J.; Burnett, M.; Goldsworthy, T.L.; Maronpot, R. Furan-induced dose-response relationships for liver cytotoxicity, cell proliferation, and tumorigenicity (furan-induced liver tumorigenicity). Exp. Toxicol. Pathol. 2009, 61, 101–111. [Google Scholar] [CrossRef]
- Dong, H.; Gill, S.; Curran, I.H.; Williams, A.; Kuo, B.; Wade, M.G.; Yauk, C.L. Toxicogenomic assessment of liver responses following subchronic exposure to furan in Fischer F344 rats. Arch. Toxicol. 2016, 90, 1351–1367. [Google Scholar] [CrossRef]
- National Toxicology Program. NTP Toxicology and Carcinogenesis Studies of Methyleugenol (CAS NO. 93-15-2) in F344/N rats and B6C3F1 mice (Gavage Studies). Natl. Toxicol. Program Tech. Rep. Ser. 2000, 491, 1–412. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T. A Note on the Use of Principal Components in Regression. J. R. Stat. Soc. Ser. C Appl. Statist. 2018, 31, 300–303. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Woo, H.J.; Yu, X.; Oyama, T.; Wallqvist, A.; Reifman, J. A strategy for evaluating pathway analysis methods. BMC Bioinform. 2017, 18, 453. [Google Scholar] [CrossRef] [PubMed]
- Schyman, P.; Printz, R.L.; Estes, S.K.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Identification of the toxicity pathways associated with thioacetamide-induced injuries in rat liver and kidney. Front. Pharmacol. 2018, 9, 1272. [Google Scholar] [CrossRef]
- Liss, K.H.; Finck, B.N. PPARs and nonalcoholic fatty liver disease. Biochimie 2017, 136, 65–74. [Google Scholar] [CrossRef]
- Qiu, Y.Y.; Zhang, J.; Zeng, F.Y.; Zhu, Y.Z. Roles of the peroxisome proliferator-activated receptors (PPARs) in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). Pharmacol. Res. 2023, 192, 106786. [Google Scholar] [CrossRef]
- Negi, C.K.; Khan, S.; Dirven, H.; Bajard, L.; Blaha, L. Flame Retardants-Mediated Interferon Signaling in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2021, 22, 4282. [Google Scholar] [CrossRef]
| Chemical | Gavage Vehicle | Concentrations * | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| ACR | Deionized water | 0.078 | 0.156 | 0.3125 | 0.625 | 1.25 | 2.5 | 5 | 10 | |
| BDCA | Deionized water | 1.25 | 2.5 | 5 | 10 | 20 | 40 | 80 | 160 | |
| COU | Corn oil | 3.125 | 6.25 | 12.5 | 25 | 50 | 100 | 20 | 400 | |
| DEHP | Corn oil | 8 | 16 | 31.25 | 62.5 | 125 | 250 | 500 | 1000 | |
| DE71 | Corn oil | 0.38 | 0.75 | 1.5 | 3 | 15 | 50 | 100 | 200 | 500 |
| EE2 | Corn oil | 0.02 | 0.067 | 0.2 | 0.6 | 1.8 | 5.4 | 16.2 | 48.6 | |
| FEN | 0.5% aqueous methylcellulose | 8 | 16 | 31.25 | 62.5 | 125 | 250 | 500 | 1000 | |
| FUR | Corn oil | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | |
| GIN | Deionized water | 39.1 | 78.125 | 156.25 | 312.5 | 625 | 1250 | 2500 | 5000 | |
| HCB | Corn oil | 0.004 | 0.015 | 0.0625 | 0.25 | 1 | 4 | 16 | 64 | |
| MET | 0.5% aqueous methylcellulose | 4.625 | 9.25 | 18.5 | 37 | 75 | 150 | 300 | 600 | |
| MTE | Corn oil | 39.1 | 78.125 | 156.25 | 312.5 | 625 | 937.5 | 1250 | 1750 | |
| PFOA | 2% Tween 80 | 0.156 | 0.3125 | 0.625 | 1.25 | 2.5 | 5 | 10 | 20 | |
| PUL | Corn oil | 2.4 | 4.7 | 9.4 | 18.75 | 37.5 | 75 | 150 | 300 | |
| TBBPA | Corn oil | 4 | 8 | 16 | 125 | 250 | 500 | 1000 | 2000 | |
| TCAB | Corn oil/acetone (99:1) | 0.1 | 0.3 | 1 | 3 | 10 | 30 | 100 | 200 | 400 |
| TCPP | 0.5% aqueous methylcellulose | 18.75 | 37.5 | 75 | 150 | 300 | 600 | 1000 | 2000 | |
| THU | 0.5% aqueous methylcellulose | 1.5 | 3 | 6.25 | 12.5 | 25 | 50 | 100 | 200 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pannala, V.R.; Balik-Meisner, M.R.; Mav, D.; Phadke, D.P.; Scholl, E.H.; Shah, R.R.; Auerbach, S.S.; Wallqvist, A. High-Throughput Transcriptomics Differentiates Toxic versus Non-Toxic Chemical Exposures Using a Rat Liver Model. Int. J. Mol. Sci. 2023, 24, 17425. https://doi.org/10.3390/ijms242417425
Pannala VR, Balik-Meisner MR, Mav D, Phadke DP, Scholl EH, Shah RR, Auerbach SS, Wallqvist A. High-Throughput Transcriptomics Differentiates Toxic versus Non-Toxic Chemical Exposures Using a Rat Liver Model. International Journal of Molecular Sciences. 2023; 24(24):17425. https://doi.org/10.3390/ijms242417425
Chicago/Turabian StylePannala, Venkat R., Michele R. Balik-Meisner, Deepak Mav, Dhiral P. Phadke, Elizabeth H. Scholl, Ruchir R. Shah, Scott S. Auerbach, and Anders Wallqvist. 2023. "High-Throughput Transcriptomics Differentiates Toxic versus Non-Toxic Chemical Exposures Using a Rat Liver Model" International Journal of Molecular Sciences 24, no. 24: 17425. https://doi.org/10.3390/ijms242417425
APA StylePannala, V. R., Balik-Meisner, M. R., Mav, D., Phadke, D. P., Scholl, E. H., Shah, R. R., Auerbach, S. S., & Wallqvist, A. (2023). High-Throughput Transcriptomics Differentiates Toxic versus Non-Toxic Chemical Exposures Using a Rat Liver Model. International Journal of Molecular Sciences, 24(24), 17425. https://doi.org/10.3390/ijms242417425






