Circadian Regulation of Apolipoproteins in the Brain: Implications in Lipid Metabolism and Disease
Abstract
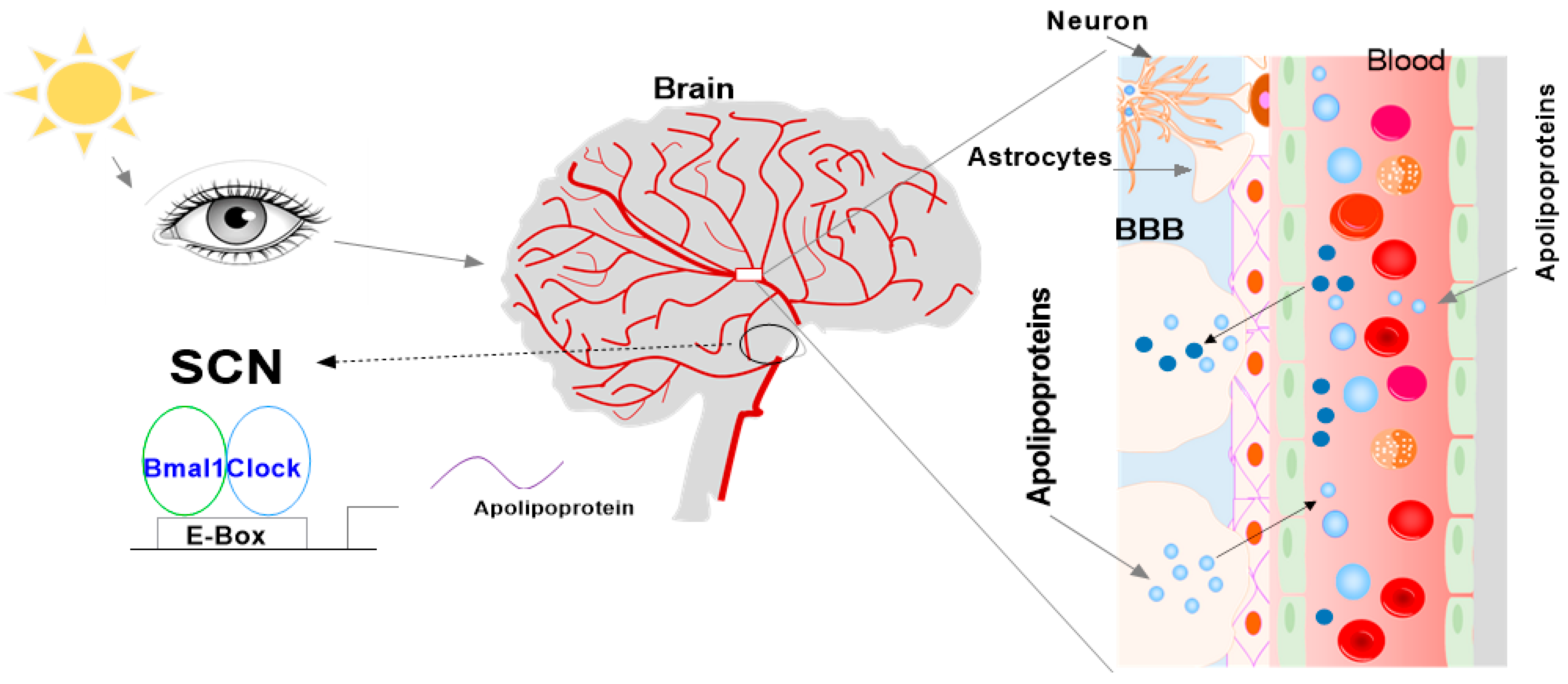
:1. Introduction
2. Function of SCN
2.1. SCN
2.2. Neurons and Hormones in the SCN
2.3. Immune Factors in the SCN
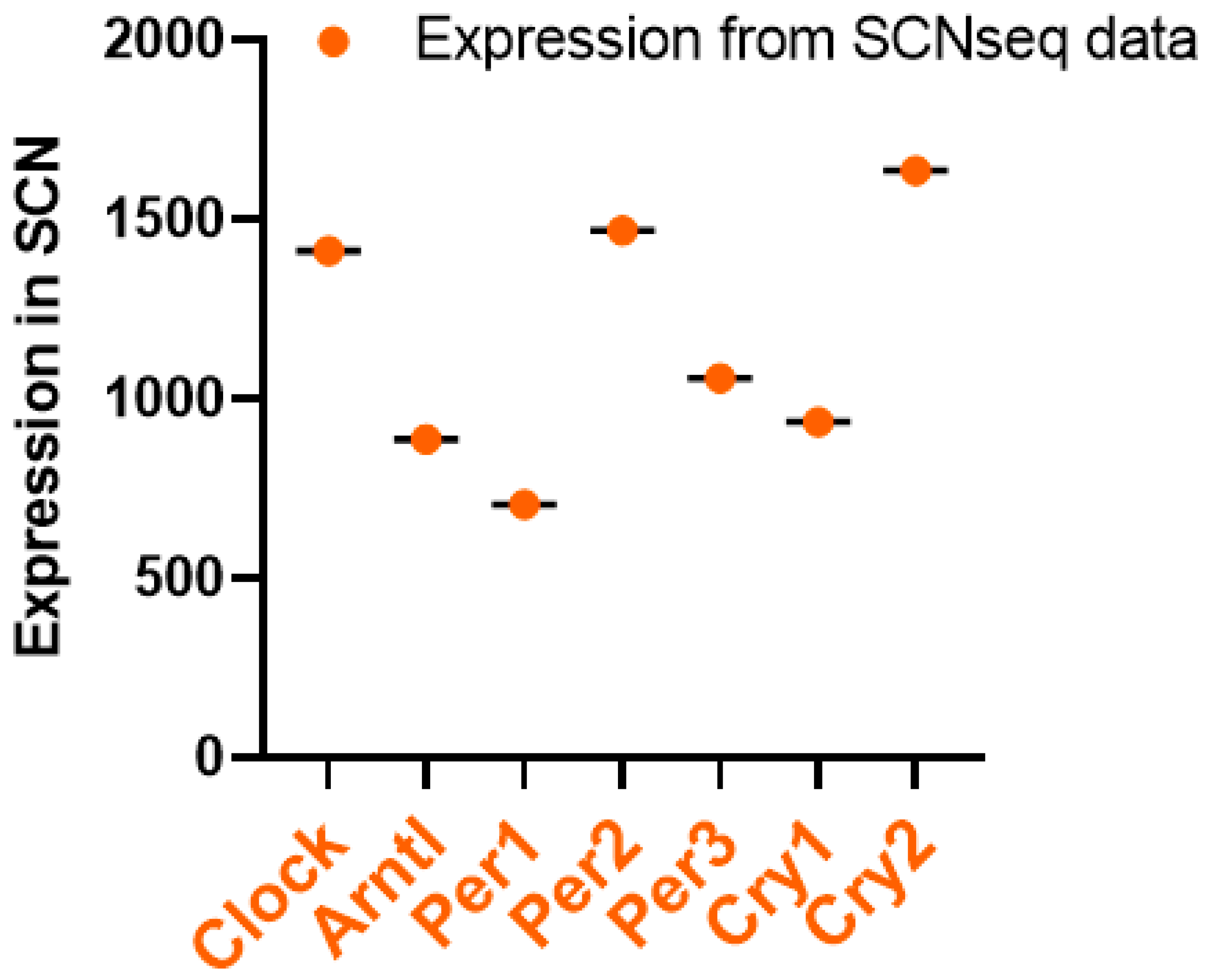
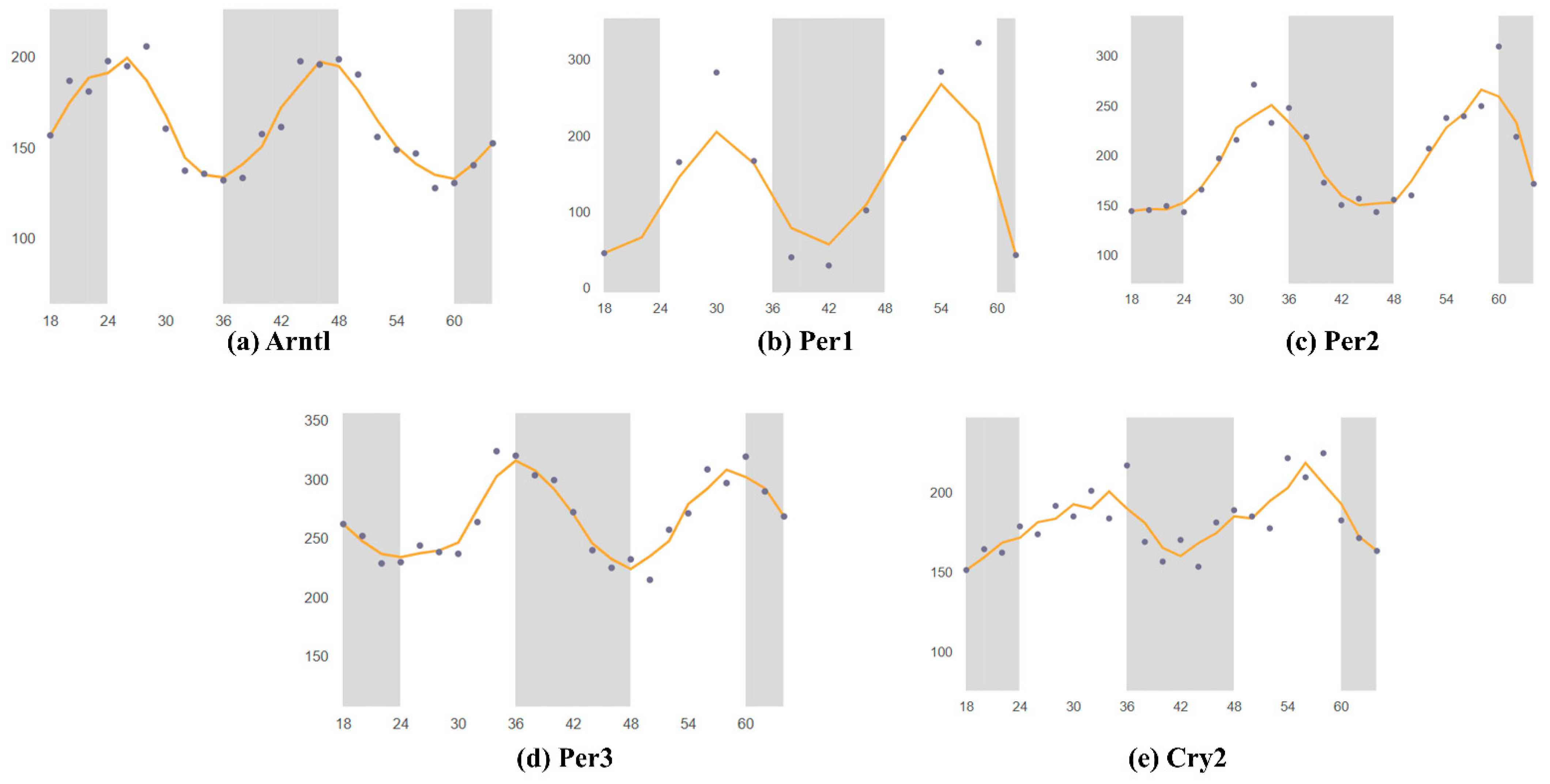
3. Circadian Rhythm of Circadian Clock Genes in the SCN
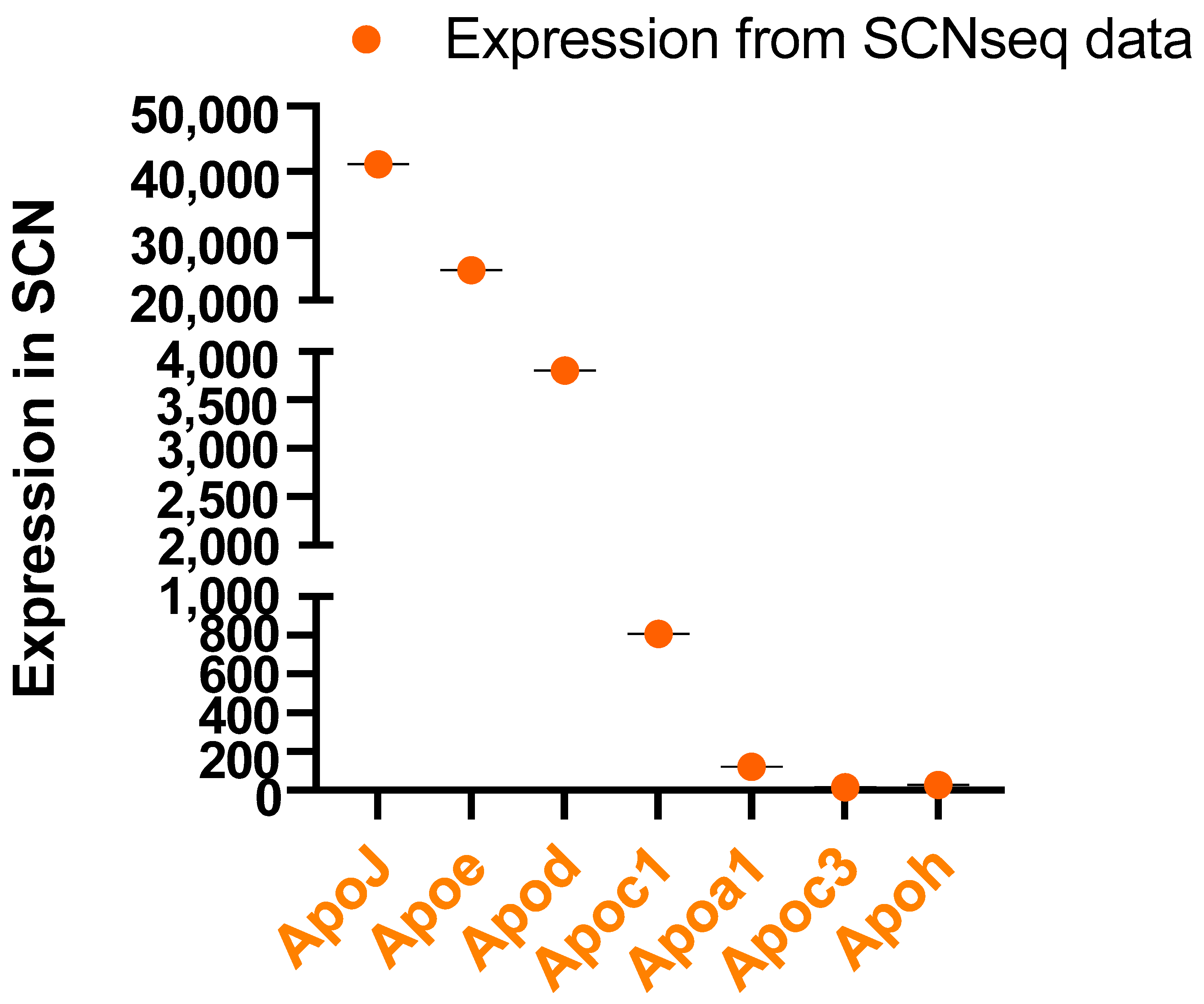
4. Apolipoproteins in the SCN and CSF
5. Circadian Rhythm of Apolipoprotein Genes in Mouse SCN
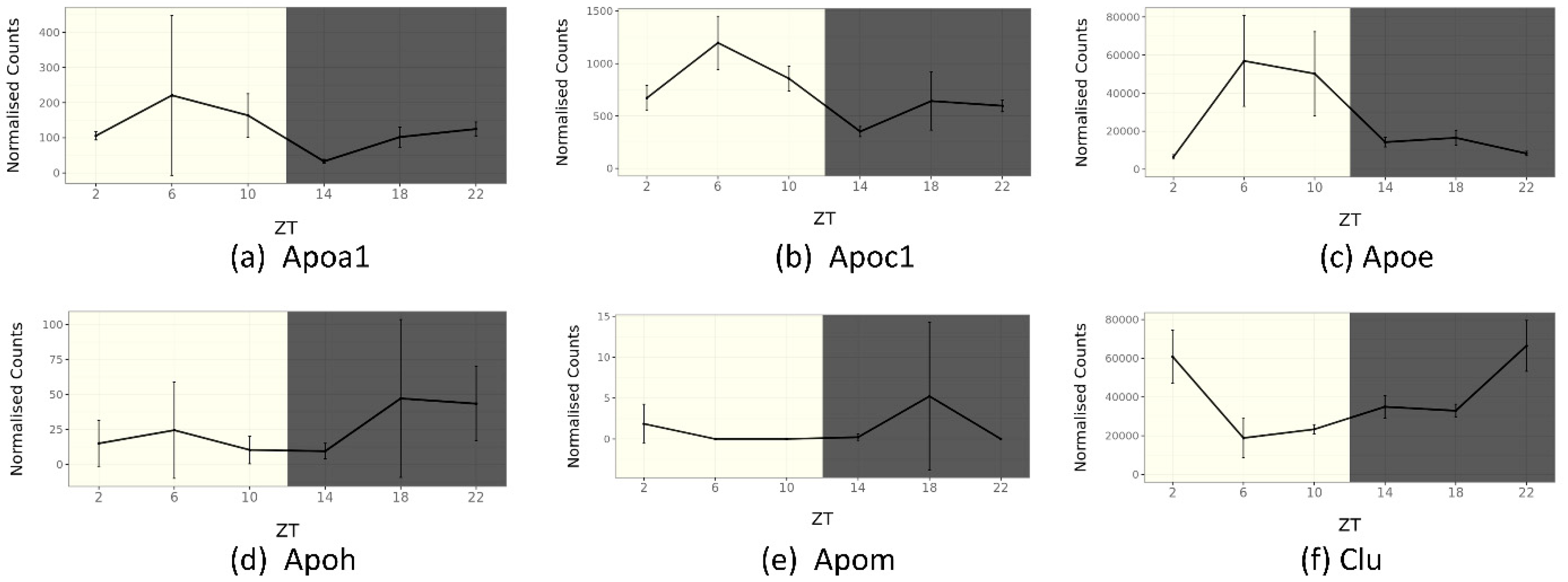
5.1. ApoA1
5.2. ApoA4
5.3. ApoB
5.4. ApoD
5.5. ApoE
5.6. ApoC1
5.7. ApoH
5.8. ApoJ
5.9. ApoM
6. Apolipoproteins and Diseases
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef]
- Green, C.B.; Takahashi, J.S.; Bass, J. The meter of metabolism. Cell 2008, 134, 728–742. [Google Scholar] [CrossRef]
- Bass, J.T. The circadian clock system’s influence in health and disease. Genome Med. 2017, 9, 94. [Google Scholar] [CrossRef]
- Lananna, B.V.; Musiek, E.S. The wrinkling of time: Aging, inflammation, oxidative stress, and the circadian clock in neurodegeneration. Neurobiol. Dis. 2020, 139, 104832. [Google Scholar] [CrossRef]
- Prasai, M.J.; George, J.T.; Scott, E.M. Molecular clocks, type 2 diabetes and cardiovascular disease. Diab Vasc. Dis. Res. 2008, 5, 89–95. [Google Scholar] [CrossRef]
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ. Res. 2010, 106, 447–462. [Google Scholar] [CrossRef]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Pan, X.; Mota, S.; Zhang, B. Circadian Clock Regulation on Lipid Metabolism and Metabolic Diseases. Adv. Exp. Med. Biol. 2020, 1276, 53–66. [Google Scholar] [CrossRef]
- Nassan, M.; Videnovic, A. Circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 2022, 18, 7–24. [Google Scholar] [CrossRef]
- Zhang, D.; Pollock, D.M. Circadian regulation of kidney function: Finding a role for Bmal1. Am. J. Physiol. Renal Physiol. 2018, 314, F675–F678. [Google Scholar] [CrossRef]
- Levi, F.; Schibler, U. Circadian rhythms: Mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 593–628. [Google Scholar] [CrossRef] [PubMed]
- Gooley, J.J. Circadian regulation of lipid metabolism. Proc. Nutr. Soc. 2016, 75, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Rana, J.N.; Choi, E.H.; Han, I. Microwave Radiation and the Brain: Mechanisms, Current Status, and Future Prospects. Int. J. Mol. Sci. 2022, 23, 9288. [Google Scholar] [CrossRef] [PubMed]
- Wardzinski, E.K.; Jauch-Chara, K.; Haars, S.; Melchert, U.H.; Scholand-Engler, H.G.; Oltmanns, K.M. Mobile Phone Radiation Deflects Brain Energy Homeostasis and Prompts Human Food Ingestion. Nutrients 2022, 14, 339. [Google Scholar] [CrossRef]
- Inskip, P.D.; Hoover, R.N.; Devesa, S.S. Brain cancer incidence trends in relation to cellular telephone use in the United States. Neuro Oncol. 2010, 12, 1147–1151. [Google Scholar] [CrossRef]
- Shimba, S.; Ishii, N.; Ohta, Y.; Ohno, T.; Watabe, Y.; Hayashi, M.; Wada, T.; Aoyagi, T.; Tezuka, M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 12071–12076. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, J.; Jiang, T.; Monslow, J.; Tang, S.Y.; Todd, L.; Pure, E.; Chen, L.; FitzGerald, G.A. Bmal1 Deletion in Myeloid Cells Attenuates Atherosclerotic Lesion Development and Restrains Abdominal Aortic Aneurysm Formation in Hyperlipidemic Mice. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1523–1532. [Google Scholar] [CrossRef]
- Douris, N.; Kojima, S.; Pan, X.; Lerch-Gaggl, A.F.; Duong, S.Q.; Hussain, M.M.; Green, C.B. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr. Biol. 2011, 21, 1347–1355. [Google Scholar] [CrossRef]
- Pan, X.; Hussain, M.M. Clock is important for food and circadian regulation of macronutrient absorption in mice. J. Lipid Res. 2009, 50, 1800–1813. [Google Scholar] [CrossRef]
- Gomez-Santos, C.; Gomez-Abellan, P.; Madrid, J.A.; Hernandez-Morante, J.J.; Lujan, J.A.; Ordovas, J.M.; Garaulet, M. Circadian rhythm of clock genes in human adipose explants. Obesity 2009, 17, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Hussain, M.M. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J. Biol. Chem. 2007, 282, 24707–24719. [Google Scholar] [CrossRef] [PubMed]
- Chua, E.C.; Shui, G.; Lee, I.T.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef]
- Brown, S.A. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol. Metab. 2016, 27, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.M.; Pan, X. Circadian regulators of intestinal lipid absorption. J. Lipid Res. 2015, 56, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Bradfield, C.A.; Hussain, M.M. Global and hepatocyte-specific ablation of Bmal1 induces hyperlipidaemia and enhances atherosclerosis. Nat. Commun. 2016, 7, 13011. [Google Scholar] [CrossRef]
- Cheng, B.; Anea, C.B.; Yao, L.; Chen, F.; Patel, V.; Merloiu, A.; Pati, P.; Caldwell, R.W.; Fulton, D.J.; Rudic, R.D. Tissue-intrinsic dysfunction of circadian clock confers transplant arteriosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 17147–17152. [Google Scholar] [CrossRef]
- Lee, J.; Dimitry, J.M.; Song, J.H.; Son, M.; Sheehan, P.W.; King, M.W.; Travis Tabor, G.; Goo, Y.A.; Lazar, M.A.; Petrucelli, L.; et al. Microglial REV-ERBalpha regulates inflammation and lipid droplet formation to drive tauopathy in male mice. Nat. Commun. 2023, 14, 5197, Correction to Nat. Commun. 2023, 14, 7760. [Google Scholar] [CrossRef]
- Bass, J.; Lazar, M.A. Circadian time signatures of fitness and disease. Science 2016, 354, 994–999. [Google Scholar] [CrossRef]
- Muenchhoff, J.; Song, F.; Poljak, A.; Crawford, J.D.; Mather, K.A.; Kochan, N.A.; Yang, Z.; Trollor, J.N.; Reppermund, S.; Maston, K.; et al. Plasma apolipoproteins and physical and cognitive health in very old individuals. Neurobiol. Aging 2017, 55, 49–60. [Google Scholar] [CrossRef]
- Pan, X.; Zhang, Y.; Wang, L.; Hussain, M.M. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010, 12, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Jiang, X.C.; Hussain, M.M. Impaired cholesterol metabolism and enhanced atherosclerosis in clock mutant mice. Circulation 2013, 128, 1758–1769. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Hussain, M.M. Bmal1 regulates production of larger lipoproteins by modulating cAMP-responsive element-binding protein H and apolipoprotein AIV. Hepatology 2022, 76, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Munshi, M.K.; Iqbal, J.; Queiroz, J.; Sirwi, A.A.; Shah, S.; Younus, A.; Hussain, M.M. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J. Biol. Chem. 2013, 288, 20464–20476. [Google Scholar] [CrossRef]
- Martins, I.J.; Hone, E.; Foster, J.K.; Sunram-Lea, S.I.; Gnjec, A.; Fuller, S.J.; Nolan, D.; Gandy, S.E.; Martins, R.N. Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer’s disease and cardiovascular disease. Mol. Psychiatry 2006, 11, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Dufouil, C.; Richard, F.; Fievet, N.; Dartigues, J.F.; Ritchie, K.; Tzourio, C.; Amouyel, P.; Alperovitch, A. APOE genotype, cholesterol level, lipid-lowering treatment, and dementia: The Three-City Study. Neurology 2005, 64, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Sparks, D.L.; Sabbagh, M.N.; Connor, D.J.; Lopez, J.; Launer, L.J.; Browne, P.; Wasser, D.; Johnson-Traver, S.; Lochhead, J.; Ziolwolski, C. Atorvastatin for the treatment of mild to moderate Alzheimer disease: Preliminary results. Arch. Neurol. 2005, 62, 753–757. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676. [Google Scholar] [CrossRef]
- Hussain, M.M.; Pan, X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol. Metab. 2009, 20, 177–185. [Google Scholar] [CrossRef]
- Froy, O. The circadian clock and metabolism. Clin. Sci. 2011, 120, 65–72. [Google Scholar] [CrossRef]
- Schnell, A.; Chappuis, S.; Schmutz, I.; Brai, E.; Ripperger, J.A.; Schaad, O.; Welzl, H.; Descombes, P.; Alberi, L.; Albrecht, U. The nuclear receptor REV-ERBalpha regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS ONE 2014, 9, e99883. [Google Scholar] [CrossRef]
- Landgraf, D.; Long, J.E.; Proulx, C.D.; Barandas, R.; Malinow, R.; Welsh, D.K. Genetic Disruption of Circadian Rhythms in the Suprachiasmatic Nucleus Causes Helplessness, Behavioral Despair, and Anxiety-like Behavior in Mice. Biol. Psychiatry 2016, 80, 827–835. [Google Scholar] [CrossRef]
- Wolff, S.E.C.; Wang, X.L.; Jiao, H.; Sun, J.; Kalsbeek, A.; Yi, C.X.; Gao, Y. The Effect of Rev-erbalpha Agonist SR9011 on the Immune Response and Cell Metabolism of Microglia. Front. Immunol. 2020, 11, 550145. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.A.; Lee, J.; Cai, Y.; Saito, T.; Saido, T.; Musiek, E.S. Astrocytes deficient in circadian clock gene Bmal1 show enhanced activation responses to amyloid-beta pathology without changing plaque burden. Sci. Rep. 2022, 12, 1796. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.E.; Griffin, P.; Sheehan, P.W.; Kim, D.H.; Musiek, E.S.; Yoon, S.Y. Inhibition of REV-ERBs stimulates microglial amyloid-beta clearance and reduces amyloid plaque deposition in the 5XFAD mouse model of Alzheimer’s disease. Aging Cell 2020, 19, e13078. [Google Scholar] [CrossRef] [PubMed]
- Hofman, A.; Ott, A.; Breteler, M.M.; Bots, M.L.; Slooter, A.J.; van Harskamp, F.; van Duijn, C.N.; Van Broeckhoven, C.; Grobbee, D.E. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet 1997, 349, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Wingo, T.S.; Cutler, D.J.; Wingo, A.P.; Le, N.A.; Rabinovici, G.D.; Miller, B.L.; Lah, J.J.; Levey, A.I. Association of Early-Onset Alzheimer Disease With Elevated Low-Density Lipoprotein Cholesterol Levels and Rare Genetic Coding Variants of APOB. JAMA Neurol. 2019, 76, 809–817. [Google Scholar] [CrossRef]
- Marchant, N.L.; Reed, B.R.; Sanossian, N.; Madison, C.M.; Kriger, S.; Dhada, R.; Mack, W.J.; DeCarli, C.; Weiner, M.W.; Mungas, D.M.; et al. The aging brain and cognition: Contribution of vascular injury and abeta to mild cognitive dysfunction. JAMA Neurol. 2013, 70, 488–495. [Google Scholar] [CrossRef]
- Power, M.C.; Rawlings, A.; Sharrett, A.R.; Bandeen-Roche, K.; Coresh, J.; Ballantyne, C.M.; Pokharel, Y.; Michos, E.D.; Penman, A.; Alonso, A.; et al. Association of midlife lipids with 20-year cognitive change: A cohort study. Alzheimers Dement. 2018, 14, 167–177. [Google Scholar] [CrossRef]
- Blue, M.L.; Williams, D.L.; Zucker, S.; Khan, S.A.; Blum, C.B. Apolipoprotein E synthesis in human kidney, adrenal gland, and liver. Proc. Natl. Acad. Sci. USA 1983, 80, 283–287. [Google Scholar] [CrossRef]
- Wang, H.; Eckel, R.H. What are lipoproteins doing in the brain? Trends Endocrinol. Metab. 2014, 25, 8–14. [Google Scholar] [CrossRef]
- Fuior, E.V.; Gafencu, A.V. Apolipoprotein C1: Its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci. 2019, 20, 5939. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Earnest, D.J.; Olschowka, J.A. Circadian regulation of c-fos expression in the suprachiasmatic pacemaker by light. J. Biol. Rhythms 1993, 8, S65–S71. [Google Scholar] [PubMed]
- Brancaccio, M.; Enoki, R.; Mazuski, C.N.; Jones, J.; Evans, J.A.; Azzi, A. Network-mediated encoding of circadian time: The suprachiasmatic nucleus (SCN) from genes to neurons to circuits, and back. J. Neurosci. 2014, 34, 15192–15199. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Ma, D.; Zhao, M.; Xie, L.; Wu, Q.; Gou, L.; Zhu, C.; Fan, Y.; Wang, H.; Yan, J. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat. Neurosci. 2020, 23, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Barca-Mayo, O.; Pons-Espinal, M.; Follert, P.; Armirotti, A.; Berdondini, L.; De Pietri Tonelli, D. Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 2017, 8, 14336. [Google Scholar] [CrossRef] [PubMed]
- Tso, C.F.; Simon, T.; Greenlaw, A.C.; Puri, T.; Mieda, M.; Herzog, E.D. Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr. Biol. 2017, 27, 1055–1061. [Google Scholar] [CrossRef]
- Brancaccio, M.; Patton, A.P.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 2017, 93, 1420–1435.e1425. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Edwards, M.D.; Patton, A.P.; Smyllie, N.J.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 2019, 363, 187–192. [Google Scholar] [CrossRef]
- Granados-Fuentes, D.; Prolo, L.M.; Abraham, U.; Herzog, E.D. The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J. Neurosci. 2004, 24, 615–619. [Google Scholar] [CrossRef]
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978. [Google Scholar] [CrossRef]
- Lehman, M.N.; Silver, R.; Gladstone, W.R.; Kahn, R.M.; Gibson, M.; Bittman, E.L. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci. 1987, 7, 1626–1638. [Google Scholar] [CrossRef]
- Harmar, A.J.; Marston, H.M.; Shen, S.; Spratt, C.; West, K.M.; Sheward, W.J.; Morrison, C.F.; Dorin, J.R.; Piggins, H.D.; Reubi, J.C.; et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002, 109, 497–508. [Google Scholar] [CrossRef]
- Welsh, D.K.; Takahashi, J.S.; Kay, S.A. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 2010, 72, 551–577. [Google Scholar] [CrossRef]
- Pittman, Q.J. Vasopressin and central control of the cardiovascular system: A 40-year retrospective. J. Neuroendocrinol. 2021, 33, e13011. [Google Scholar] [CrossRef]
- Smith, M.C.; Dunn, M.J. The role of prostaglandins in human hypertension. Am. J. Kidney Dis. 1985, 5, A32–A39. [Google Scholar] [CrossRef]
- Reghunandanan, V. Vasopressin in circadian function of SCN. J. Biosci. 2020, 45, 140. [Google Scholar] [CrossRef]
- Olszewski, P.K.; Wirth, M.M.; Shaw, T.J.; Grace, M.K.; Levine, A.S. Peptides that regulate food intake: Effect of peptide histidine isoleucine on consummatory behavior in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R1445–R1453. [Google Scholar] [CrossRef]
- Shinohara, K.; Funabashi, T.; Mitushima, D.; Kimura, F. Effects of gap junction blocker on vasopressin and vasoactive intestinal polypeptide rhythms in the rat suprachiasmatic nucleus in vitro. Neurosci. Res. 2000, 38, 43–47. [Google Scholar] [CrossRef]
- Kruijver, F.P.; Swaab, D.F. Sex hormone receptors are present in the human suprachiasmatic nucleus. Neuroendocrinology 2002, 75, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.; Silver, R. Sex differences in circadian timing systems: Implications for disease. Front. Neuroendocrinol. 2014, 35, 111–139. [Google Scholar] [CrossRef] [PubMed]
- Alvord, V.M.; Kantra, E.J.; Pendergast, J.S. Estrogens and the circadian system. Semin. Cell Dev. Biol. 2022, 126, 56–65. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.R.; Howarth, C.; Sibson, N.R. The role of astrocytes in CNS tumors: Pre-clinical models and novel imaging approaches. Front. Cell. Neurosci. 2013, 7, 40. [Google Scholar] [CrossRef]
- Brierley, D.I.; Holt, M.K.; Singh, A.; de Araujo, A.; McDougle, M.; Vergara, M.; Afaghani, M.H.; Lee, S.J.; Scott, K.; Maske, C.; et al. Central and peripheral GLP-1 systems independently suppress eating. Nat. Metab. 2021, 3, 258–273. [Google Scholar] [CrossRef]
- Rowlands, J.; Heng, J.; Newsholme, P.; Carlessi, R. Pleiotropic Effects of GLP-1 and Analogs on Cell Signaling, Metabolism, and Function. Front. Endocrinol. 2018, 9, 672. [Google Scholar] [CrossRef]
- Nauck, M.A.; Meier, J.J. GLP-1 receptor agonists and SGLT2 inhibitors: A couple at last? Lancet Diabetes Endocrinol. 2016, 4, 963–964. [Google Scholar] [CrossRef]
- Hussein, H.; Zaccardi, F.; Khunti, K.; Davies, M.J.; Patsko, E.; Dhalwani, N.N.; Kloecker, D.E.; Ioannidou, E.; Gray, L.J. Efficacy and tolerability of sodium-glucose co-transporter-2 inhibitors and glucagon-like peptide-1 receptor agonists: A systematic review and network meta-analysis. Diabetes Obes. Metab. 2020, 22, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.J.; Marpegan, L.; Bekinschtein, T.A.; Costas, M.A.; Golombek, D.A. Suprachiasmatic astrocytes as an interface for immune-circadian signalling. J. Neurosci. Res. 2006, 84, 1521–1527. [Google Scholar] [CrossRef] [PubMed]
- Hainich, E.C.; Pizzio, G.A.; Golombek, D.A. Constitutive activation of the ERK-MAPK pathway in the suprachiasmatic nuclei inhibits circadian resetting. FEBS Lett. 2006, 580, 6665–6668. [Google Scholar] [CrossRef]
- Liu, X.; Quan, N. Microglia and CNS Interleukin-1: Beyond Immunological Concepts. Front. Neurol. 2018, 9, 8. [Google Scholar] [CrossRef]
- Lundkvist, G.B.; Hill, R.H.; Kristensson, K. Disruption of circadian rhythms in synaptic activity of the suprachiasmatic nuclei by African trypanosomes and cytokines. Neurobiol. Dis. 2002, 11, 20–27. [Google Scholar] [CrossRef]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Musiek, E.S.; Hu, K.; Cappuccio, F.P.; Yaffe, K. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol. 2019, 18, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Musiek, E.S.; Holtzman, D.M. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 2016, 354, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Kress, G.J.; Liao, F.; Dimitry, J.; Cedeno, M.R.; FitzGerald, G.A.; Holtzman, D.M.; Musiek, E.S. Regulation of amyloid-beta dynamics and pathology by the circadian clock. J. Exp. Med. 2018, 215, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, P.W.; Nadarajah, C.J.; Kanan, M.F.; Patterson, J.N.; Novotny, B.; Lawrence, J.H.; King, M.W.; Brase, L.; Inman, C.E.; Yuede, C.M.; et al. An astrocyte BMAL1-BAG3 axis protects against alpha-synuclein and tau pathology. Neuron 2023, 111, 2383–2398.e7. [Google Scholar] [CrossRef] [PubMed]
- Spisska, V.; Pacesova, D.; Mikova, H.; Pohanova, P.; Telensky, P.; Novotny, J.; Bendova, Z. Prenatal exposure to lipopolysaccharide induces changes in the circadian clock in the SCN and AA-NAT activity in the pineal gland. Brain Res. 2020, 1743, 146952. [Google Scholar] [CrossRef]
- Huo, M.; Huang, Y.; Qu, D.; Zhang, H.; Wong, W.T.; Chawla, A.; Huang, Y.; Tian, X.Y. Myeloid Bmal1 deletion increases monocyte recruitment and worsens atherosclerosis. FASEB J. 2017, 31, 1097–1106. [Google Scholar] [CrossRef]
- Ma, Z.; Jiang, W.; Zhang, E.E. Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci. Rep. 2016, 6, 36035. [Google Scholar] [CrossRef]
- Chen, Q.; Peng, X.D.; Huang, C.Q.; Hu, X.Y.; Zhang, X.M. Association between ARNTL (BMAL1) rs2278749 polymorphism T >C and susceptibility to Alzheimer disease in a Chinese population. Genet. Mol. Res. 2015, 14, 18515–18522. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Ogawa, T.; Hitosugi, S.; Ichihashi, Y.; Nakadaira, Y.; Kobayashi, M.; Tezuka, M.; Kosuge, Y.; Ishige, K.; Ito, Y.; et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE 2011, 6, e25231. [Google Scholar] [CrossRef]
- Nikolaeva, S.; Ansermet, C.; Centeno, G.; Pradervand, S.; Bize, V.; Mordasini, D.; Henry, H.; Koesters, R.; Maillard, M.; Bonny, O.; et al. Nephron-Specific Deletion of Circadian Clock Gene Bmal1 Alters the Plasma and Renal Metabolome and Impairs Drug Disposition. J. Am. Soc. Nephrol. 2016, 27, 2997–3004. [Google Scholar] [CrossRef] [PubMed]
- Vitali, C.; Wellington, C.L.; Calabresi, L. HDL and cholesterol handling in the brain. Cardiovasc. Res. 2014, 103, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R. Introduction to Lipids and Lipoproteins. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Koch, S.; Donarski, N.; Goetze, K.; Kreckel, M.; Stuerenburg, H.J.; Buhmann, C.; Beisiegel, U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J. Lipid Res. 2001, 42, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Borghini, I.; Barja, F.; Pometta, D.; James, R.W. Characterization of subpopulations of lipoprotein particles isolated from human cerebrospinal fluid. Biochim. Biophys. Acta 1995, 1255, 192–200. [Google Scholar] [CrossRef]
- Rhea, E.M.; Banks, W.A. Interactions of Lipids, Lipoproteins, and Apolipoproteins with the Blood-Brain Barrier. Pharm. Res. 2021, 38, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Button, E.B.; Robert, J.; Caffrey, T.M.; Fan, J.; Zhao, W.; Wellington, C.L. HDL from an Alzheimer’s disease perspective. Curr. Opin. Lipidol. 2019, 30, 224–234. [Google Scholar] [CrossRef]
- Hartmann, H.; Ho, W.Y.; Chang, J.C.; Ling, S.C. Cholesterol dyshomeostasis in amyotrophic lateral sclerosis: Cause, consequence, or epiphenomenon? FEBS J. 2022, 289, 7688–7709. [Google Scholar] [CrossRef]
- Colardo, M.; Petraroia, M.; Lerza, L.; Pensabene, D.; Martella, N.; Pallottini, V.; Segatto, M. NGF Modulates Cholesterol Metabolism and Stimulates ApoE Secretion in Glial Cells Conferring Neuroprotection against Oxidative Stress. Int. J. Mol. Sci. 2022, 23, 4842. [Google Scholar] [CrossRef]
- Elliott, D.A.; Weickert, C.S.; Garner, B. Apolipoproteins in the brain: Implications for neurological and psychiatric disorders. Clin. Lipidol. 2010, 51, 555–573. [Google Scholar] [CrossRef]
- Koch, M.; Aroner, S.A.; Fitzpatrick, A.L.; Longstreth, W.T., Jr.; Furtado, J.D.; Mukamal, K.J.; Jensen, M.K. HDL (High-Density Lipoprotein) Subspecies, Prevalent Covert Brain Infarcts, and Incident Overt Ischemic Stroke: Cardiovascular Health Study. Stroke 2022, 53, 1292–1300. [Google Scholar] [CrossRef]
- Delk, S.C.; Chattopadhyay, A.; Escola-Gil, J.C.; Fogelman, A.M.; Reddy, S.T. Apolipoprotein mimetics in cancer. Semin. Cancer Biol. 2021, 73, 158–168. [Google Scholar] [CrossRef]
- Mockel, B.; Zinke, H.; Flach, R.; Weiss, B.; Weiler-Guttler, H.; Gassen, H.G. Expression of apolipoprotein A-I in porcine brain endothelium in vitro. J. Neurochem. 1994, 62, 788–798. [Google Scholar] [CrossRef]
- Weiler-Guttler, H.; Sommerfeldt, M.; Papandrikopoulou, A.; Mischek, U.; Bonitz, D.; Frey, A.; Grupe, M.; Scheerer, J.; Gassen, H.G. Synthesis of apolipoprotein A-1 in pig brain microvascular endothelial cells. J. Neurochem. 1990, 54, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Endres, K. Apolipoprotein A1, the neglected relative of Apolipoprotein E and its potential role in Alzheimer’s disease. Neural Regen. Res. 2021, 16, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.R.; Li, K.; Unger, T.L.; Gallagher, M.D.; Van Deerlin, V.M.; Agarwal, P.; Leverenz, J.; Roberts, J.; Samii, A.; Gross, R.G.; et al. Lower plasma apolipoprotein A1 levels are found in Parkinson’s disease and associate with apolipoprotein A1 genotype. Mov. Disord. 2015, 30, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Smach, M.A.; Edziri, H.; Charfeddine, B.; Ben Othman, L.; Lammouchi, T.; Ltaief, A.; Nafati, S.; Dridi, H.; Bennamou, S.; Limem, K. Polymorphism in apoA1 Influences High-Density Lipoprotein Cholesterol Levels but Is Not a Major Risk Factor of Alzheimer’s Disease. Dement. Geriatr. Cogn. Dis. Extra 2011, 1, 249–257. [Google Scholar] [CrossRef]
- Vollbach, H.; Heun, R.; Morris, C.M.; Edwardson, J.A.; McKeith, I.G.; Jessen, F.; Schulz, A.; Maier, W.; Kolsch, H. APOA1 polymorphism influences risk for early-onset nonfamiliar AD. Ann. Neurol. 2005, 58, 436–441. [Google Scholar] [CrossRef]
- Wang, F.; Kohan, A.B.; Lo, C.M.; Liu, M.; Howles, P.; Tso, P. Apolipoprotein A-IV: A protein intimately involved in metabolism. J. Lipid Res. 2015, 56, 1403–1418. [Google Scholar] [CrossRef]
- Shen, L.; Pearson, K.J.; Xiong, Y.; Lo, C.M.; Tso, P.; Woods, S.C.; Davidson, W.S.; Liu, M. Characterization of apolipoprotein A-IV in brain areas involved in energy homeostasis. Physiol. Behav. 2008, 95, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Doi, T.; Shen, L.; Woods, S.C.; Seeley, R.J.; Zheng, S.; Jackman, A.; Tso, P. Intestinal satiety protein apolipoprotein AIV is synthesized and regulated in rat hypothalamus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R1382–R1387. [Google Scholar] [CrossRef] [PubMed]
- Fagan, A.M.; Holtzman, D.M.; Munson, G.; Mathur, T.; Schneider, D.; Chang, L.K.; Getz, G.S.; Reardon, C.A.; Lukens, J.; Shah, J.A.; et al. Unique lipoproteins secreted by primary astrocytes from wild type, apoE (-/-), and human apoE transgenic mice. J. Biol. Chem. 1999, 274, 30001–30007. [Google Scholar] [CrossRef] [PubMed]
- Okumura, T.; Fukagawa, K.; Tso, P.; Taylor, I.L.; Pappas, T.N. Intracisternal injection of apolipoprotein A-IV inhibits gastric secretion in pylorus-ligated conscious rats. Gastroenterology 1994, 107, 1861–1864. [Google Scholar] [CrossRef] [PubMed]
- Tso, P.; Chen, Q.; Fujimoto, K.; Fukagawa, K.; Sakata, T. Apolipoprotein A-IV: A circulating satiety signal produced by the small intestine. Obes. Res. 1995, 3 (Suppl. S5), 689S–695S. [Google Scholar] [CrossRef] [PubMed]
- Roula, D.; Theiler, A.; Luschnig, P.; Sturm, G.J.; Tomazic, P.V.; Marsche, G.; Heinemann, A.; Sturm, E.M. Apolipoprotein A-IV acts as an endogenous anti-inflammatory protein and is reduced in treatment-naive allergic patients and allergen-challenged mice. Allergy 2020, 75, 392–402. [Google Scholar] [CrossRef]
- Csaszar, A.; Kalman, J.; Szalai, C.; Janka, Z.; Romics, L. Association of the apolipoprotein A-IV codon 360 mutation in patients with Alzheimer’s disease. Neurosci. Lett. 1997, 230, 151–154. [Google Scholar] [CrossRef]
- Cui, Y.; Huang, M.; He, Y.; Zhang, S.; Luo, Y. Genetic ablation of apolipoprotein A-IV accelerates Alzheimer’s disease pathogenesis in a mouse model. Am. J. Pathol. 2011, 178, 1298–1308. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, J.; Li, Y.; Chen, X.; Zhang, T.; You, W.; Xue, Y.; Shen, C. The Variants at APOA1 and APOA4 Contribute to the Susceptibility of Schizophrenia With Inhibiting mRNA Expression in Peripheral Blood Leukocytes. Front. Mol. Biosci. 2021, 8, 785445. [Google Scholar] [CrossRef]
- Deng, X.; Walker, R.G.; Morris, J.; Davidson, W.S.; Thompson, T.B. Role of Conserved Proline Residues in Human Apolipoprotein A-IV Structure and Function. J. Biol. Chem. 2015, 290, 10689–10702. [Google Scholar] [CrossRef]
- Haas, M.E.; Attie, A.D.; Biddinger, S.B. The regulation of ApoB metabolism by insulin. Trends Endocrinol. Metab. 2013, 24, 391–397. [Google Scholar] [CrossRef]
- Picard, C.; Nilsson, N.; Labonte, A.; Auld, D.; Rosa-Neto, P.; Alzheimer’s Disease Neuroimaging Initiative; Ashton, N.J.; Zetterberg, H.; Blennow, K.; Breitner, J.C.B.; et al. Apolipoprotein B is a novel marker for early tau pathology in Alzheimer’s disease. Alzheimers Dement. 2022, 18, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Dassati, S.; Waldner, A.; Schweigreiter, R. Apolipoprotein D takes center stage in the stress response of the aging and degenerative brain. Neurobiol. Aging 2014, 35, 1632–1642. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.; Ganfornina, M.D. The Lipocalin Apolipoprotein D Functional Portrait: A Systematic Review. Front. Physiol. 2021, 12, 738991. [Google Scholar] [CrossRef] [PubMed]
- Belloir, B.; Kovari, E.; Surini-Demiri, M.; Savioz, A. Altered apolipoprotein D expression in the brain of patients with Alzheimer disease. J. Neurosci. Res. 2001, 64, 61–69. [Google Scholar] [CrossRef]
- Kalman, J.; McConathy, W.; Araoz, C.; Kasa, P.; Lacko, A.G. Apolipoprotein D in the aging brain and in Alzheimer’s dementia. Neurol. Res. 2000, 22, 330–336. [Google Scholar] [CrossRef]
- Thomas, E.A.; Laws, S.M.; Sutcliffe, J.G.; Harper, C.; Dean, B.; McClean, C.; Masters, C.; Lautenschlager, N.; Gandy, S.E.; Martins, R.N. Apolipoprotein D levels are elevated in prefrontal cortex of subjects with Alzheimer’s disease: No relation to apolipoprotein E expression or genotype. Biol. Psychiatry 2003, 54, 136–141. [Google Scholar] [CrossRef]
- Terrisse, L.; Poirier, J.; Bertrand, P.; Merched, A.; Visvikis, S.; Siest, G.; Milne, R.; Rassart, E. Increased levels of apolipoprotein D in cerebrospinal fluid and hippocampus of Alzheimer’s patients. J. Neurochem. 1998, 71, 1643–1650. [Google Scholar] [CrossRef]
- Paasila, P.J.; Aramideh, J.A.; Sutherland, G.T.; Graeber, M.B. Synapses, Microglia, and Lipids in Alzheimer’s Disease. Front. Neurosci. 2021, 15, 778822. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Jeon, S.; Baik, K.; Lee, Y.H.; Chung, S.J.; Yoo, H.S.; Jeong, S.H.; Sohn, Y.H.; Lee, P.H.; Ye, B.S. Apolipoprotein E4, amyloid, and cognition in Alzheimer’s and Lewy body disease. Neurobiol. Aging 2021, 106, 45–54. [Google Scholar] [CrossRef]
- Zhao, J.; Lu, W.; Ren, Y.; Fu, Y.; Martens, Y.A.; Shue, F.; Davis, M.D.; Wang, X.; Chen, K.; Li, F.; et al. Apolipoprotein E regulates lipid metabolism and alpha-synuclein pathology in human iPSC-derived cerebral organoids. Acta Neuropathol. 2021, 142, 807–825. [Google Scholar] [CrossRef]
- Pohlkamp, T.; Xian, X.; Wong, C.H.; Durakoglugil, M.S.; Werthmann, G.C.; Saido, T.C.; Evers, B.M.; White, C.L., 3rd; Connor, J.; Hammer, R.E.; et al. NHE6 depletion corrects ApoE4-mediated synaptic impairments and reduces amyloid plaque load. eLife 2021, 10, e72034. [Google Scholar] [CrossRef]
- Montagne, A.; Nation, D.A.; Sagare, A.P.; Barisano, G.; Sweeney, M.D.; Chakhoyan, A.; Pachicano, M.; Joe, E.; Nelson, A.R.; D’Orazio, L.M.; et al. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature 2020, 581, 71–76. [Google Scholar] [CrossRef]
- Kotredes, K.P.; Oblak, A.; Pandey, R.S.; Lin, P.B.; Garceau, D.; Williams, H.; Uyar, A.; O’Rourke, R.; O’Rourke, S.; Ingraham, C.; et al. Uncovering Disease Mechanisms in a Novel Mouse Model Expressing Humanized APOEepsilon4 and Trem2*R47H. Front. Aging Neurosci. 2021, 13, 735524. [Google Scholar] [CrossRef]
- Vervoordt, S.M.; Arnett, P.; Engeland, C.; Rabinowitz, A.R.; Hillary, F.G. Depression associated with APOE status and hippocampal volume but not cognitive decline in older adults aging with traumatic brain injury. Neuropsychology 2021, 35, 863–875. [Google Scholar] [CrossRef]
- Main, B.S.; Villapol, S.; Sloley, S.S.; Barton, D.J.; Parsadanian, M.; Agbaegbu, C.; Stefos, K.; McCann, M.S.; Washington, P.M.; Rodriguez, O.C.; et al. Apolipoprotein E4 impairs spontaneous blood brain barrier repair following traumatic brain injury. Mol. Neurodegener. 2018, 13, 17. [Google Scholar] [CrossRef]
- Kundu, P.; Zimmerman, B.; Perez, R.; Whitlow, C.T.; Cline, J.M.; Olson, J.D.; Andrews, R.N.; Raber, J. Apolipoprotein E levels in the amygdala and prefrontal cortex predict relative regional brain volumes in irradiated Rhesus macaques. Sci. Rep. 2021, 11, 22130. [Google Scholar] [CrossRef]
- Kloske, C.M.; Wilcock, D.M. The Important Interface Between Apolipoprotein E and Neuroinflammation in Alzheimer’s Disease. Front. Immunol. 2020, 11, 754. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Meltzer, J.C.; Nguyen, H.; Commins, C.; Bennett, R.E.; Hudry, E.; Hyman, B.T. APOE4 derived from astrocytes leads to blood-brain barrier impairment. Brain 2022, 145, 3582–3593. [Google Scholar] [CrossRef] [PubMed]
- den Hoedt, S.; Crivelli, S.M.; Leijten, F.P.J.; Losen, M.; Stevens, J.A.A.; Mane-Damas, M.; de Vries, H.E.; Walter, J.; Mirzaian, M.; Sijbrands, E.J.G.; et al. Effects of Sex, Age, and Apolipoprotein E Genotype on Brain Ceramides and Sphingosine-1-Phosphate in Alzheimer’s Disease and Control Mice. Front. Aging Neurosci. 2021, 13, 765252. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.S.; Watson, K.Q.; Rebeck, G.W. High-fat diet increases gliosis and immediate early gene expression in APOE3 mice, but not APOE4 mice. J. Neuroinflamm. 2021, 18, 214. [Google Scholar] [CrossRef] [PubMed]
- Asaro, A.; Sinha, R.; Bakun, M.; Kalnytska, O.; Carlo-Spiewok, A.S.; Rubel, T.; Rozeboom, A.; Dadlez, M.; Kaminska, B.; Aronica, E.; et al. ApoE4 disrupts interaction of sortilin with fatty acid-binding protein 7 essential to promote lipid signaling. J. Cell Sci. 2021, 134, jcs258894. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.S.; Tensaouti, Y.; Stephanz, E.P.; Chintamen, S.; Rafikian, E.E.; Yang, M.; Kernie, S.G. Astrocytic ApoE underlies maturation of hippocampal neurons and cognitive recovery after traumatic brain injury in mice. Commun. Biol. 2021, 4, 1303. [Google Scholar] [CrossRef] [PubMed]
- Pankiewicz, J.E.; Lizinczyk, A.M.; Franco, L.A.; Diaz, J.R.; Marta-Ariza, M.; Sadowski, M.J. Absence of Apolipoprotein E is associated with exacerbation of prion pathology and promotes microglial neurodegenerative phenotype. Acta Neuropathol. Commun. 2021, 9, 157. [Google Scholar] [CrossRef]
- Konings, S.C.; Torres-Garcia, L.; Martinsson, I.; Gouras, G.K. Astrocytic and Neuronal Apolipoprotein E Isoforms Differentially Affect Neuronal Excitability. Front. Neurosci. 2021, 15, 734001. [Google Scholar] [CrossRef]
- Zhang, S.H.; Reddick, R.L.; Piedrahita, J.A.; Maeda, N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 1992, 258, 468–471. [Google Scholar] [CrossRef]
- Wang, H.; Kulas, J.A.; Wang, C.; Holtzman, D.M.; Ferris, H.A.; Hansen, S.B. Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc. Natl. Acad. Sci. USA 2021, 118, e2102191118. [Google Scholar] [CrossRef]
- Weisgraber, K.H.; Mahley, R.W.; Kowal, R.C.; Herz, J.; Goldstein, J.L.; Brown, M.S. Apolipoprotein C-I modulates the interaction of apolipoprotein E with beta-migrating very low density lipoproteins (beta-VLDL) and inhibits binding of beta-VLDL to low density lipoprotein receptor-related protein. J. Biol. Chem. 1990, 265, 22453–22459. [Google Scholar] [CrossRef]
- Kowal, R.C.; Herz, J.; Weisgraber, K.H.; Mahley, R.W.; Brown, M.S.; Goldstein, J.L. Opposing effects of apolipoproteins E and C on lipoprotein binding to low density lipoprotein receptor-related protein. J. Biol. Chem. 1990, 265, 10771–10779. [Google Scholar] [CrossRef]
- Sehayek, E.; Eisenberg, S. Mechanisms of inhibition by apolipoprotein C of apolipoprotein E-dependent cellular metabolism of human triglyceride-rich lipoproteins through the low density lipoprotein receptor pathway. J. Biol. Chem. 1991, 266, 18259–18267. [Google Scholar] [CrossRef]
- Petit-Turcotte, C.; Stohl, S.M.; Beffert, U.; Cohn, J.S.; Aumont, N.; Tremblay, M.; Dea, D.; Yang, L.; Poirier, J.; Shachter, N.S. Apolipoprotein C-I expression in the brain in Alzheimer’s disease. Neurobiol. Dis. 2001, 8, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Drigalenko, E.; Poduslo, S.; Elston, R. Interaction of the apolipoprotein E and CI loci in predisposing to late-onset Alzheimer’s disease. Neurology 1998, 51, 131–135. [Google Scholar] [CrossRef]
- Jong, M.C.; Hofker, M.H.; Havekes, L.M. Role of ApoCs in lipoprotein metabolism: Functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 472–484. [Google Scholar] [CrossRef] [PubMed]
- van der Hoogt, C.C.; Berbee, J.F.; Espirito Santo, S.M.; Gerritsen, G.; Krom, Y.D.; van der Zee, A.; Havekes, L.M.; van Dijk, K.W.; Rensen, P.C. Apolipoprotein CI causes hypertriglyceridemia independent of the very-low-density lipoprotein receptor and apolipoprotein CIII in mice. Biochim. Biophys. Acta 2006, 1761, 213–220. [Google Scholar] [CrossRef]
- Jong, M.C.; Dahlmans, V.E.; van Gorp, P.J.; van Dijk, K.W.; Breuer, M.L.; Hofker, M.H.; Havekes, L.M. In the absence of the low density lipoprotein receptor, human apolipoprotein C1 overexpression in transgenic mice inhibits the hepatic uptake of very low density lipoproteins via a receptor-associated protein-sensitive pathway. J. Clin. Investig. 1996, 98, 2259–2267. [Google Scholar] [CrossRef]
- Abildayeva, K.; Berbee, J.F.; Blokland, A.; Jansen, P.J.; Hoek, F.J.; Meijer, O.; Lutjohann, D.; Gautier, T.; Pillot, T.; De Vente, J.; et al. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J. Lipid Res. 2008, 49, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Cudaback, E.; Li, X.; Yang, Y.; Yoo, T.; Montine, K.S.; Craft, S.; Montine, T.J.; Keene, C.D. Apolipoprotein C-I is an APOE genotype-dependent suppressor of glial activation. J. Neuroinflamm. 2012, 9, 192. [Google Scholar] [CrossRef]
- Song, F.; Poljak, A.; Crawford, J.; Kochan, N.A.; Wen, W.; Cameron, B.; Lux, O.; Brodaty, H.; Mather, K.; Smythe, G.A.; et al. Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS ONE 2012, 7, e34078. [Google Scholar] [CrossRef]
- Mather, K.A.; Thalamuthu, A.; Oldmeadow, C.; Song, F.; Armstrong, N.J.; Poljak, A.; Holliday, E.G.; McEvoy, M.; Kwok, J.B.; Assareh, A.A.; et al. Genome-wide significant results identified for plasma apolipoprotein H levels in middle-aged and older adults. Sci. Rep. 2016, 6, 23675. [Google Scholar] [CrossRef]
- Hoekstra, M.; Chen, H.Y.; Rong, J.; Dufresne, L.; Yao, J.; Guo, X.; Tsai, M.Y.; Tsimikas, S.; Post, W.S.; Vasan, R.S.; et al. Genome-Wide Association Study Highlights APOH as a Novel Locus for Lipoprotein(a) Levels-Brief Report. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 458–464. [Google Scholar] [CrossRef]
- Vogrinc, Z.; Trbojevic-Cepe, M.; Coen, D.; Vitale, K.; Stavljenic-Rukavina, A. Apolipoprotein H (apoH)-dependent autoantibodies and apoH protein polymorphism in selected patients showing lupus anticoagulant activity. Clin. Chem. Lab. Med. 2005, 43, 17–21. [Google Scholar] [CrossRef]
- Gelissen, I.C.; Hochgrebe, T.; Wilson, M.R.; Easterbrook-Smith, S.B.; Jessup, W.; Dean, R.T.; Brown, A.J. Apolipoprotein J (clusterin) induces cholesterol export from macrophage-foam cells: A potential anti-atherogenic function? Biochem. J. 1998, 331 Pt 1, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.R.; Yerbury, J.J.; Wilson, M.R. Structural characterization of clusterin-chaperone client protein complexes. J. Biol. Chem. 2009, 284, 21920–21927. [Google Scholar] [CrossRef] [PubMed]
- Giannakopoulos, P.; Kovari, E.; French, L.E.; Viard, I.; Hof, P.R.; Bouras, C. Possible neuroprotective role of clusterin in Alzheimer’s disease: A quantitative immunocytochemical study. Acta Neuropathol. 1998, 95, 387–394. [Google Scholar] [CrossRef]
- DeMattos, R.B.; Cirrito, J.R.; Parsadanian, M.; May, P.C.; O’Dell, M.A.; Taylor, J.W.; Harmony, J.A.; Aronow, B.J.; Bales, K.R.; Paul, S.M.; et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: Evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron 2004, 41, 193–202. [Google Scholar] [CrossRef]
- Freixes, M.; Puig, B.; Rodriguez, A.; Torrejon-Escribano, B.; Blanco, R.; Ferrer, I. Clusterin solubility and aggregation in Creutzfeldt-Jakob disease. Acta Neuropathol. 2004, 108, 295–301. [Google Scholar] [CrossRef]
- Sasaki, K.; Doh-ura, K.; Ironside, J.W.; Iwaki, T. Increased clusterin (apolipoprotein J) expression in human and mouse brains infected with transmissible spongiform encephalopathies. Acta Neuropathol. 2002, 103, 199–208. [Google Scholar] [CrossRef]
- Liu, D.; Pan, J.M.; Pei, X.; Li, J.S. Interaction Between Apolipoprotein M Gene Single-Nucleotide Polymorphisms and Obesity and its Effect on Type 2 Diabetes Mellitus Susceptibility. Sci. Rep. 2020, 10, 7859. [Google Scholar] [CrossRef]
- Katz, P.; Pedro, S.; Trupin, L.; Yelin, E.; Michaud, K. The Impact of Asthma and Chronic Obstructive Pulmonary Disease (COPD) on Patient-Reported Outcomes in Systemic Lupus Erythematosus (SLE). ACR Open Rheumatol. 2021, 3, 221–230. [Google Scholar] [CrossRef]
- Du, W.; Shen, T.; Li, H.; Liu, Y.; He, L.; Tan, L.; Hu, M.; Ren, Y. Low apolipoprotein M serum levels correlate with Systemic lupus erythematosus disease activity and apolipoprotein M gene polymorphisms with Lupus. Lipids Health Dis. 2017, 16, 88. [Google Scholar] [CrossRef]
- Liu, M.; Frej, C.; Langefeld, C.D.; Divers, J.; Bowden, D.W.; Carr, J.J.; Gebre, A.K.; Xu, J.; Larsson, B.; Dahlback, B.; et al. Plasma apoM and S1P levels are inversely associated with mortality in African Americans with type 2 diabetes mellitus. J. Lipid Res. 2019, 60, 1425–1431. [Google Scholar] [CrossRef]
- Christoffersen, C. Apolipoprotein M-A Marker or an Active Player in Type II Diabetes? Front. Endocrinol. 2021, 12, 665393. [Google Scholar] [CrossRef] [PubMed]
- Mathiesen Janiurek, M.; Soylu-Kucharz, R.; Christoffersen, C.; Kucharz, K.; Lauritzen, M. Apolipoprotein M-bound sphingosine-1-phosphate regulates blood-brain barrier paracellular permeability and transcytosis. eLife 2019, 8, e49405. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, C.; Jauhiainen, M.; Moser, M.; Porse, B.; Ehnholm, C.; Boesl, M.; Dahlback, B.; Nielsen, L.B. Effect of apolipoprotein M on high density lipoprotein metabolism and atherosclerosis in low density lipoprotein receptor knock-out mice. J. Biol. Chem. 2008, 283, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Raulin, A.C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.C. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef]
- Pizarro, A.; Hayer, K.; Lahens, N.F.; Hogenesch, J.B. CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013, 41, D1009–D1013. [Google Scholar] [CrossRef]
- Pembroke, W.G.; Babbs, A.; Davies, K.E.; Ponting, C.P.; Oliver, P.L. Temporal transcriptomics suggest that twin-peaking genes reset the clock. eLife 2015, 4, e10518. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.H.; Murrell, C.E.; Chu, A.; Pan, X. Circadian Regulation of Apolipoproteins in the Brain: Implications in Lipid Metabolism and Disease. Int. J. Mol. Sci. 2023, 24, 17415. https://doi.org/10.3390/ijms242417415
Lee CH, Murrell CE, Chu A, Pan X. Circadian Regulation of Apolipoproteins in the Brain: Implications in Lipid Metabolism and Disease. International Journal of Molecular Sciences. 2023; 24(24):17415. https://doi.org/10.3390/ijms242417415
Chicago/Turabian StyleLee, Chaeeun Hannah, Charlotte Ellzabeth Murrell, Alexander Chu, and Xiaoyue Pan. 2023. "Circadian Regulation of Apolipoproteins in the Brain: Implications in Lipid Metabolism and Disease" International Journal of Molecular Sciences 24, no. 24: 17415. https://doi.org/10.3390/ijms242417415
APA StyleLee, C. H., Murrell, C. E., Chu, A., & Pan, X. (2023). Circadian Regulation of Apolipoproteins in the Brain: Implications in Lipid Metabolism and Disease. International Journal of Molecular Sciences, 24(24), 17415. https://doi.org/10.3390/ijms242417415





