Structure and Expression Analysis of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Gene Families in Populus trichocarpa Torr. and Gray
Abstract
:1. Introduction
2. Results
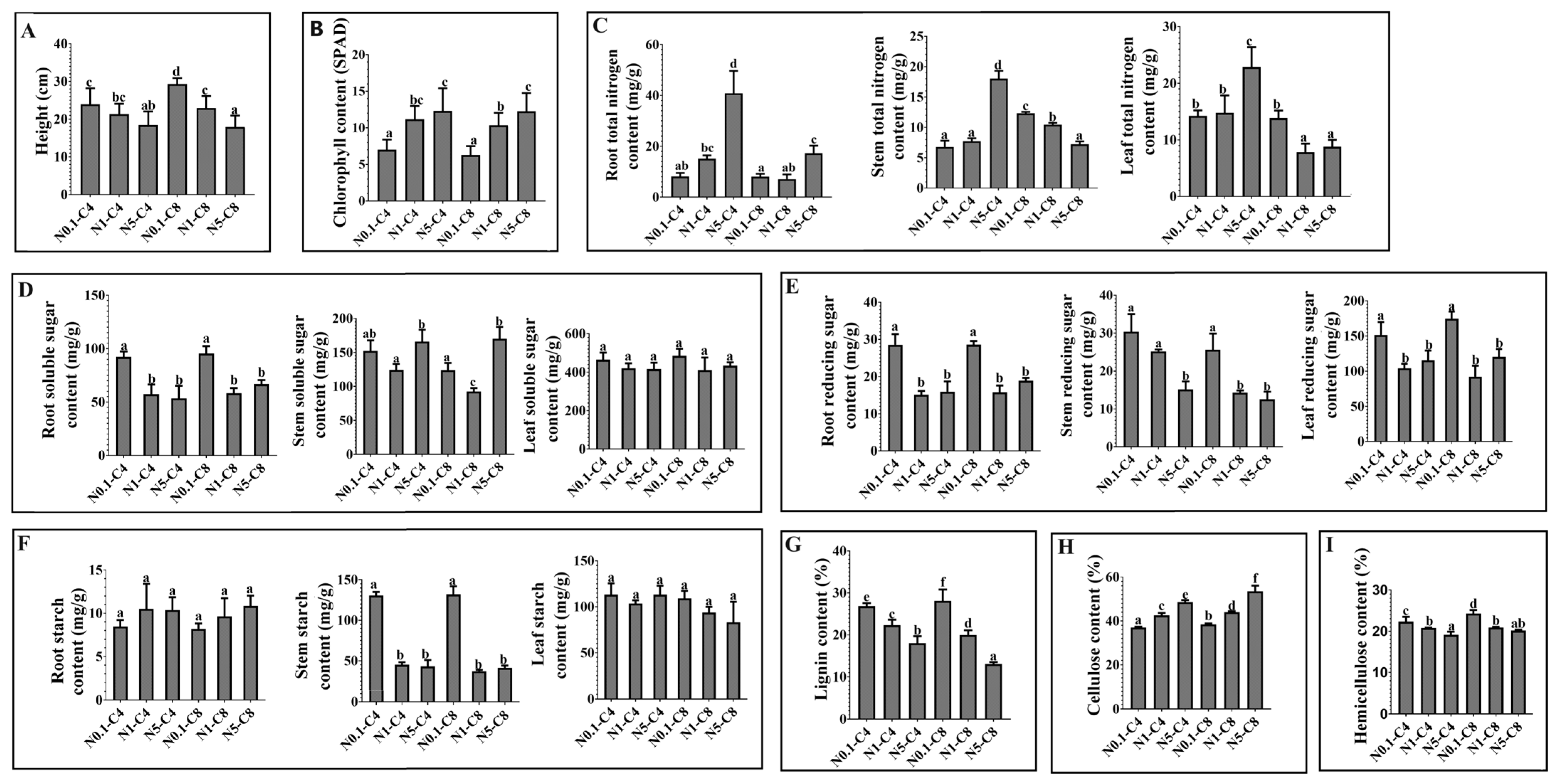
2.1. Changes in Plant Growth and Development in Response to Exogenous C and N Treatments
2.2. Analysis of the PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Family Members in P. trichocarpa
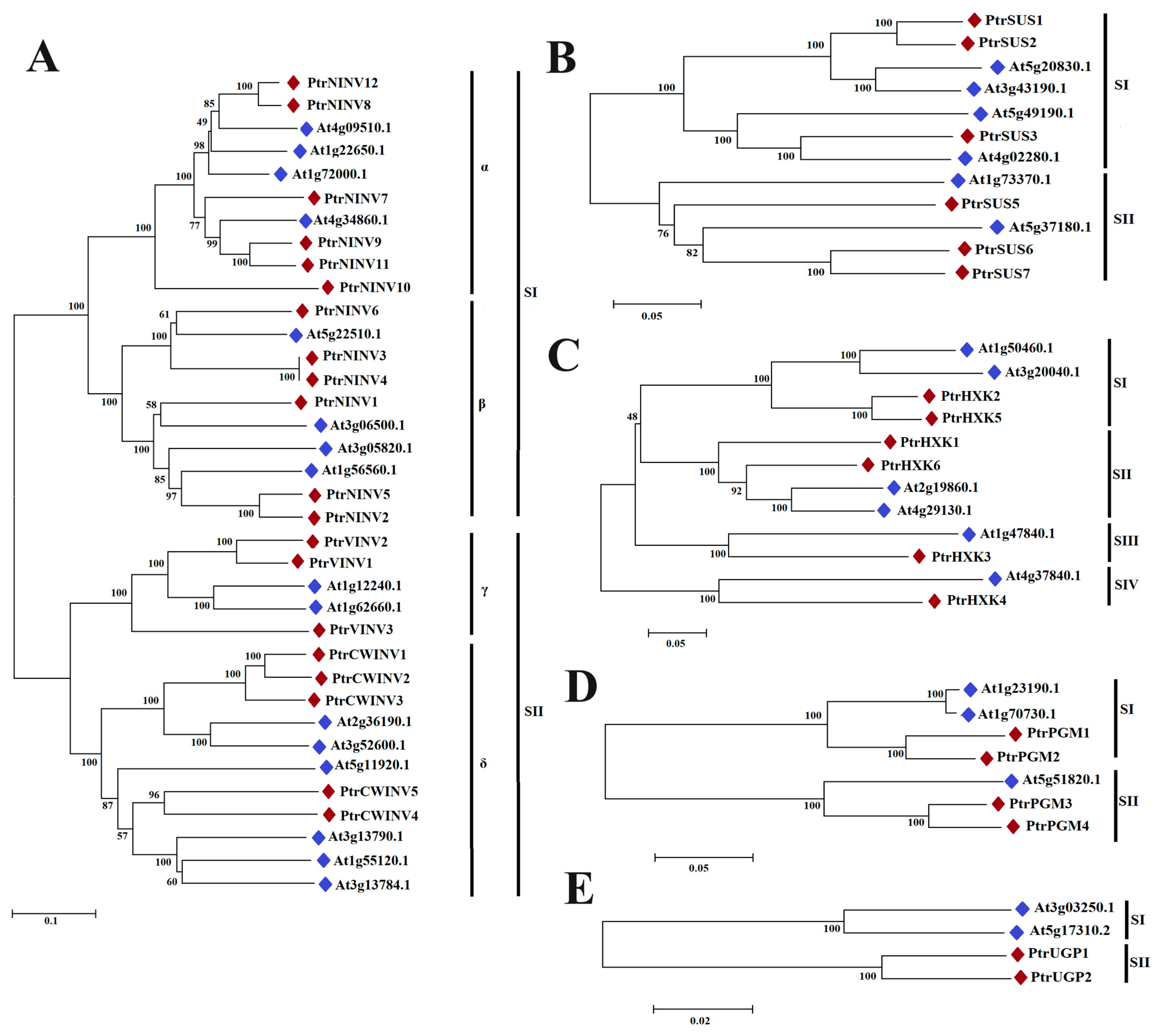
2.3. Gene Structure and Phylogenetic Analysis
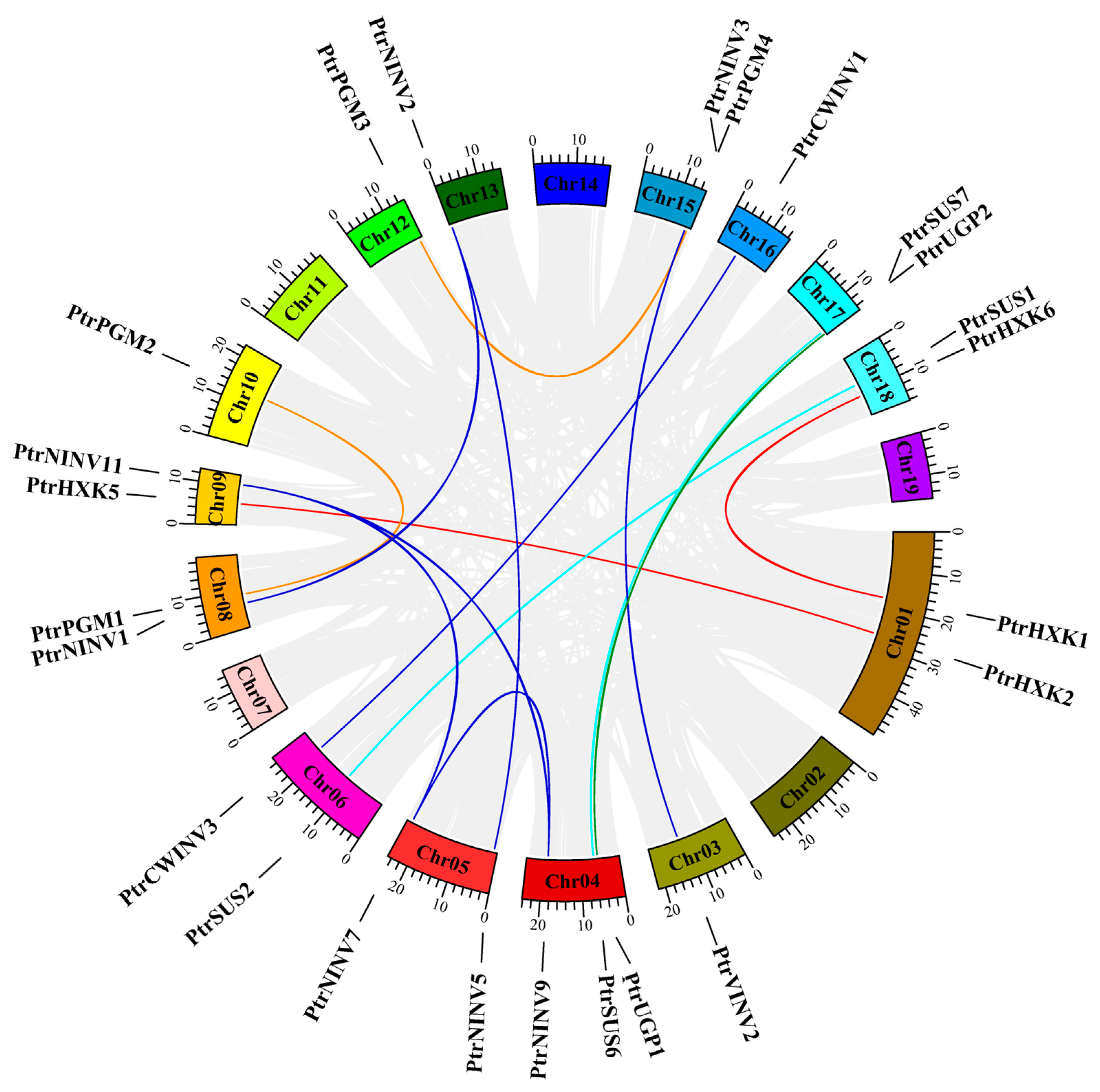
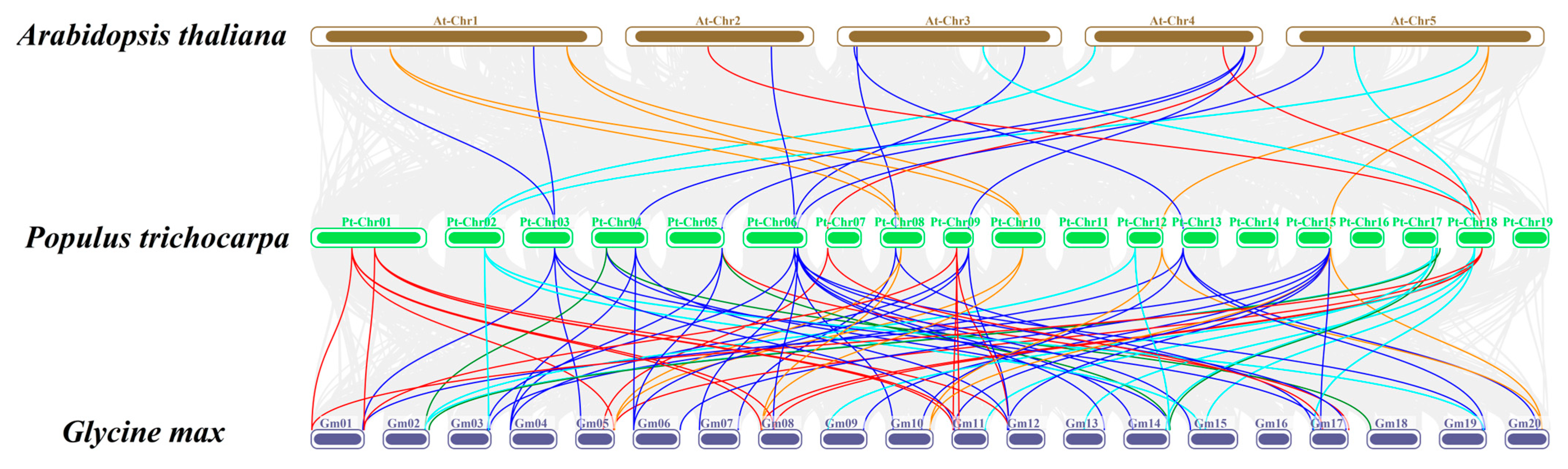
2.4. Chromosome Distribution and Collinearity Analysis
2.5. Secondary and Tertiary Structure Prediction
2.6. Tissue-Specific Expression Analysis
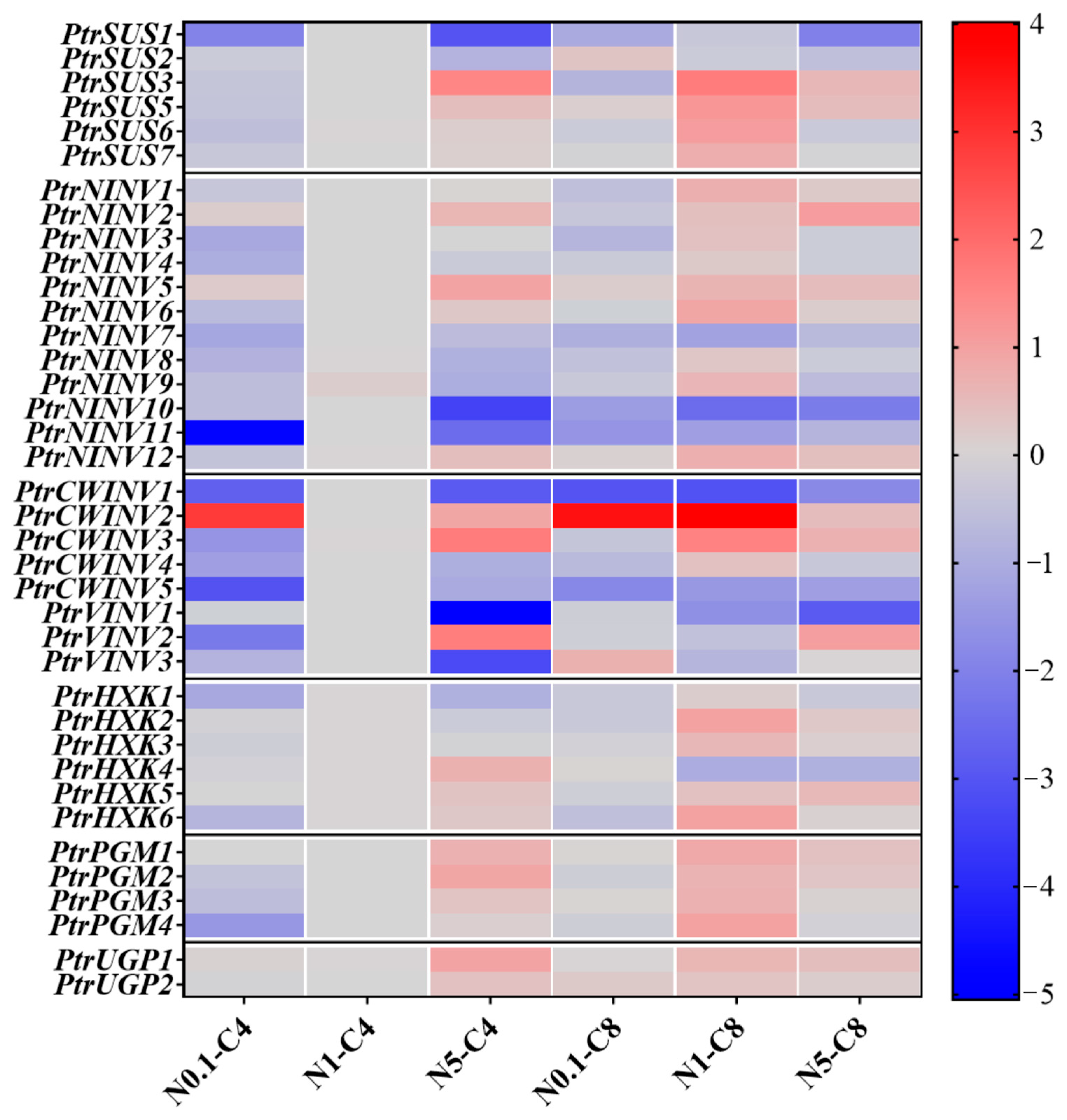
2.7. Effects of Different Treatments on Expression Patterns of PtrSUSs, PtrINVs, PtrHXKs, PtrPGMs, and PtrUGPs
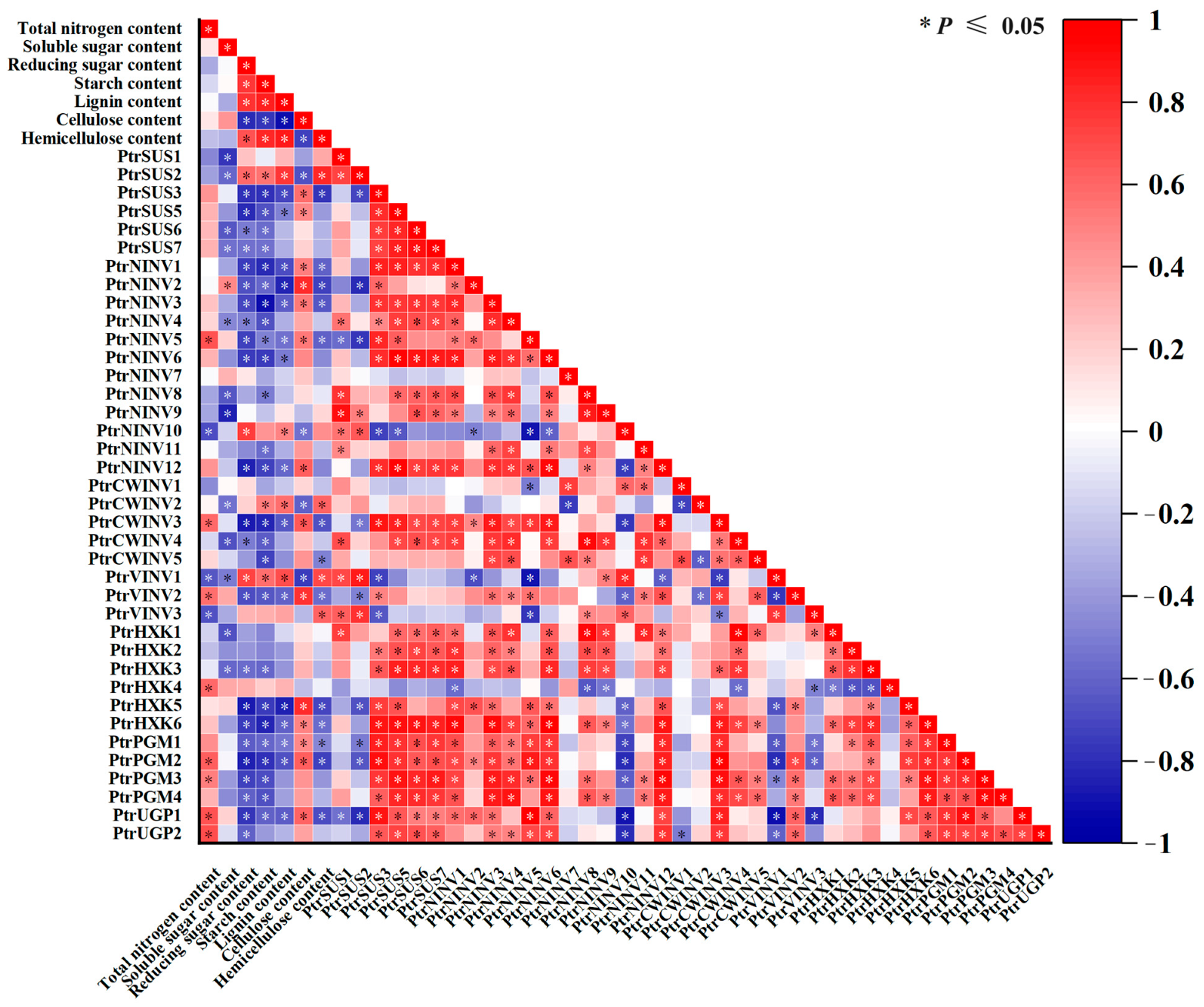
2.8. Correlation Analysis between Expression Levels of PtrSUSs, PtrINVs, PtrHXKs, PtrPGMs, and PtrUGPs and Physiological Characteristics under Different Treatments
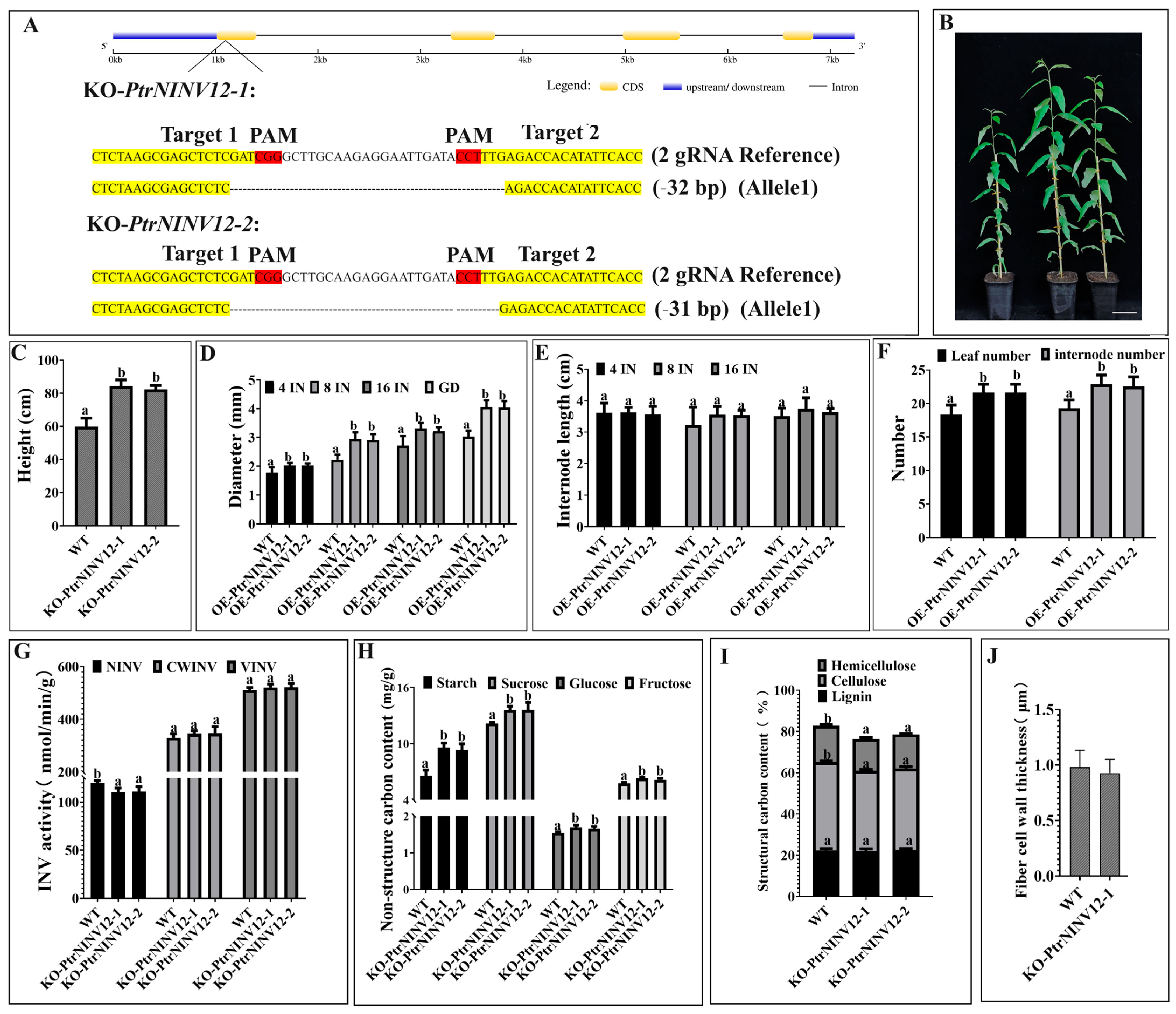
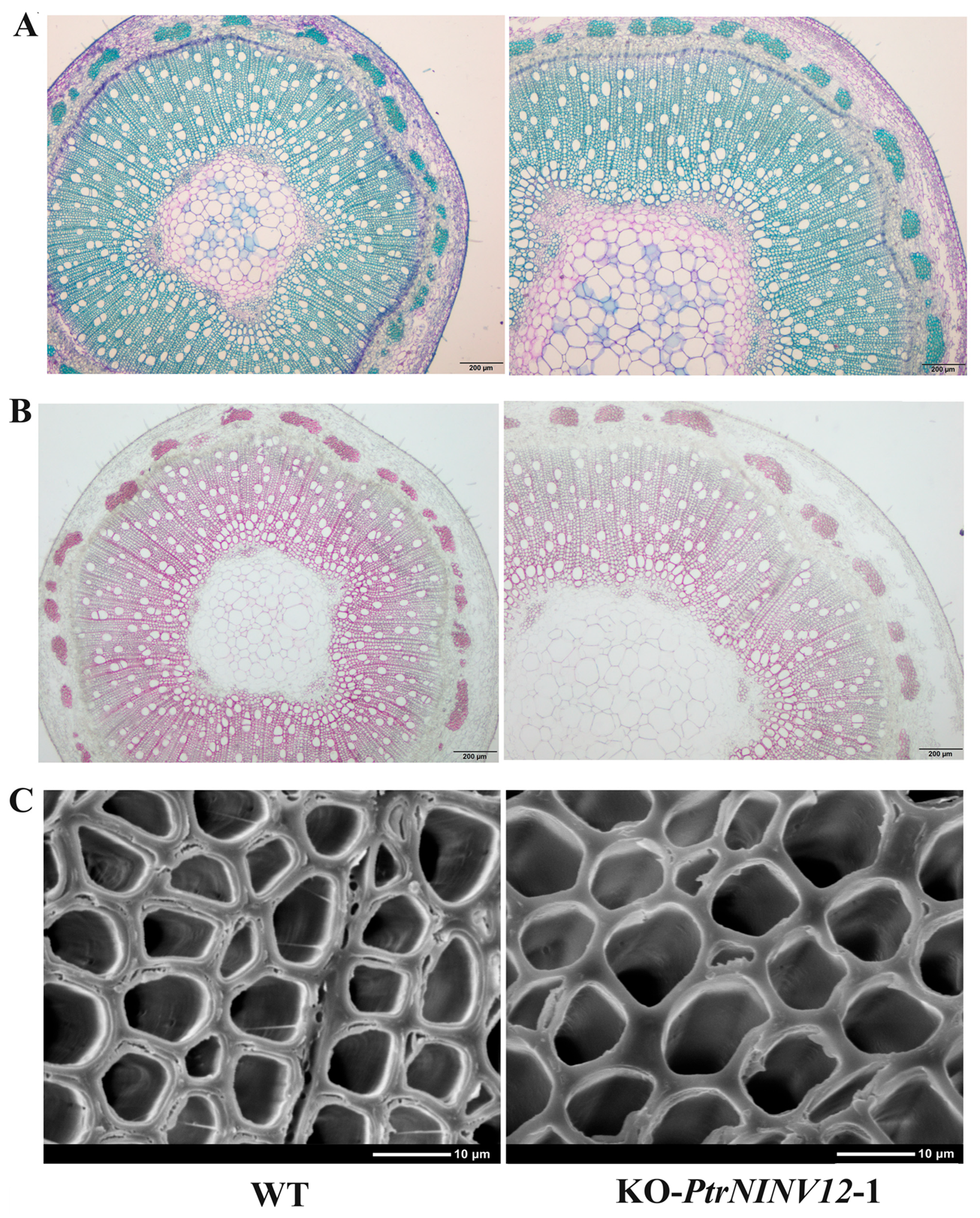
2.9. Effects of PtrNINV12-knockout on Growth and Cell Wall Development of Poplar Seedlings
3. Discussion
3.1. Evolution and Structure
3.2. Expression Patterns of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Genes
3.3. Changes in Structural C Content and Gene Expression Patterns under N and C Treatments
3.4. Effects of PtrNINV12-Knockout on Contents of Structural C and Non-Structural C
3.5. PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Family Genes Participate in the Process of Exogenous N and C Affecting Structural C Synthesis
4. Materials and Methods
4.1. Analysis of SUS, INV, HXK, PGM, and UGP Family Members in P. trichocarpa
4.2. Analysis of Gene Structure, Conserved Motifs, Multiple Sequence Alignment, Phylogenetic Trees
4.3. Protein Secondary, Tertiary Structure Prediction, and Chromosome Distribution
4.4. Plant Materials, Growing Conditions, and Treatments
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Biochemical Analyses
4.7. The Acquisition of Transgenic Plants
4.8. Microscopy Analyses
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verbancic, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef]
- Sandhu, A.P.; Randhawa, G.S.; Dhugga, K.S. Plant cell wall matrix polysaccharide biosynthesis. Mol. Plant 2009, 2, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Geisler, M.; Ciereszko, I.; Johansson, H. UDP-glucose pyrophosphorylase. An old protein with new tricks. Plant Physiol. 2004, 134, 912–918. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Chen, Z.; Wang, J.; Ye, M.; Ji, L.; Wang, J.; Liao, W.; Ma, H. Identification and characterization of the Populus sucrose synthase gene family. Gene 2014, 539, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wei, P.; Wu, M.; Xu, Y.; Li, F.; Luo, Z.; Zhang, J.; Chen, A.; Xie, X.; Cao, P.; et al. Analysis of the sucrose synthase gene family in tobacco: Structure, phylogeny, and expression patterns. Planta 2015, 242, 153–166. [Google Scholar] [CrossRef]
- Chen, A.; He, S.; Li, F.; Li, Z.; Ding, M.; Liu, Q.; Rong, J. Analyses of the sucrose synthase gene family in cotton: Structure, phylogeny and expression patterns. BMC Plant Biol. 2012, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Bieniawska, Z.; Paul Barratt, D.H.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef]
- Koramutla, M.K.; Ram, C.; Bhatt, D.; Annamalai, M.; Bhattacharya, R. Genome-wide identification and expression analysis of sucrose synthase genes in allotetraploid Brassica juncea. Gene 2019, 707, 126–135. [Google Scholar] [CrossRef]
- Xiao, X.; Tang, C.; Fang, Y.; Yang, M.; Zhou, B.; Qi, J.; Zhang, Y. Structure and expression profile of the sucrose synthase gene family in the rubber tree: Indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014, 281, 291–305. [Google Scholar] [CrossRef]
- Hirose, T.; Scofield, G.N.; Terao, T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008, 174, 534–543. [Google Scholar] [CrossRef]
- Poovaiah, C.R.; Mazarei, M.; Decker, S.R.; Turner, G.B.; Sykes, R.W.; Davis, M.F.; Stewart, C.N., Jr. Transgenic switchgrass (Panicum virgatum L.) biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol. J. 2015, 10, 552–563. [Google Scholar] [CrossRef]
- Xu, S.M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.L. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Baroja-Fernandez, E.; Bahaji, A.; Munoz, F.J.; Ovecka, M.; Montero, M.; Sesma, M.T.; Alonso-Casajus, N.; Almagro, G.; Sanchez-Lopez, A.M.; et al. Enhancing sucrose synthase activity results in increased levels of starch and ADP-glucose in maize (Zea mays L.) seed endosperms. Plant Cell Physiol. 2013, 54, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Qu, Z.; Zhang, L.; Zhao, S.; Bi, Z.; Ji, X.; Wang, X.; Wei, H. Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height. PLoS ONE 2015, 10, e0120669. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L.; Llewellyn, D.J.; Liu, Q.; Xu, S.M.; Wu, L.M.; Wang, L.; Furbank, R.T. Expression of sucrose synthase in the developing endosperm is essential for early seed development in cotton. Funct. Plant Biol. 2008, 35, 382–393. [Google Scholar] [CrossRef]
- Gerber, L.; Zhang, B.; Roach, M.; Rende, U.; Gorzsas, A.; Kumar, M.; Burgert, I.; Niittyla, T.; Sundberg, B. Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers. New Phytol. 2014, 203, 1220–1230. [Google Scholar] [CrossRef]
- Barratt, D.H.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef]
- Roitsch, T.; Gonzalez, M.C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef]
- Haouazine-Takvorian, N.; Tymowska-Lalanne, Z.; Takvorian, A.; Tregear, J.; Lejeune, B.; Lecharny, A.; Kreis, M. Characterization of two members of the Arabidopsis thaliana gene family, Atβfruct3 and Atβfruct4, coding for vacuolar invertases. Gene 1997, 197, 239–251. [Google Scholar] [CrossRef]
- Sherson, S.M.; Alford, H.L.; Forbes, S.M.; Wallace, G.; Smith, S.M. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 2003, 54, 525–531. [Google Scholar] [CrossRef]
- Ji, X.; van den Ende, W.; van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Bocock, P.N.; Morse, A.M.; Dervinis, C.; Davis, J.M. Evolution and diversity of invertase genes in Populus trichocarpa. Planta 2008, 227, 565–576. [Google Scholar] [CrossRef]
- Canam, T.; Park, J.Y.; Yu, K.Y.; Campbell, M.M.; Ellis, D.D.; Mansfield, S.D. Varied growth, biomass and cellulose content in tobacco expressing yeast-derived invertases. Planta 2006, 224, 1315–1327. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.R.; Lian, H.; Ni, D.A.; He, Y.K.; Chen, X.Y.; Ruan, Y.L. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiol. 2010, 154, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cook, A.; Patrick, J.W.; Chen, X.Y.; Ruan, Y.L. Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 2014, 78, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Barnes, W.J.; Anderson, C.T. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J. 2018, 94, 956–974. [Google Scholar] [CrossRef] [PubMed]
- Welham, T.; Pike, J.; Horst, I.; Flemetakis, E.; Katinakis, P.; Kaneko, T.; Sato, S.; Tabata, S.; Perry, J.; Parniske, M.; et al. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J. Exp. Bot. 2009, 60, 3353–3365. [Google Scholar] [CrossRef] [PubMed]
- Rende, U.; Wang, W.; Gandla, M.L.; Jonsson, L.J.; Niittyla, T. Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol. 2017, 214, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.; Rauh, B.L.; Xia, X.; Kandasamy, M.; Meagher, R.B.; Sheen, J.; Moore, B.D. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 2008, 228, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Kandel-Kfir, M.; Damari-Weissler, H.; German, M.A.; Gidoni, D.; Mett, A.; Belausov, E.; Petreikov, M.; Adir, N.; Granot, D. Two newly identified membrane-associated and plastidic tomato HXKs: Characteristics, predicted structure and intracellular localization. Planta 2006, 224, 1341–1352. [Google Scholar] [CrossRef]
- Giese, J.O.; Herbers, K.; Hoffmann, M.; Klosgen, R.B.; Sonnewald, U. Isolation and functional characterization of a novel plastidic hexokinase from Nicotiana tabacum. FEBS Lett. 2005, 579, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Ryoo, N.; Ko, S.; Lee, S.K.; Lee, J.; Jung, K.H.; Lee, Y.H.; Bhoo, S.H.; Winderickx, J.; An, G.; et al. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 2006, 224, 598–611. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, Y.; Zhang, Q.; Wu, R.; Wang, X.; Feng, S.; Chen, S.; Lu, C.; Du, L. Genome-wide identification and characterization of hexokinase genes in moso bamboo (Phyllostachys edulis). Front. Plant Sci. 2020, 11, 600. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Ding, L.; Wei, J. Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 2014, 41, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Qi, K.; Yi, X.; Chen, G.; Liu, X.; Qi, X.; Zhang, S. Identification of hexokinase family members in pear (Pyrus × bretschneideri) and functional exploration of PbHXK1 in modulating sugar content and plant growth. Gene 2019, 711, 143932. [Google Scholar] [CrossRef] [PubMed]
- Dai, N.; Schaffer, A.; Petreikov, M.; Shahak, Y.; Giller, Y.; Ratner, K.; Levine, A.; Granot, D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 1999, 11, 1253–1266. [Google Scholar] [CrossRef]
- Kim, M.; Lim, J.H.; Ahn, C.S.; Park, K.; Kim, G.T.; Kim, W.T.; Pai, H.S. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 2006, 18, 2341–2355. [Google Scholar] [CrossRef]
- Cho, J.I.; Ryoo, N.; Eom, J.S.; Lee, D.W.; Kim, H.B.; Jeong, S.W.; Lee, Y.H.; Kwon, Y.K.; Cho, M.H.; Bhoo, S.H.; et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009, 149, 745–759. [Google Scholar] [CrossRef]
- Herbert, M.; Burkhard, C.; Schnarrenberger, C. Survey for isoenzymes of glucosephosphate isomerase, phosphoglucomutase, glucose-6-phosphatedehydrogenase and 6-phosphogluconate dehydrogenase in C3-, C4-metabolism and crassulacean-acid-metabolism plants, and green algae. Planta 1979, 145, 95–104. [Google Scholar] [CrossRef]
- Gottlieb, L.D. Conservation and duplication of isozymes in plants. Science 1982, 216, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.R.; McHale, N.A. A starchless mutant of nicotiana sylvestris containing a modified plastid phosphoglucomutase. Plant Physiol. 1988, 88, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Egli, B.; Kolling, K.; Kohler, C.; Zeeman, S.C.; Streb, S. Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol. 2010, 154, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Muhlbach, H.; Schnarrenberger, C. Properties and intracellular distribution of two phosphoglucomutases from spinach leaves. Planta 1978, 141, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Tauberger, E.; Lytovchenko, A.; Roessner, U.; Willmitzer, L.; Trethewey, R.N. Antisense repression of cytosolic phosphoglucomutase in potato (Solanum tuberosum) results in severe growth retardation, reduction in tuber number and altered carbon metabolism. Planta 2002, 214, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sun, L.; Zhong, M.; Zhou, Q.; Gong, Z.; Li, P.; Tai, P.; Li, X. Cadmium-induced DNA damage and mutations in Arabidopsis plantlet shoots identified by DNA fingerprinting. Chemosphere 2012, 89, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Lytovchenko, A.; Sweetlove, L.; Pauly, M.; Fernie, A.R. The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 2002, 215, 1013–1021. [Google Scholar] [CrossRef]
- Malinova, I.; Kunz, H.H.; Alseekh, S.; Herbst, K.; Fernie, A.R.; Gierth, M.; Fettke, J. Reduction of the cytosolic phosphoglucomutase in Arabidopsis reveals impact on plant growth, seed and root development, and carbohydrate partitioning. PLoS ONE 2014, 9, e112468. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Smith, S.M.; Smith, A.M. The diurnal metabolism of leaf starch. Biochem. J. 2007, 401, 13–28. [Google Scholar]
- Turesson, H.; Andersson, M.; Marttila, S.; Thulin, I.; Hofvander, P. Starch biosynthetic genes and enzymes are expressed and active in the absence of starch accumulation in sugar beet tap-root. BMC Plant Biol. 2014, 14, 104. [Google Scholar] [CrossRef]
- Caspar, T.; Huber, S.C.; Somerville, C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985, 79, 11–17. [Google Scholar] [CrossRef]
- Tauberger, E.; Fernie, A.R.; Emmermann, M.; Renz, A.; Kossmann, J.; Willmitzer, L.; Trethewey, R.N. Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J. 2000, 23, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Ishimizu, T.; Suwabe, K.; Sudo, K.; Masuko, H.; Hakozaki, H.; Nou, I.S.; Suzuki, G.; Watanabe, M. UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 981–996. [Google Scholar] [CrossRef]
- Katsube, T.; Kazuta, Y.; Mori, H.; Nakano, K.; Tanizawa, K.; Fukui, T. UDP-glucose pyrophosphorylase from potato tuber: cDNA cloning and sequencing. J. Biochem. 1990, 108, 321–326. [Google Scholar] [CrossRef]
- Abe, T.; Niiyama, H.; Sasahara, T. Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor. Appl. Genet. 2002, 105, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Pua, E.C.; Lim, S.W.; Liu, P.; Liu, J.Z. Expression of a UDP glucose pyrophosphorylase cDNA during fruit ripening of banana (Musa acuminata). Funct. Plant Biol. 2000, 27, 1151–1159. [Google Scholar] [CrossRef]
- Zrenner, R.; Willmitzer, L.; Sonnewald, U. Analysis of the expression of potato uridinediphosphate-glucose pyrophosphorylase and its inhibition by antisense RNA. Planta 1993, 190, 247–252. [Google Scholar] [CrossRef]
- Spychalla, J.P.; Scheffler, B.E.; Sowokinos, J.R.; Bevan, M.W. Cloning, antisense RNA inhibition and the coordinated expression of UDP-glucose pyrophosphorylase with starch biosynthetic genes in potato tubers. J. Plant Physiol. 1994, 144, 444–453. [Google Scholar] [CrossRef]
- Borovkov, A.Y.; Mcclean, P.E.; Sowokinos, J.R.; Ruud, S.H.; Secor, G.A. Effect of expression of UDP-glucose pyrophosphorylase ribozyme and antisense RNAs on the enzyme activity and carbohydrate composition of field-grown transgenic potato plants. J. Plant Physiol. 1996, 147, 644–652. [Google Scholar] [CrossRef]
- Coleman, H.D.; Ellis, D.D.; Gilbert, M.; Mansfield, S.D. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol. J. 2006, 4, 87–101. [Google Scholar] [CrossRef]
- Chen, R.; Zhao, X.; Shao, Z.; Wei, Z.; Wang, Y.; Zhu, L.; Zhao, J.; Sun, M.; He, R.; He, G. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 2007, 19, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Woo, M.O.; Ham, T.H.; Ji, H.S.; Choi, M.S.; Jiang, W.; Chu, S.H.; Piao, R.; Chin, J.H.; Kim, J.A.; Park, B.S.; et al. Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). Plant J. 2008, 54, 190–204. [Google Scholar] [CrossRef]
- Sahrawy, M.; Avila, C.; Chueca, A.; Canovas, F.M.; Lopez-Gorge, J. Increased sucrose level and altered nitrogen metabolism in Arabidopsis thaliana transgenic plants expressing antisense chloroplastic fructose-1,6-bisphosphatase. J. Exp. Bot. 2004, 55, 2495–2503. [Google Scholar] [CrossRef]
- Gao, P.; Xin, Z.; Zheng, Z.L. The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE 2008, 3, e1387. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef] [PubMed]
- Euring, D.; Bai, H.; Janz, D.; Polle, A. Nitrogen-driven stem elongation in poplar is linked with wood modification and gene clusters for stress, photosynthesis and cell wall formation. BMC Plant Biol. 2014, 14, 391. [Google Scholar] [CrossRef]
- Araya, T.; Noguchi, K.; Terashima, I. Effect of nitrogen nutrition on the carbohydrate repression of photosynthesis in leaves of Phaseolus vulgaris L. J. Plant Res. 2010, 123, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Canton, F.R.; Suarez, M.F.; Canovas, F.M. Molecular aspects of nitrogen mobilization and recycling in trees. Photosynth. Res. 2005, 83, 265–278. [Google Scholar] [CrossRef]
- Luo, Z.B.; Calfapietra, C.; Liberloo, M.; Scarascia-Mugnozza, G.; Polle, A. Carbon partitioning to mobile and structural fractions in poplar wood under elevated CO2 (EUROFACE) and N fertilization. Glob. Change Biol. 2010, 12, 272–283. [Google Scholar] [CrossRef]
- Castro-Rodriguez, V.; Garcia-Gutierrez, A.; Canales, J.; Canas, R.A.; Kirby, E.G.; Avila, C.; Canovas, F.M. Poplar trees for phytoremediation of high levels of nitrate and applications in bioenergy. Plant Biotechnol. J. 2016, 14, 299–312. [Google Scholar] [CrossRef]
- Sun, G.; Zhao, P.; Zeng, X.; Peng, S. Influence of elevated atmospheric CO2 concentration on photosynthesis and leaf nitrogen partition in process of photosynthetic carbon cycle in Musa paradisiaca. Ying Yong Sheng Tai Xue Bao 2001, 12, 429–434. [Google Scholar] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- van Raemdonck D, Pesquet E, Cloquet S, Beeckman H, Boerjan W, Goffner D, El Jaziri M, Baucher M: Molecular changes associated with the setting up of secondary growth in aspen. J. Exp. Bot. 2005, 56, 2211–2227. [CrossRef]
- Katz, M.E.; Masoumi, A.; Burrows, S.R.; Shirtliff, C.G.; Cheetham, B.F. The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of extracellular proteases. Genetics 2000, 156, 1559–1571. [Google Scholar] [CrossRef]
- Chen, J.; Qu, C.; Chang, R.; Suo, J.; Yu, J.; Sun, X.; Liu, G.; Xu, Z. Genome-wide identification of BXL genes in Populus trichocarpa and their expression under different nitrogen treatments. 3 Biotech 2020, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Redfern, O.C.; Dessailly, B.H.; Dallman, T.J.; Sillitoe, I.; Orengo, C.A. FLORA: A novel method to predict protein function from structure in diverse superfamilies. PLoS Comput. Biol. 2009, 5, e1000485. [Google Scholar] [CrossRef]
- Kraiser, T.; Gras, D.E.; Gutierrez, A.G.; Gonzalez, B.; Gutierrez, R.A. A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 2011, 62, 1455–1466. [Google Scholar] [CrossRef]
- Huber, S.C.; Huber, J.L.; Liao, P.C.; Gage, D.A.; McMichael, R.W., Jr.; Chourey, P.S.; Hannah, L.C.; Koch, K. Phosphorylation of serine-15 of maize leaf sucrose synthase. Occurrence in vivo and possible regulatory significance. Plant Physiol. 1996, 112, 793–802. [Google Scholar] [CrossRef]
- Bork, P.; Sander, C.; Valencia, A. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993, 2, 31–40. [Google Scholar] [CrossRef]
- Periappuram, C.; Steinhauer, L.; Barton, D.L.; Taylor, D.C.; Chatson, B.; Zou, J. The plastidic phosphoglucomutase from Arabidopsis. A reversible enzyme reaction with an important role in metabolic control. Plant Physiol. 2000, 122, 1193–1199. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Han, M.; Min, J.; Chen, P.; Mao, Y.; Huang, Q.; Tong, Q.; Liu, Q.; Fang, Y. Genome-wide survey of invertase encoding genes and functional characterization of an extracellular fungal pathogen-responsive invertase in Glycine max. Int. J. Mol. Sci. 2018, 19, 2395. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Yang, L.; Zhu, H.; Zhou, J.; Gong, H. Structure and expression analysis of sucrose phosphate synthase, sucrose synthase and invertase gene families in Solanum lycopersicum. Int. J. Mol. Sci. 2021, 22, 4698. [Google Scholar] [CrossRef]
- Xu, X.; Yang, Y.; Liu, C.; Sun, Y.; Zhang, T.; Hou, M. The evolutionary history of the sucrose synthase gene family in higher plants. BMC Plant Biol. 2019, 19, 566. [Google Scholar] [CrossRef]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.B.; Yao, Y.; He, H.; Qin, Y.L.; Liu, Z.J.; Liu, W.X.; Qi, Z.Q.; Yang, L.J.; Cao, Z.M.; Yang, Y. Genome-wide identification, expression, and functional analysis of the alkaline/neutral invertase gene family in pepper. Int. J. Mol. Sci. 2018, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.G.; Kodama, R.; Wang, H.; Ichii, M.; Taketa, S.; Yoshida, H. Analysis of the rice SHORT-ROOT5 gene revealed functional diversification of plant neutral/alkaline invertase family. Plant Sci. 2009, 176, 627–634. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, K.; Su, X.; Rao, P.; An, X. Genome-wide identification of the invertase gene family in Populus. PLoS ONE 2015, 10, e0138540. [Google Scholar]
- Guo, X.; Chen, H.; Liu, Y.; Chen, W.; Ying, Y.; Han, J.; Gui, R.; Zhang, H. The acid invertase gene family is involved in internode elongation in Phyllostachys heterocycla cv. pubescens. Tree Physiol. 2020, 40, 1217–1231. [Google Scholar] [CrossRef]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Lou, Y.; Gou, J.Y.; Xue, H.W. PIP5K9, an phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 2007, 19, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Sorek, H.; Wemmer, D.E.; Somerville, C.R. Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol. 2016, 171, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, S.; Zhang, H.; Cao, L.; Chang, R.; Liu, Z.; Zhang, H.; Xu, Z.; Liu, G. Identification and expression pattern analysis of PtCarA and PtCarB genes in Populus trichocarpa under different nitrogen treatments. Plant Biol. 2023, 25, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- McKown, A.D.; Guy, R.D.; Klápste, J.; Geraldes, A.; Friedmann, M.; Cronk, Q.C.B.; El-Kassaby, Y.A.; Mansfield, S.D.; Douglas, C.J. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 2014, 201, 1263–1276. [Google Scholar] [CrossRef]
- Liu, B.; Rennenberg, H.; Kreuzwieser, J. Hypoxia affects nitrogen uptake and distribution in young poplar (Populus × canescens) Trees. PLoS ONE 2015, 10, e0136579. [Google Scholar] [CrossRef]
- Kim, K.W.; Oh, C.Y.; Lee, J.C.; Lee, S.; Kim, P.G. Alteration of leaf surface structures of poplars under elevated air temperature and carbon dioxide concentration. Appl. Microsc. 2013, 43, 110–116. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, H.; Qu, C.; Xu, Z.; Li, W.; Hao, B.; Yang, C.; Sun, G.; Liu, G. Sequence and expression analysis of the AMT gene family in poplar. Front. Plant Sci. 2015, 6, 337. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chengjie, C.; Rui, X.; Hao, C.; Yehua, H. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Foster, C.E.; Martin, T.M.; Pauly, M. Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: Lignin. J. Vis. Exp. 2010, 37, e1745. [Google Scholar]
- Foster, C.E.; Martin, T.M.; Pauly, M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: Carbohydrates. J. Vis. Exp. 2010, 37, e1837. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Wang, W.; Chang, R.; Yu, J.; Yan, J.; Yu, W.; Li, C.; Xu, Z. Structure and Expression Analysis of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Gene Families in Populus trichocarpa Torr. and Gray. Int. J. Mol. Sci. 2023, 24, 17277. https://doi.org/10.3390/ijms242417277
Zhang S, Wang W, Chang R, Yu J, Yan J, Yu W, Li C, Xu Z. Structure and Expression Analysis of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Gene Families in Populus trichocarpa Torr. and Gray. International Journal of Molecular Sciences. 2023; 24(24):17277. https://doi.org/10.3390/ijms242417277
Chicago/Turabian StyleZhang, Shuang, Wenjie Wang, Ruhui Chang, Jiajie Yu, Junxin Yan, Wenxi Yu, Chunming Li, and Zhiru Xu. 2023. "Structure and Expression Analysis of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Gene Families in Populus trichocarpa Torr. and Gray" International Journal of Molecular Sciences 24, no. 24: 17277. https://doi.org/10.3390/ijms242417277
APA StyleZhang, S., Wang, W., Chang, R., Yu, J., Yan, J., Yu, W., Li, C., & Xu, Z. (2023). Structure and Expression Analysis of PtrSUS, PtrINV, PtrHXK, PtrPGM, and PtrUGP Gene Families in Populus trichocarpa Torr. and Gray. International Journal of Molecular Sciences, 24(24), 17277. https://doi.org/10.3390/ijms242417277






