Fluorescence of the Retinal Chromophore in Microbial and Animal Rhodopsins
Abstract
:1. Introduction
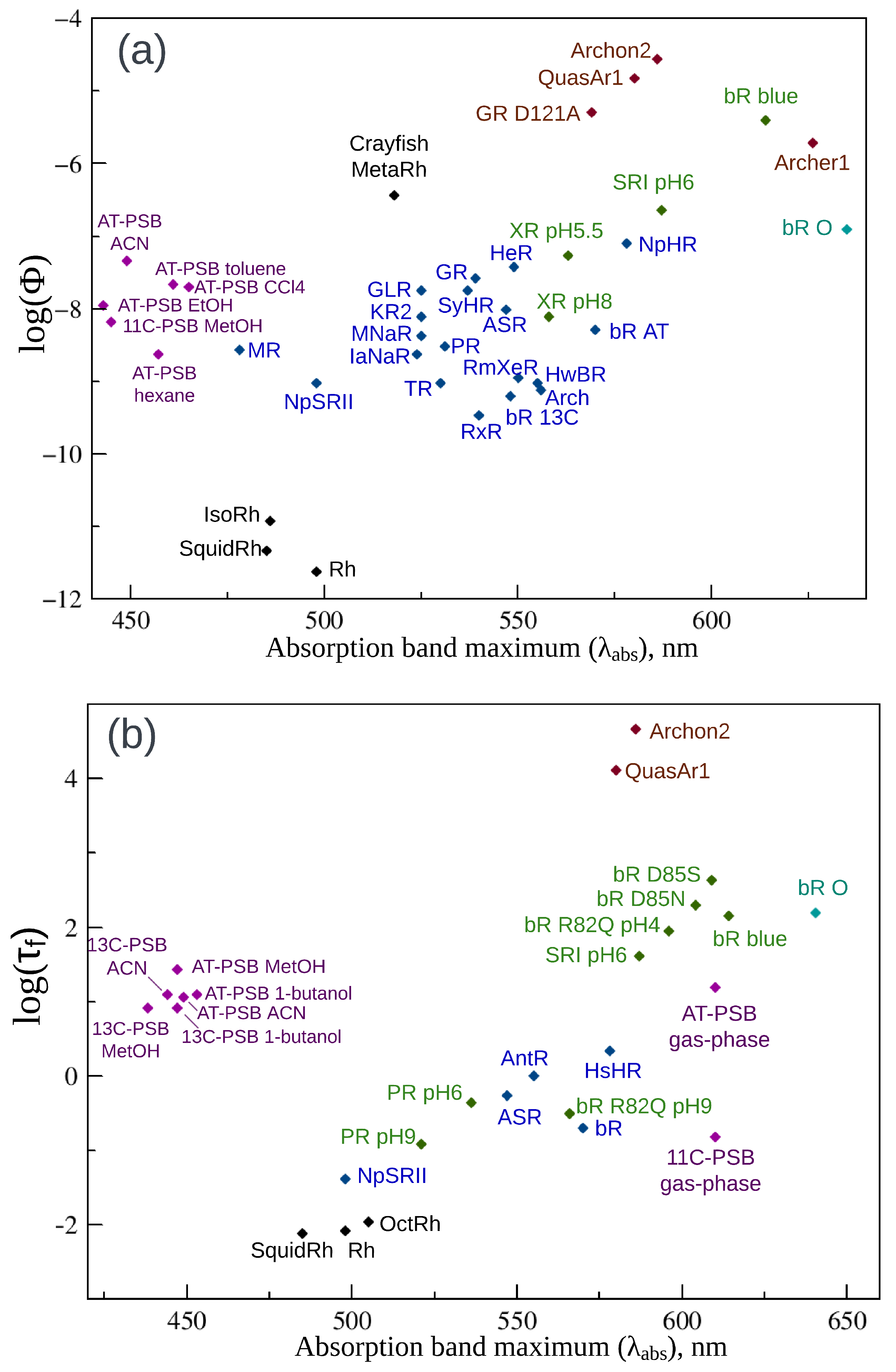
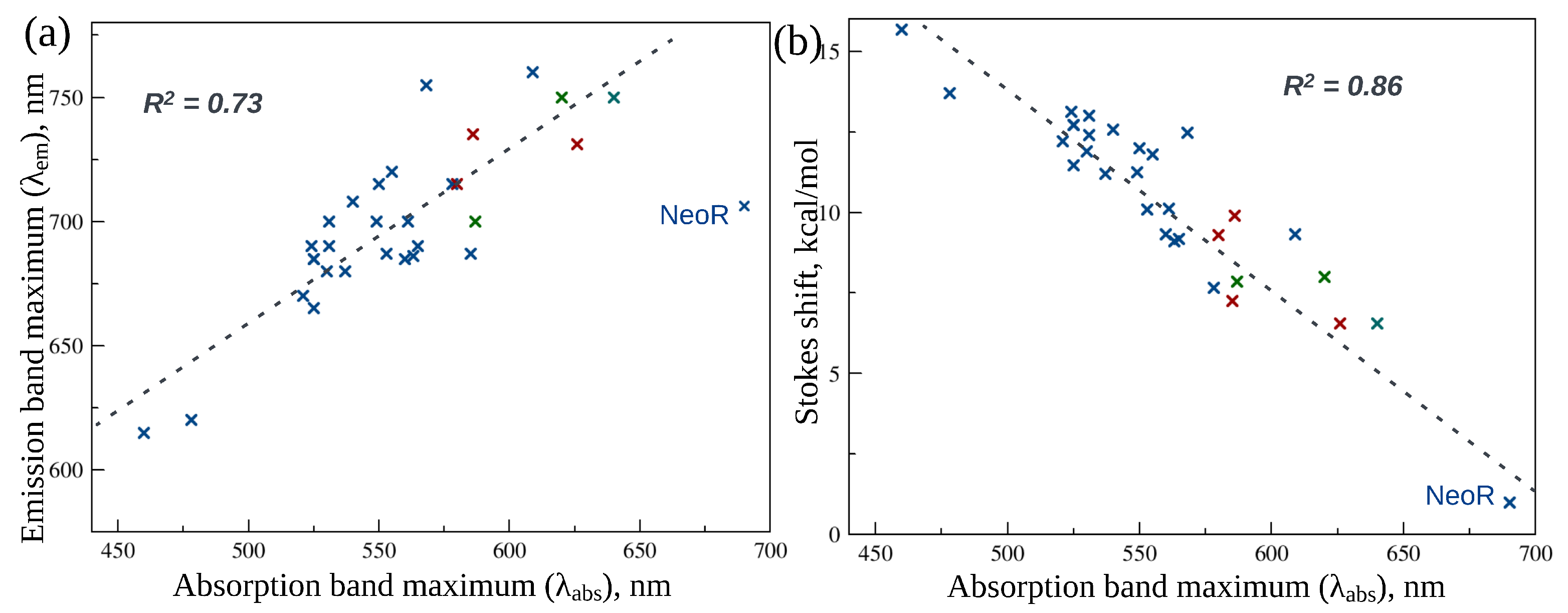
2. Fluorescence Properties of Microbial Rhodopsins
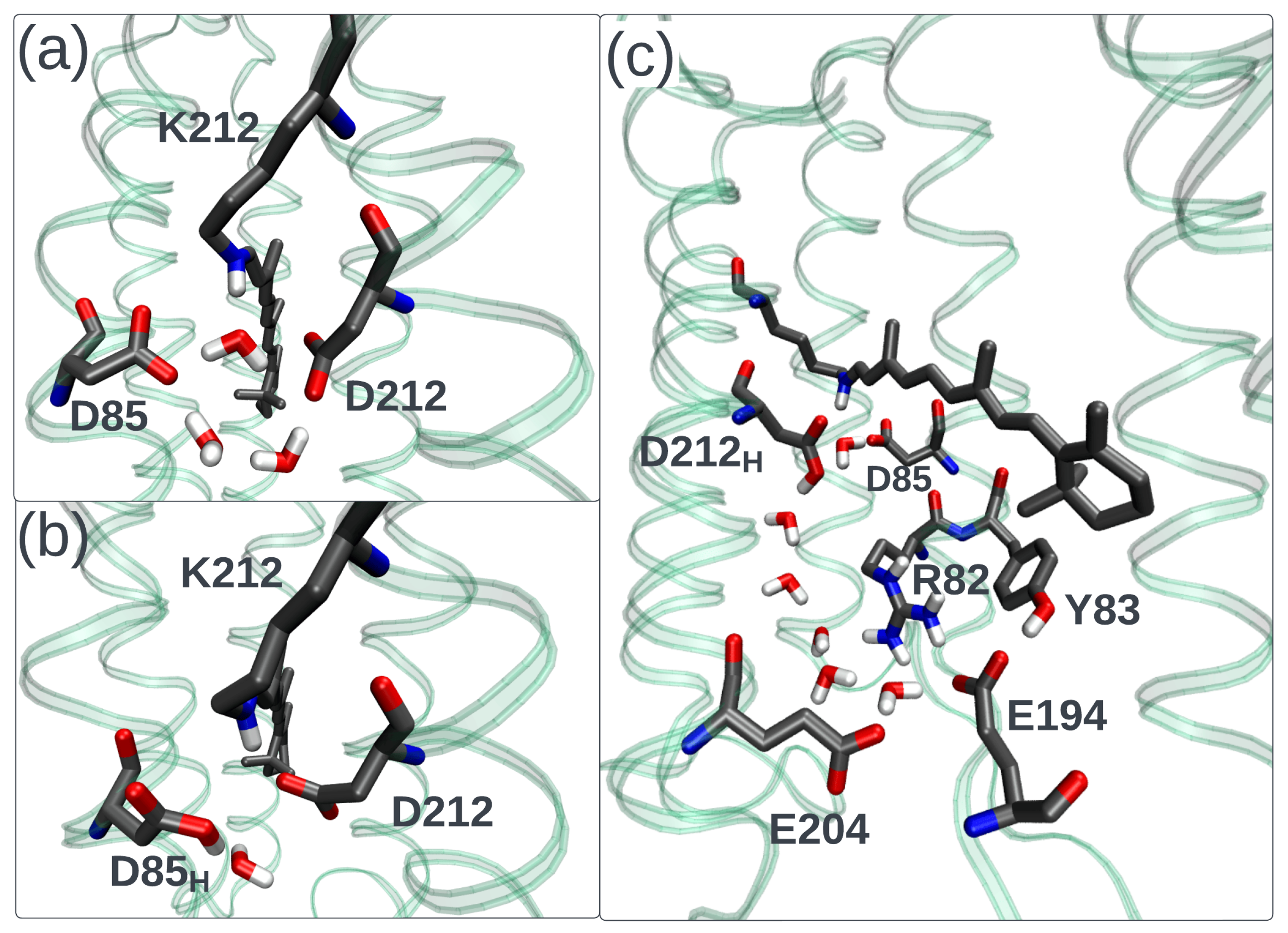
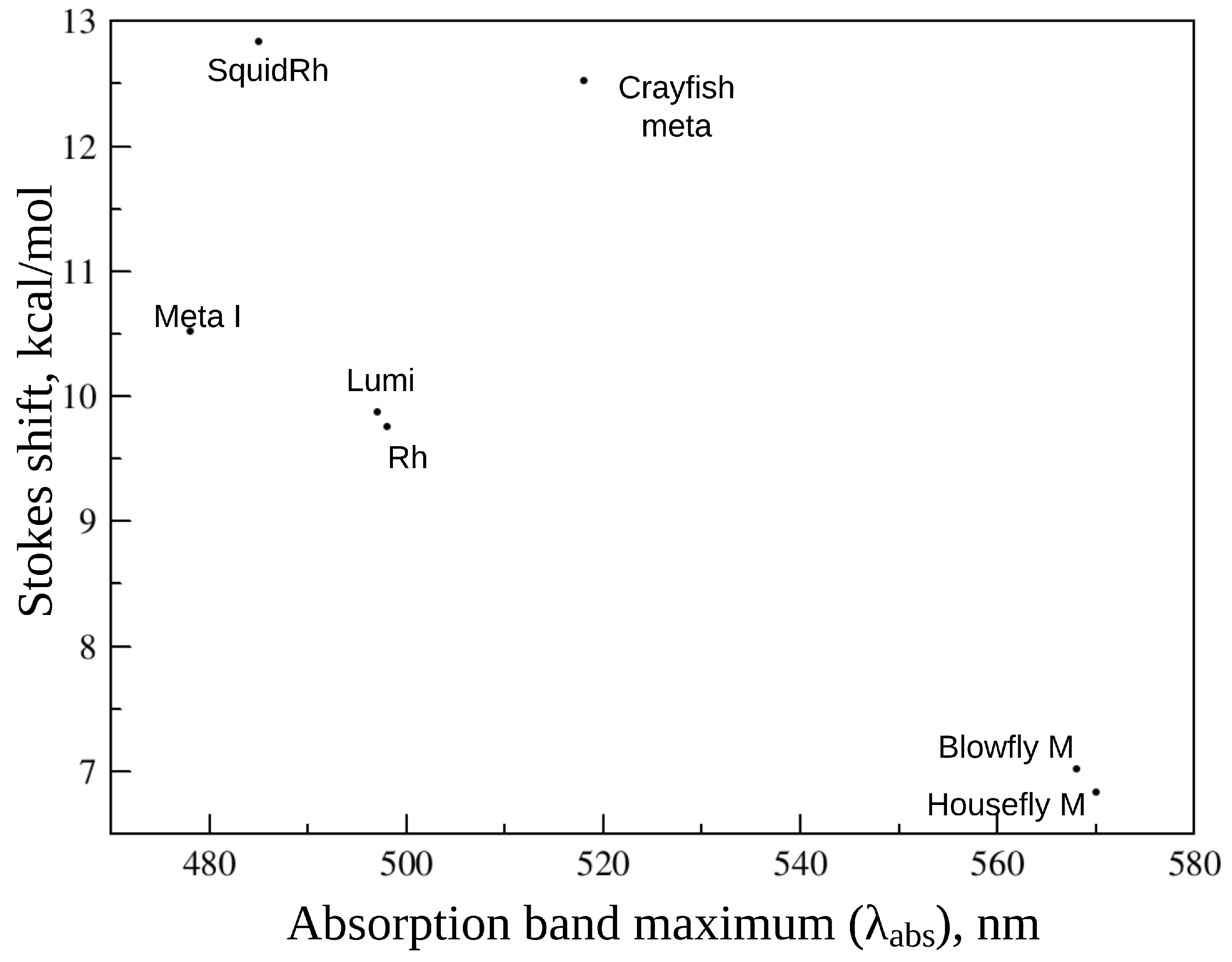
3. Animal Opsin-Based Photoreceptors
| Protein | , nm | , nm | , nm | Stokes Shift, nm | , ps | |
|---|---|---|---|---|---|---|
| Rh | 498 [76] | - | 600 [76] | 102 | [76] [78] | 0.05 [78] 0.146, 1.5 [75] 0.125, 1 [83] |
| IsoRh | 485 [78] | - | ≈620 [78] | 135 | [78] | - |
| SquidRh | 485 [76] | - | 620 [76] | 135 | [76] | 0.12 [76] |
| OctRh | 505 [82] | - | - | - | - | 0.14 [82] |
| Batho | 543 [84] | - | - | - | <10 [84] | - |
| Lumi | 497 [77] | - | 600 | 103 | <10 [77] | - |
| MetaI | 478 [77] | - | 580 | 102 | <10 [77] | - |
| MetaII | 380 [77] | - | 500–515 | 120 | <10 [77] | - |
| Crayfish meta | - | 518 [81] | 660–670 [81] | 142 | [81] | - |
| Blowfly meta | 584 (M) 568 (M) [85] | - | 660 [85] | 92 | (M)> 3(M) [85] | - |
| Housefly meta | 580 (M) 570 (M) [86] | - | 660 [86] | 90 | - | - |
| Droso meta | - | 570 [87] | >646 [87] | - | - | - |
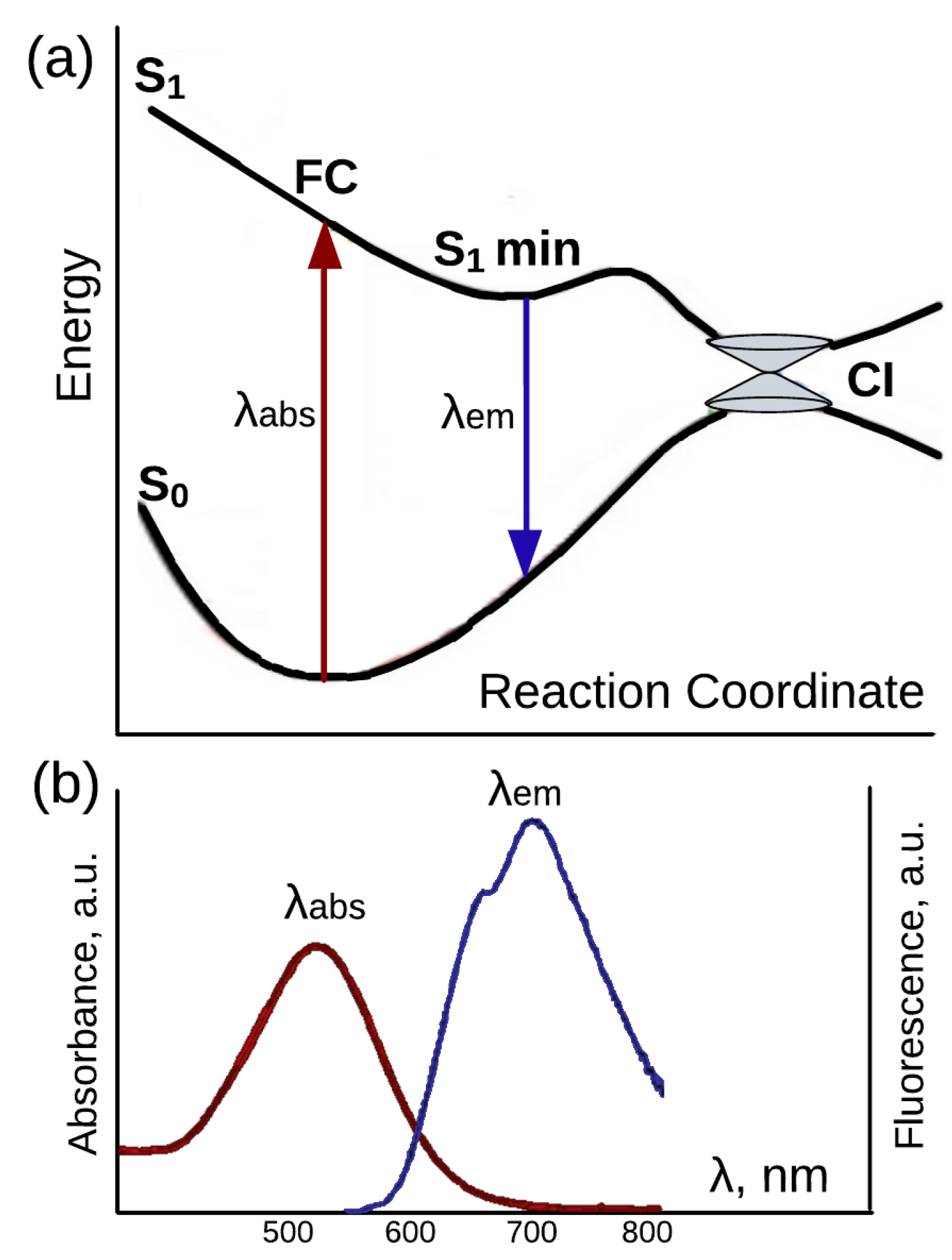
4. Retinal Protonated Schiff Based in the Gas Phase and Solvents
5. Neorhodopsin
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagata, T.; Inoue, K. Rhodopsins at a glance. J. Cell Sci. 2021, 134, jcs258989. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, A.; Inoue, K.; Kandori, H.; Béjà, O. Microbial rhodopsins: The last two decades. Annu. Rev. Microbiol. 2021, 75, 427–447. [Google Scholar] [CrossRef]
- Bratanov, D.; Kovalev, K.; Machtens, J.P.; Astashkin, R.; Chizhov, I.; Soloviov, D.; Volkov, D.; Polovinkin, V.; Zabelskii, D.; Mager, T.; et al. Unique structure and function of viral rhodopsins. Nat. Commun. 2019, 10, 4939. [Google Scholar] [CrossRef] [PubMed]
- Andrew, S.M.; Moreno, C.M.; Plumb, K.; Hassanzadeh, B.; Gomez-Consarnau, L.; Smith, S.N.; Schofield, O.; Yoshizawa, S.; Fujiwara, T.; Sunda, W.G.; et al. Widespread use of proton-pumping rhodopsin in Antarctic phytoplankton. Proc. Natl. Acad. Sci. USA 2023, 120, e2307638120. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K. Diversity, mechanism, and optogenetic application of light-driven ion pump rhodopsins. In Optogenetics: Light-Sensing Proteins and Their Applications in Neuroscience and Beyond; Springer: Singapore, 2021; pp. 89–126. [Google Scholar]
- De Grip, W.J.; Ganapathy, S. Rhodopsins: An excitingly versatile protein species for research, development and creative engineering. Front. Chem. 2022, 10, 879609. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K. Photochemistry of the Retinal Chromophore in Microbial Rhodopsins. J. Phys. Chem. B 2023, 127, 9215–9222. [Google Scholar] [CrossRef] [PubMed]
- Ostrovsky, M.A.; Smitienko, O.A.; Bochenkova, A.V.; Feldman, T.B. Similarities and Differences in Photochemistry of Type I and Type II Rhodopsins. Biochemistry 2023, 88, 1528–1543. [Google Scholar] [CrossRef]
- Sumita, M.; Ryazantsev, M.N.; Saito, K. Acceleration of the Z to E photoisomerization of penta-2, 4-dieniminium by hydrogen out-of-plane motion: Theoretical study on a model system of retinal protonated Schiff base. Phys. Chem. Chem. Phys. 2009, 11, 6406–6414. [Google Scholar] [CrossRef]
- Sugiura, M.; Ishikawa, K.; Katayama, K.; Sumii, Y.; Abe-Yoshizumi, R.; Tsunoda, S.P.; Furutani, Y.; Shibata, N.; Brown, L.S.; Kandori, H. Unusual photoisomerization pathway in a near-Infrared light absorbing enzymerhodopsin. J. Phys. Chem. Lett. 2022, 13, 9539–9543. [Google Scholar] [CrossRef]
- Broser, M. Far-red absorbing rhodopsins, insights from heterodimeric rhodopsin-cyclases. Front. Mol. Biosci. 2022, 8, 806922. [Google Scholar] [CrossRef]
- Broser, M.; Busse, W.; Spreen, A.; Reh, M.; Bernal Sierra, Y.A.; Hwang, S.; Utesch, T.; Sun, H.; Hegemann, P. Diversity of rhodopsin cyclases in zoospore-forming fungi. Proc. Natl. Acad. Sci. USA 2023, 120, e2310600120. [Google Scholar] [CrossRef] [PubMed]
- Lenz, M.O.; Huber, R.; Schmidt, B.; Gilch, P.; Kalmbach, R.; Engelhard, M.; Wachtveitl, J. First steps of retinal photoisomerization in proteorhodopsin. Biophys. J. 2006, 91, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.Z.; Kim, D.K.; Jennings, J.H.; Tian, H.; Wang, P.Y.; Ramakrishnan, C.; Randles, S.; Sun, Y.; Thadhani, E.; Kim, Y.S.; et al. All-optical physiology resolves a synaptic basis for behavioral timescale plasticity. Cell 2023, 186, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.P.; Brinks, D.; Testa-Silva, G.; Tian, H.; Phil Brooks, F., III; Adam, Y.; Bloxham, B.; Gmeiner, B.; Kheifets, S.; Cohen, A.E. Photoactivated voltage imaging in tissue with an archaerhodopsin-derived reporter. Sci. Adv. 2021, 7, eabe3216. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Boyden, E.S. Optogenetic Control of Neural Activity: The Biophysics of Microbial rhodopsins in Neuroscience. Q. Rev. Biophys. 2023, 1–81. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, M.; Naskar, S.; Garcia, J.P.; Sommer, M.; Kim, E.; Adam, Y.; Haydon, P.G.; Boyden, E.S.; Cohen, A.E.; Dulla, C.G. Neuronal activity drives pathway-specific depolarization of peripheral astrocyte processes. Nat. Neurosci. 2022, 25, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Bensussen, S.; Tseng, H.A.; Shroff, S.N.; Lopez-Huerta, V.G.; Park, D.; Jung, E.E.; Shemesh, O.A.; Straub, C.; Gritton, H.J.; et al. Population imaging of neural activity in awake behaving mice. Nature 2019, 574, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Moreaux, L.C.; Yatsenko, D.; Sacher, W.D.; Choi, J.; Lee, C.; Kubat, N.J.; Cotton, R.J.; Boyden, E.S.; Lin, M.Z.; Tian, L.; et al. Integrated neurophotonics: Toward dense volumetric interrogation of brain circuit activity—At depth and in real time. Neuron 2020, 108, 66–92. [Google Scholar] [CrossRef]
- Zhang, X.M.; Yokoyama, T.; Sakamoto, M. Imaging voltage with microbial rhodopsins. Front. Mol. Biosci. 2021, 8, 738829. [Google Scholar] [CrossRef]
- Nikolaev, D.M.; Mironov, V.N.; Shtyrov, A.A.; Kvashnin, I.D.; Mereshchenko, A.S.; Vasin, A.V.; Panov, M.S.; Ryazantsev, M.N. Fluorescence Imaging of Cell Membrane Potential: From Relative Changes to Absolute Values. Int. J. Mol. Sci. 2023, 24, 2435. [Google Scholar] [CrossRef]
- Meng, X.; Ganapathy, S.; van Roemburg, L.; Post, M.; Brinks, D. Voltage Imaging with Engineered Proton-Pumping Rhodopsins: Insights from the Proton Transfer Pathway. ACS Phys. Chem. Au 2023, 3, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Silapetere, A.; Hwang, S.; Hontani, Y.; Fernandez Lahore, R.G.; Balke, J.; Escobar, F.V.; Tros, M.; Konold, P.E.; Matis, R.; Croce, R.; et al. QuasAr Odyssey: The origin of fluorescence and its voltage sensitivity in microbial rhodopsins. Nat. Commun. 2022, 13, 5501. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Kurihara, R.; Sakamoto, M.; Takanashi, T.; Kuramochi, H.; Zhang, X.M.; Bito, H.; Tahara, T.; Sudo, Y. Comparative studies of the fluorescence properties of microbial rhodopsins: Spontaneous emission versus photointermediate fluorescence. J. Phys. Chem. B 2020, 124, 7361–7367. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, N.; Ishikawa, M.; Kasahara, K.; Kaneko, M.; Yamamoto, N.; Ohtani, H. Picosecond fluorescence spectroscopy of the purple membrane of Halobacterium halobium in alkaline suspension. Chem. Phys. Lett. 1997, 265, 595–599. [Google Scholar] [CrossRef]
- Ohtani, H.; Ishikawa, M.; Itoh, H.; Takiguchi, Y.; Urakami, T.; Tsuchiya, Y. Picosecond fluorescence spectroscopy of purple membrane in Halobacterium halobium with a photon-counting streak camera. Chem. Phys. Lett. 1990, 168, 493–498. [Google Scholar] [CrossRef]
- Shapiro, S.; Campillo, A.; Lewis, A.; Perreault, G.; Spoonhower, J.; Clayton, R.; Stoeckenius, W. Picosecond and steady state, variable intensity and variable temperature emission spectroscopy of bacteriorhodopsin. Biophys. J. 1978, 23, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H.; Tsukamoto, Y.; Sakoda, Y.; Hamaguchi, H.o. Fluorescence spectra of bacteriorhodopsin and the intermediates O and Q at room temperature. FEBS Lett. 1995, 359, 65–68. [Google Scholar] [CrossRef]
- Kouyama, T.; Kinosita, K.; Ikegami, A. Excited-state dynamics of bacteriorhodopsin. Biophys. J. 1985, 47, 43–54. [Google Scholar] [CrossRef]
- Chang, C.F.; Kuramochi, H.; Singh, M.; Abe-Yoshizumi, R.; Tsukuda, T.; Kandori, H.; Tahara, T. A unified view on varied ultrafast dynamics of the primary process in microbial rhodopsins. Angew. Chem. Int. Ed. 2022, 61, e202111930. [Google Scholar] [CrossRef]
- Ohtani, H.; Kobayashi, T.; Iwai, J.; Ikegami, A. Picosecond and nanosecond spectroscopies of the photochemical cycles of acidified bacteriorhodopsin. Biochemistry 1986, 25, 3356–3363. [Google Scholar] [CrossRef]
- Kobayashi, T.; Terauchi, M.; Kouyama, T.; Yoshizawa, M.; Taiji, M. Femtosecond spectroscopy of acidified and neutral bacteriorhodopsin. In Proceedings of the Laser Applications in Life Sciences, Moscow, Russia, 1–31 August 1990; Volume 1403, pp. 407–416. [Google Scholar]
- Gillbro, T.; Kriebel, A.N. Emission from secondary intermediates in the photocycle of bacteriorhodopsin at 77° K. FEBS Lett. 1977, 79, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H.; Kaneko, M.; Ishikawa, M.; Kamiya, N.; Yamamoto, N. Picosecond-millisecond dual-time-base spectroscopy of fluorescent photointermediates formed in the purple membrane of Halobacterium halobium. Chem. Phys. Lett. 1999, 299, 571–575. [Google Scholar] [CrossRef]
- Kennis, J.T.; Larsen, D.S.; Ohta, K.; Facciotti, M.T.; Glaeser, R.M.; Fleming, G.R. Ultrafast protein dynamics of bacteriorhodopsin probed by photon echo and transient absorption spectroscopy. J. Phys. Chem. B 2002, 106, 6067–6080. [Google Scholar] [CrossRef]
- Song, L.; El-Sayed, M.; Lanyi, J. Protein catalysis of the retinal subpicosecond photoisomerization in the primary process of bacteriorhodopsin photosynthesis. Science 1993, 261, 891–894. [Google Scholar] [CrossRef]
- Kralj, J.M.; Douglass, A.D.; Hochbaum, D.R.; Maclaurin, D.; Cohen, A.E. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 2012, 9, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Penzkofer, A.; Silapetere, A.; Hegemann, P. Absorption and emission spectroscopic investigation of the thermal dynamics of the Archaerhodopsin 3 based fluorescent voltage sensor QuasAr1. Int. J. Mol. Sci. 2019, 20, 4086. [Google Scholar] [CrossRef] [PubMed]
- Hochbaum, D.R.; Zhao, Y.; Farhi, S.L.; Klapoetke, N.; Werley, C.A.; Kapoor, V.; Zou, P.; Kralj, J.M.; Maclaurin, D.; Smedemark-Margulies, N.; et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 2014, 11, 825–833. [Google Scholar] [CrossRef]
- Penzkofer, A.; Silapetere, A.; Hegemann, P. Absorption and emission spectroscopic investigation of the thermal dynamics of the Archaerhodopsin 3 based fluorescent voltage sensor Archon2. Int. J. Mol. Sci. 2020, 21, 6576. [Google Scholar] [CrossRef]
- McIsaac, R.S.; Engqvist, M.K.; Wannier, T.; Rosenthal, A.Z.; Herwig, L.; Flytzanis, N.C.; Imasheva, E.S.; Lanyi, J.K.; Balashov, S.P.; Gradinaru, V.; et al. Directed evolution of a far-red fluorescent rhodopsin. Proc. Natl. Acad. Sci. USA 2014, 111, 13034–13039. [Google Scholar] [CrossRef]
- Nikolaev, D.M.; Mironov, V.N.; Metelkina, E.M.; Shtyrov, A.A.; Mereshchenko, A.S.; Demidov, N.A.; Moskalenko, S.E.; Bondarev, S.A.; Zhouravleva, G.A.; Vasin, A.V.; et al. Rational design of far-red archaerhodopsin3-based fluorescent genetically-encoded voltage indicators: From elucidation of fluorescence mechanism in Archers to novel red-shifted variants. ACS Phys. Chem. Au 2023. submitted. [Google Scholar]
- Huber, R.; Köhler, T.; Lenz, M.O.; Bamberg, E.; Kalmbach, R.; Engelhard, M.; Wachtveitl, J. pH-dependent photoisomerization of retinal in proteorhodopsin. Biochemistry 2005, 44, 1800–1806. [Google Scholar] [CrossRef] [PubMed]
- Balashov, S.P.; Imasheva, E.S.; Wang, J.M.; Lanyi, J.K. Excitation energy-transfer and the relative orientation of retinal and carotenoid in xanthorhodopsin. Biophys. J. 2008, 95, 2402–2414. [Google Scholar] [CrossRef] [PubMed]
- Imasheva, E.S.; Balashov, S.P.; Choi, A.R.; Jung, K.H.; Lanyi, J.K. Reconstitution of Gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry 2009, 48, 10948–10955. [Google Scholar] [CrossRef] [PubMed]
- Engqvist, M.K.; McIsaac, R.S.; Dollinger, P.; Flytzanis, N.C.; Abrams, M.; Schor, S.; Arnold, F.H. Directed evolution of Gloeobacter violaceus rhodopsin spectral properties. J. Mol. Biol. 2015, 427, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Balashov, S.; Petrovskaya, L.; Lukashev, E.; Imasheva, E.; Dioumaev, A.; Wang, J.; Sychev, S.; Dolgikh, D.; Rubin, A.; Kirpichnikov, M.; et al. Aspartate–Histidine interaction in the retinal Schiff base counterion of the light-driven proton pump of Exiguobacterium sibiricum. Biochemistry 2012, 51, 5748–5762. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, S.; Meng, X.; Mossel, D.; Jagt, M.; Brinks, D. Expanding the family of genetically encoded voltage indicators with a candidate Heliorhodopsin exhibiting near-infrared fluorescence. J. Biol. Chem. 2023, 299, 104771. [Google Scholar] [CrossRef] [PubMed]
- Rozin, R.; Wand, A.; Jung, K.H.; Ruhman, S.; Sheves, M. pH dependence of Anabaena sensory rhodopsin: Retinal isomer composition, rate of dark adaptation, and photochemistry. J. Phys. Chem. B 2014, 118, 8995–9006. [Google Scholar] [CrossRef] [PubMed]
- Cheminal, A.; Léonard, J.; Kim, S.; Jung, K.H.; Kandori, H.; Haacke, S. Steady state emission of the fluorescent intermediate of Anabaena Sensory Rhodopsin as a function of light adaptation conditions. Chem. Phys. Lett. 2013, 587, 75–80. [Google Scholar] [CrossRef]
- Cheminal, A.; Léonard, J.; Kim, S.Y.; Jung, K.H.; Kandori, H.; Haacke, S. 100 fs photo-isomerization with vibrational coherences but low quantum yield in Anabaena Sensory Rhodopsin. Phys. Chem. Chem. Phys. 2015, 17, 25429–25439. [Google Scholar] [CrossRef]
- Kandori, H.; Yoshihara, K.; Tomioka, H.; Sasabe, H. Primary photochemical events in halorhodopsin studied by subpicosecond time-resolved spectroscopy. J. Phys. Chem. 1992, 96, 6066–6071. [Google Scholar] [CrossRef]
- Polland, H.J.; Franz, M.; Zinth, W.; Kaiser, W.; Hegemann, P.; Oesterhelt, D. Picosecond events in the photochemical cycle of the light-driven chloride-pump halorhodopsin. Biophys. J. 1985, 47, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Arlt, T.; Schmidt, S.; Zinth, W.; Haupts, U.; Oesterhelt, D. The initial reaction dynamics of the light-driven chloride pump halorhodopsin. Chem. Phys. Lett. 1995, 241, 559–565. [Google Scholar] [CrossRef]
- Lutz, I.; Sieg, A.; Wegener, A.; Engelhard, M.; Boche, I.; Otsuka, M.; Oesterhelt, D.; Wachtveitl, J.; Zinth, W. Primary reactions of sensory rhodopsins. Proc. Natl. Acad. Sci. USA 2001, 98, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Das, I.; Bhattacharya, S.; Harris, A.; Sheves, M.; Brown, L.S.; Ruhman, S. Bidirectional Photochemistry of Antarctic Microbial Rhodopsin: Emerging Trend of Ballistic Photoisomerization from the 13-cis Resting State. J. Phys. Chem. Lett. 2022, 13, 8134–8140. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.J. Electronic Couplings and Electrostatic Interactions Behind the Light Absorption of Retinal Proteins. Front. Mol. Biosci. 2021, 8, 752700. [Google Scholar] [CrossRef] [PubMed]
- Tsujimura, M.; Ishikita, H. Insights into the protein functions and absorption wavelengths of microbial rhodopsins. J. Phys. Chem. B 2020, 124, 11819–11826. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Chuon, K.; Cho, S.G.; Choi, A.; Meas, S.; Cho, H.S.; Jung, K.H. Color-tuning of natural variants of heliorhodopsin. Sci. Rep. 2021, 11, 854. [Google Scholar] [CrossRef] [PubMed]
- Shtyrov, A.A.; Nikolaev, D.M.; Mironov, V.N.; Vasin, A.V.; Panov, M.S.; Tveryanovich, Y.S.; Ryazantsev, M.N. Simple models to study spectral properties of microbial and animal rhodopsins: Evaluation of the electrostatic effect of charged and polar residues on the first absorption band maxima. Int. J. Mol. Sci. 2021, 22, 3029. [Google Scholar] [CrossRef]
- Ryazantsev, M.N.; Altun, A.; Morokuma, K. Color tuning in rhodopsins: The origin of the spectral shift between the chloride-bound and anion-free forms of halorhodopsin. J. Am. Chem. Soc. 2012, 134, 5520–5523. [Google Scholar] [CrossRef]
- Brown, L.; Bonet, L.; Needleman, R.; Lanyi, J. Estimated acid dissociation constants of the Schiff base, Asp-85, and Arg-82 during the bacteriorhodopsin photocycle. Biophys. J. 1993, 65, 124–130. [Google Scholar] [CrossRef]
- Chang, C.F.; Kuramochi, H.; Singh, M.; Abe-Yoshizumi, R.; Tsukuda, T.; Kandori, H.; Tahara, T. Acid–base equilibrium of the chromophore counterion results in distinct photoisomerization reactivity in the primary event of proteorhodopsin. Phys. Chem. Chem. Phys. 2019, 21, 25728–25734. [Google Scholar] [CrossRef] [PubMed]
- Tahara, S.; Takeuchi, S.; Abe-Yoshizumi, R.; Inoue, K.; Ohtani, H.; Kandori, H.; Tahara, T. Origin of the reactive and nonreactive excited states in the primary reaction of rhodopsins: pH dependence of femtosecond absorption of light-driven sodium ion pump rhodopsin KR2. J. Phys. Chem. B 2018, 122, 4784–4792. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Sudo, Y. Convergent evolution of animal and microbial rhodopsins. RSC Adv. 2023, 13, 5367–5381. [Google Scholar] [CrossRef] [PubMed]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef] [PubMed]
- Maag, D.; Mast, T.; Elstner, M.; Cui, Q.; Kubař, T. O to bR transition in bacteriorhodopsin occurs through a proton hole mechanism. Proc. Natl. Acad. Sci. USA 2021, 118, e2024803118. [Google Scholar] [CrossRef] [PubMed]
- Balashov, S.P.; Imasheva, E.S.; Govindjee, R.; Ebrey, T.G. Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys. J. 1996, 70, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Maclaurin, D.; Venkatachalam, V.; Lee, H.; Cohen, A.E. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proc. Natl. Acad. Sci. USA 2013, 110, 5939–5944. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, D.M.; Shtyrov, A.A.; Mereshchenko, A.S.; Panov, M.S.; Tveryanovich, Y.S.; Ryazantsev, M.N. An assessment of water placement algorithms in quantum mechanics/molecular mechanics modeling: The case of rhodopsins’ first spectral absorption band maxima. Phys. Chem. Chem. Phys. 2020, 22, 18114–18123. [Google Scholar] [CrossRef]
- Nikolaev, D.M.; Shtyrov, A.A.; Panov, M.S.; Jamal, A.; Chakchir, O.B.; Kochemirovsky, V.A.; Olivucci, M.; Ryazantsev, M.N. A comparative study of modern homology modeling algorithms for rhodopsin structure prediction. ACS Omega 2018, 3, 7555–7566. [Google Scholar] [CrossRef]
- Wang, W.W.; Sineshchekov, O.A.; Spudich, E.N.; Spudich, J.L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Bio. Chem. 2003, 278, 33985–33991. [Google Scholar] [CrossRef]
- Sharaabi, Y.; Brumfeld, V.; Sheves, M. Binding of Anions to Proteorhodopsin Affects the Asp97 p K a. Biochemistry 2010, 49, 4457–4465. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.O. Mechanism of Activation of the Visual Receptor Rhodopsin. Annu. Rev. Biophys. 2023, 52, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Chosrowjan, H.; Mataga, N.; Shibata, Y.; Tachibanaki, S.; Kandori, H.; Shichida, Y.; Okada, T.; Kouyama, T. Rhodopsin emission in real time: A new aspect of the primary event in vision. J. Am. Chem. Soc. 1998, 120, 9706–9707. [Google Scholar] [CrossRef]
- Doukas, A.; Junnarkar, M.; Alfano, R.; Callender, R.; Kakitani, T.; Honig, B. Fluorescence quantum yield of visual pigments: Evidence for subpicosecond isomerization rates. Proc. Natl. Acad. Sci. USA 1984, 81, 4790–4794. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, A.; Pool, G. Fluorescence spectra of the intermediates of rhodopsin bleaching. Photochem. Photobiol. 1969, 9, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Kochendoerfer, G.G.; Mathies, R.A. Spontaneous emission study of the femtosecond isomerization dynamics of rhodopsin. J. Phys. Chem. 1996, 100, 14526–14532. [Google Scholar] [CrossRef]
- Yamashita, T. Unexpected molecular diversity of vertebrate nonvisual opsin Opn5. Biophys. Rev. 2020, 12, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, M.; Saito, T.; Wada, S.; Nagata, T.; Kawano-Yamashita, E.; Terakita, A. Optogenetic potentials of diverse animal opsins: Parapinopsin, peropsin, LWS bistable opsin. In Optogenetics: Light-Sensing Proteins and Their Applications in Neuroscience and Beyond; Springer: Singapore, 2021; pp. 141–151. [Google Scholar]
- Cronin, T.; Goldsmith, T. Fluorescence of crayfish metarhodopsin studied in single rhabdoms. Biophys. J. 1981, 35, 653–664. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kim, M.; Taiji, M.; Iwasa, T.; Nakagawa, M.; Tsuda, M. Femtosecond Spectroscopy of Halorhodopsin and Rhodopsin in a Broad Spectral Range of 400–1000 nm. J. Phys. Chem. B 1998, 102, 272–280. [Google Scholar] [CrossRef]
- Kandori, H.; Furutani, Y.; Nishimura, S.; Shichida, Y.; Chosrowjan, H.; Shibata, Y.; Mataga, N. Excited-state dynamics of rhodopsin probed by femtosecond fluorescence spectroscopy. Chem. Phys. Lett. 2001, 334, 271–276. [Google Scholar] [CrossRef]
- Doukas, A.; Junnarkar, M.; Alfano, R.; Callender, R.; Balogh-Nair, V. The primary event in vision investigated by time-resolved fluorescence spectroscopy. Biophys. J. 1985, 47, 795–798. [Google Scholar] [CrossRef]
- Kruizinga, B.; Stavenga, D. Fluorescence spectra of blowfly metaxanthopsins. Photochem. Photobiol. 1990, 51, 197–201. [Google Scholar] [CrossRef]
- Stavenga, D.; Franceschini, N.; Kirschfeld, K. Fluorescence of housefly visual pigment. Photochem. Photobiol. 1984, 40, 653–659. [Google Scholar] [CrossRef]
- Miller, G.; Itoku, K.; Fleischer, A.; Stark, W. Studies of fluorescence in Drosophila compound eyes: Changes induced by intense light and vitamin A deprivation. J. Comp. Physiol. A 1984, 154, 297–305. [Google Scholar] [CrossRef]
- Matsuyama, T.; Yamashita, T.; Imamoto, Y.; Shichida, Y. Photochemical properties of mammalian melanopsin. Biochemistry 2012, 51, 5454–5462. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Tauber, M.J.; Mathies, R.A. Wavelength dependent cis-trans isomerization in vision. Biochemistry 2001, 40, 13774–13778. [Google Scholar] [CrossRef] [PubMed]
- Ehrenberg, D.; Varma, N.; Deupi, X.; Koyanagi, M.; Terakita, A.; Schertler, G.F.; Heberle, J.; Lesca, E. The two-photon reversible reaction of the bistable jumping spider rhodopsin-1. Biophys. J. 2019, 116, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Varma, N.; Mutt, E.; Mühle, J.; Panneels, V.; Terakita, A.; Deupi, X.; Nogly, P.; Schertler, G.F.; Lesca, E. Crystal structure of jumping spider rhodopsin-1 as a light sensitive GPCR. Proc. Natl. Acad. Sci. USA 2019, 116, 14547–14556. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, H.V.; Gruber, E.; Langeland, J.; Kusochek, P.A.; Bochenkova, A.V.; Andersen, L.H. Intrinsic photoisomerization dynamics of protonated Schiff-base retinal. Nat. Commun. 2019, 10, 1210. [Google Scholar] [CrossRef]
- Rasmussen, A.P.; Gruber, E.; Teiwes, R.; Sheves, M.; Andersen, L.H. Spectroscopy and photoisomerization of protonated Schiff-base retinal derivatives in vacuo. Phys. Chem. Chem. Phys. 2021, 23, 27227–27233. [Google Scholar] [CrossRef]
- Kandori, H.; Sasabe, H. Excited-state dynamics of a protonated Schiff base of all-trans retinal in methanol probed by femtosecond fluorescence measurement. Chem. Phys. Lett. 1993, 216, 126–172. [Google Scholar] [CrossRef]
- Bachilo, S.; Bondarev, S.; Gillbro, T. Fluorescence properties of protonated and unprotonated Schiff bases of retinal at room temperature. J. Photochem. Photobiol. B Biol. 1996, 34, 39–46. [Google Scholar] [CrossRef]
- Logunov, S.L.; Song, L.; El-Sayed, M.A. Excited-state dynamics of a protonated retinal Schiff base in solution. J. Phys. Chem. 1996, 100, 18586–18591. [Google Scholar] [CrossRef]
- Kandori, H.; Katsuta, Y.; Ito, M.; Sasabe, H. Femtosecond fluorescence study of the rhodopsin chromophore in solution. J. Am. Chem. Soc. 1995, 117, 2669–2670. [Google Scholar] [CrossRef]
- Tahara, S.; Singh, M.; Kuramochi, H.; Shihoya, W.; Inoue, K.; Nureki, O.; Béjà, O.; Mizutani, Y.; Kandori, H.; Tahara, T. Ultrafast dynamics of heliorhodopsins. J. Phys. Chem. B 2019, 123, 2507–2512. [Google Scholar] [CrossRef]
- Ryazantsev, M.N.; Nikolaev, D.M.; Struts, A.V.; Brown, M.F. Quantum mechanical and molecular mechanics modeling of membrane-embedded rhodopsins. J. Membr. Biol. 2019, 252, 425–449. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, V.; Entcheva, E.; Hedrich, R.; Hegemann, P.; Konrad, K.R.; Lüscher, C.; Mahn, M.; Pan, Z.H.; Sims, R.R.; Vierock, J.; et al. Optogenetics for light control of biological systems. Nat. Rev. Methods Prim. 2022, 2, 55. [Google Scholar] [CrossRef]
- Chen, W.; Li, C.; Liang, W.; Li, Y.; Zou, Z.; Xie, Y.; Liao, Y.; Yu, L.; Lin, Q.; Huang, M.; et al. The roles of optogenetics and technology in neurobiology: A review. Front. Aging Neurosci. 2022, 14, 867863. [Google Scholar] [CrossRef]
- Nikolaev, D.M.; Panov, M.S.; Shtyrov, A.A.; Boitsov, V.M.; Vyazmin, S.Y.; Chakchir, O.B.; Yakovlev, I.P.; Ryazantsev, M.N. Perspective tools for optogenetics and photopharmacology: From design to implementation. In Progress in Photon Science: Recent Advances; Springer: Cham, Switzerland, 2019; pp. 139–172. [Google Scholar]
- Bregestovski, P.; Maleeva, G. Photopharmacology: A brief review using the control of potassium channels as an example. Neurosci. Behav. Physiol. 2019, 49, 184–191. [Google Scholar] [CrossRef]
- Kobauri, P.; Dekker, F.J.; Szymanski, W.; Feringa, B.L. Rational Design in Photopharmacology with Molecular Photoswitches. Angew. Chem. 2023, 135, e202300681. [Google Scholar] [CrossRef]
- Broichhagen, J.; Levitz, J. Advances in tethered photopharmacology for precise optical control of signaling proteins. Curr. Opin. Pharmacol. 2022, 63, 102196. [Google Scholar] [CrossRef]
- Ryazantsev, M.N.; Strashkov, D.M.; Nikolaev, D.M.; Shtyrov, A.A.; Panov, M.S. Photopharmacological compounds based on azobenzenes and azoheteroarenes: Principles of molecular design, molecular modelling, and synthesis. Russ. Chem. Rev. 2021, 90, 868. [Google Scholar] [CrossRef]


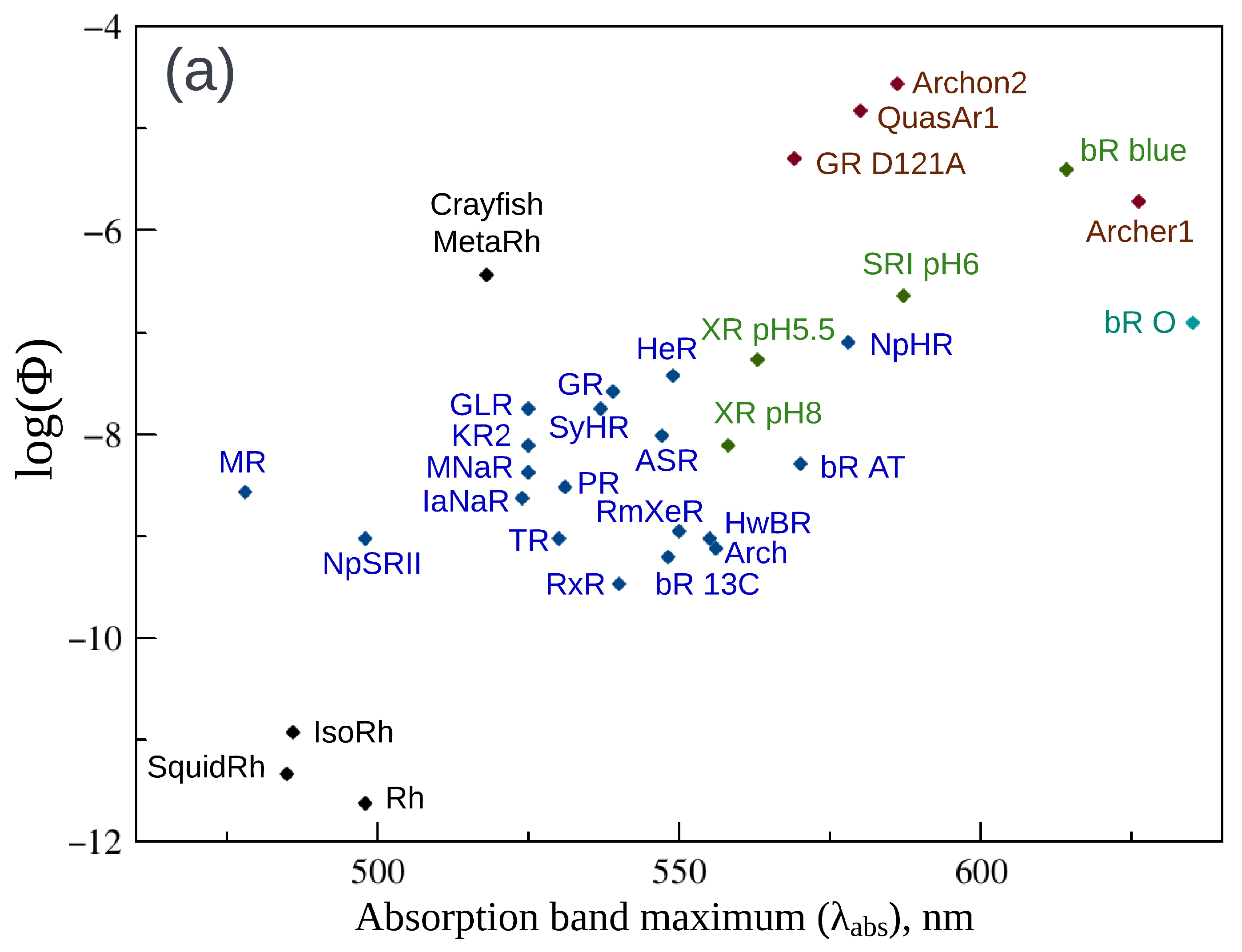

| Protein | , nm | , nm | , nm | Stokes Shift, nm | , ps | |
|---|---|---|---|---|---|---|
| bR | 568 [25,26] | 570 [27] | 755 ± 10 [28] | 187 ± 10 | 2.5–2.7 × 10 (AT) 0.7–1.2 × 10 (13C) [29] | 0.5 [26] 0.56 [30] |
| bR blue | 610–620 [31] | 613–614 [31] | 750 [29] | 130-140 | [31] | 1.5, 8.6 [32] 7.8 [30] |
| bR O | 635–640 [33] | - | [28] | 110–120 | ≈1 [34] | [34] |
| bR Q | - | - | <720 [28] | - | (bR) [25] [34] | [34] |
| bR D85S | 609 [35] | - | 760 [35] | 161 | - | 3.6, 14 [35] |
| bR D85N | 604 [36] | - | - | - | - | 2.0, 10 [36] |
| bR R82Q, pH4.4 | 596 [36] | - | - | - | - | 2.0, 7 [36] |
| bR R82Q, pH9.6 | 556 [36] | - | - | - | - | 0.6 [36] |
| Arch | 553 [24] | 553 [24] | 687 [24] | 134 | [24] [37] | 5.9, 60 (intense illumination) [24] |
| Arch D95N | 585 [37] | - | 687 [37] | 102 | [37] | - |
| QuasAr1 | 580 [23,38] | 590 [39] 585 [38] | 715 [39] 740 [38] | 135, 160 | [39] [38] | 61.5 [38] |
| Archon2 | 586 [40] | - | 735 [40] | 149 | [40] | 106 [40] |
| Archer1 | 626 [41] | 627 [42] | 731 [41] | 103 | [41] | - |
| PR | 531 [13] | 565 [13] | 700 [13] | 169 | [13] | 1.4 [13] 0.7, 15 (pH6) 0.4, 8 (pH9) [43] |
| XR | 560 [44] | - | 685 [44] | 125 | (pH8) (pH5.5) [44] | - |
| GR | 541 [45] | 568 (pH4.5) [45] 550 [24] | 670 (pH7.2) 680 (pH4.5) [45] 723 [24] | 129 (pH7.2) 135 (pH4.5) 173 [24] | F(pH4.5)= 4F(pH7.2) [45] [24] | 9.6, 47 (intense illumination) [24] |
| GR D121A | ≈568 [46] | - | - | - | [46] | - |
| ESR | 546 (pH2) 534 (pH5) 531 (pH7) 521 (pH10.5) [47] | 565 (pH5) 556 (pH7) 538 (pH8.8) [47] | ≈690 [47] | F(pH5) = 2F(pH7) [47] | - | |
| ESR D85N | 563 (pH5) [47] | 564 (pH5) [47] | 686 [47] | 123 | F=7F(ESR) [47] | - |
| ESR H57M | 565 (pH5) 517 (pH8.5) [47] | 568 (pH4.5) 520 (pH8.8) [47] | 690 (pH4.5) 650 (pH7.5) [47] | 125 (pH4.5) 133 (pH8) | F(pH4.5)≈ 100F(pH9) [47] | - |
| HeR | 549 [48] | - | ≈700 [48] | 151 | [48] | - |
| ASR | 541 (pH7) [24] 547 (AT) [49] 533 (13C) [49] | 540 (pH7) [24] | 700 [24] | 160 | (AT) (13C) [50] | 0.1, 0.77 [51] |
| KR2 | 525 [24] | 525 [24] | 685 [24] | 160 | [24] | 8.2, 46 (intense illumination) [24] |
| RxR | 540 [24] | 540 [24] | 708 [24] | 168 | [24] | 5.2, 54 (intense illumination) [24] |
| NpHR | 576 [24] | 576 [24] | 715 [24,52] ≈750 [53] | 115 150 | [24] [53] | 6.7, 24 (intense illumination) [24] 0.17, 1.5, 8.5 [54] 2.3 [52] |
| GLR | 525 [24] | 520 [24] | 685 [24] | 165 | [24] | - |
| HwBR | 555 [24] | 550 [24] | 720 [24] | 170 [24] | [24] | - |
| IaNaR | 524 [24] | 525 [24] | 690 [24] | 165 [24] | [24] | - |
| MNaR | 525 [24] | 500 [24] | 665 [24] | 165 [24] | [24] | - |
| MR | 478 [24] | 460 [24] | 620 [24] | 160 [24] | [24] | - |
| HsHR | 578 [54] | - | - | - | - | 1.5. 8.5 [54] |
| NpSRII | 498 [24,52] 460 [52] | 460 [24] | 615 [24] 630 [52] | 155 [24] 132 [52] | [24,55] | 0.25, 3.0 [52] |
| SRI pH6 | 587 [55] | - | 700 [55] | 113 | ≈1.3 [55] | 5, 33 [55] |
| RmXeR | 550 [24] | 545 [24] | 715 [24] | 170 [24] | [24] | - |
| SyHR | 537 [24] | 535 [24] | 680 [24] | 145 [24] | [24] | - |
| TR | 530 [24] | 518 [24] | 680 [24] | 162 [24] | [24] | - |
| AntR | 555 [56] | - | - | - | - | 1, 5 [56] |
| NeoR | 690 [11] | - | 707 [11] | 17 | 0.2 [11] | 1100 [11] |
| Species | Solvent | , nm | , nm | Stokes Shift, nm | ||
|---|---|---|---|---|---|---|
| 11C-PSB | gas-phase | 610 [92] | - | - | - | 0.442 ± 0.121 ps [92,93] |
| AT-PSB | gas-phase | 610 [92] | - | - | - | 3.3 ± 1 ps [92] |
| AT-PSB | methanol | 445 [94] 442 [95] 447 [96] | 655 [94] 675 [95] 630 [96] | 210 [94] 233 [95] 183 [96] | - | 90 fs (525) 0.5, 2.8 ps (605) 3.7 ps (762) [94] <0.2, 4.2 ps [96] |
| 13C-PSB | methanol | 438 [96] | 630 [96] | 192 | - | 2.5 ps |
| 11C-PSB | methanol | 445 [97] | 660 [97] | 215 | [97] | 0.5, 2.0 ps (605) 3.1 ps (695) [97] |
| AT-PSB | acetonitrile | 449 [96] | 690 [95] 650 [96] | 241 201 | [95] | <5 ps [95] 2.9 ps [96] |
| 13C-PSB | acetonitrile | 444 [96] | 652 [96] | 208 | - | 3.0 ps [96] |
| AT-PSB | 1-butanol | 453 [96] | 610 [96] | 157 | - | 3.0 ps [96] |
| 13C-PSB | 1-butanol | 447 [96] | 603 [96] | 156 | - | 2.5 ps [96] |
| AT-PSB | hexane | 457 [95] | 620 [95] | 163 | [95] | <5 ps [95] |
| AT-PSB | carbon tetrachloride | 465 [95] | 650 [95] | 185 | [95] | - |
| AT-PSB | toluene | 461 [95] | 660 [95] | 199 | [95] | - |
| AT-PSB | ethyl acetate | 432 [95] | 650 [95] | 218 | - | - |
| AT-PSB | acetone | 447 [95] | 680 [95] | 233 | - | - |
| AT-PSB | ethanol | 443 [95] | 660 [95] | 217 | [95] | <6ps [95] |
| AT-PSB | propanol | 449 [95] | 650 [95] | 201 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaev, D.M.; Shtyrov, A.A.; Vyazmin, S.Y.; Vasin, A.V.; Panov, M.S.; Ryazantsev, M.N. Fluorescence of the Retinal Chromophore in Microbial and Animal Rhodopsins. Int. J. Mol. Sci. 2023, 24, 17269. https://doi.org/10.3390/ijms242417269
Nikolaev DM, Shtyrov AA, Vyazmin SY, Vasin AV, Panov MS, Ryazantsev MN. Fluorescence of the Retinal Chromophore in Microbial and Animal Rhodopsins. International Journal of Molecular Sciences. 2023; 24(24):17269. https://doi.org/10.3390/ijms242417269
Chicago/Turabian StyleNikolaev, Dmitrii M., Andrey A. Shtyrov, Sergey Yu. Vyazmin, Andrey V. Vasin, Maxim S. Panov, and Mikhail N. Ryazantsev. 2023. "Fluorescence of the Retinal Chromophore in Microbial and Animal Rhodopsins" International Journal of Molecular Sciences 24, no. 24: 17269. https://doi.org/10.3390/ijms242417269
APA StyleNikolaev, D. M., Shtyrov, A. A., Vyazmin, S. Y., Vasin, A. V., Panov, M. S., & Ryazantsev, M. N. (2023). Fluorescence of the Retinal Chromophore in Microbial and Animal Rhodopsins. International Journal of Molecular Sciences, 24(24), 17269. https://doi.org/10.3390/ijms242417269







