Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease
Abstract
:1. Introduction
2. Results
2.1. Identification of Anoxybacillus flavithermus Cas9
2.2. Obtaining AnoCas9 Recombinant Protein
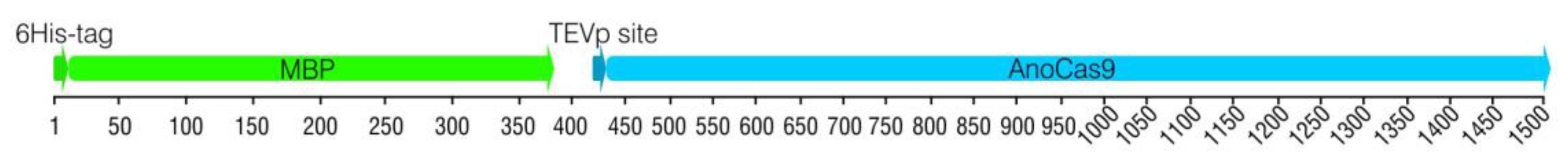
2.2.1. Cloning anoCas9 Gene into pETm Vector
2.2.2. 5′-End anoCas9 Codon Optimization
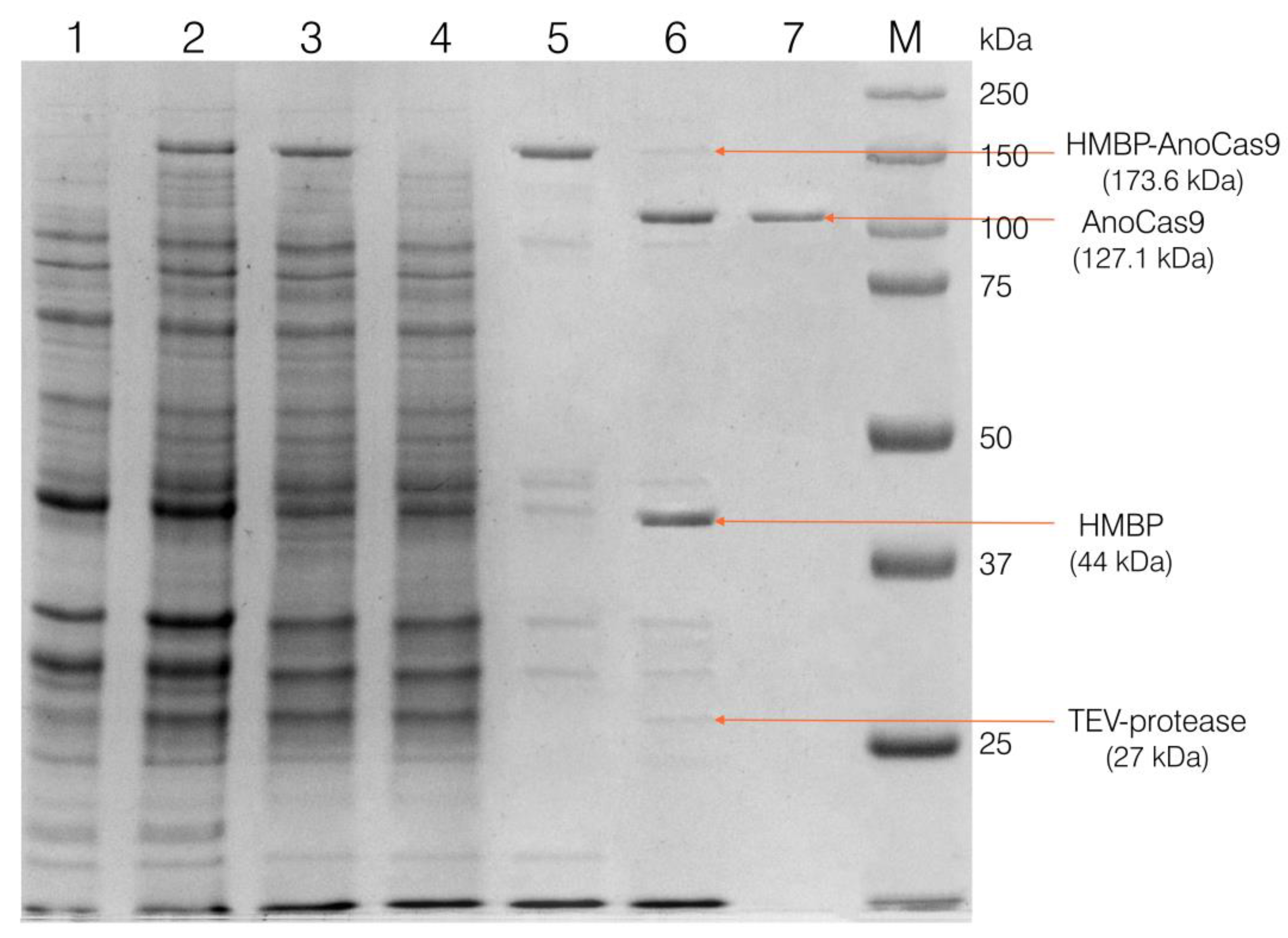
2.2.3. Cloning Partially Optimized anoCas9 Gene into pD441-HMBP and Producing Recombinant AnoCas9
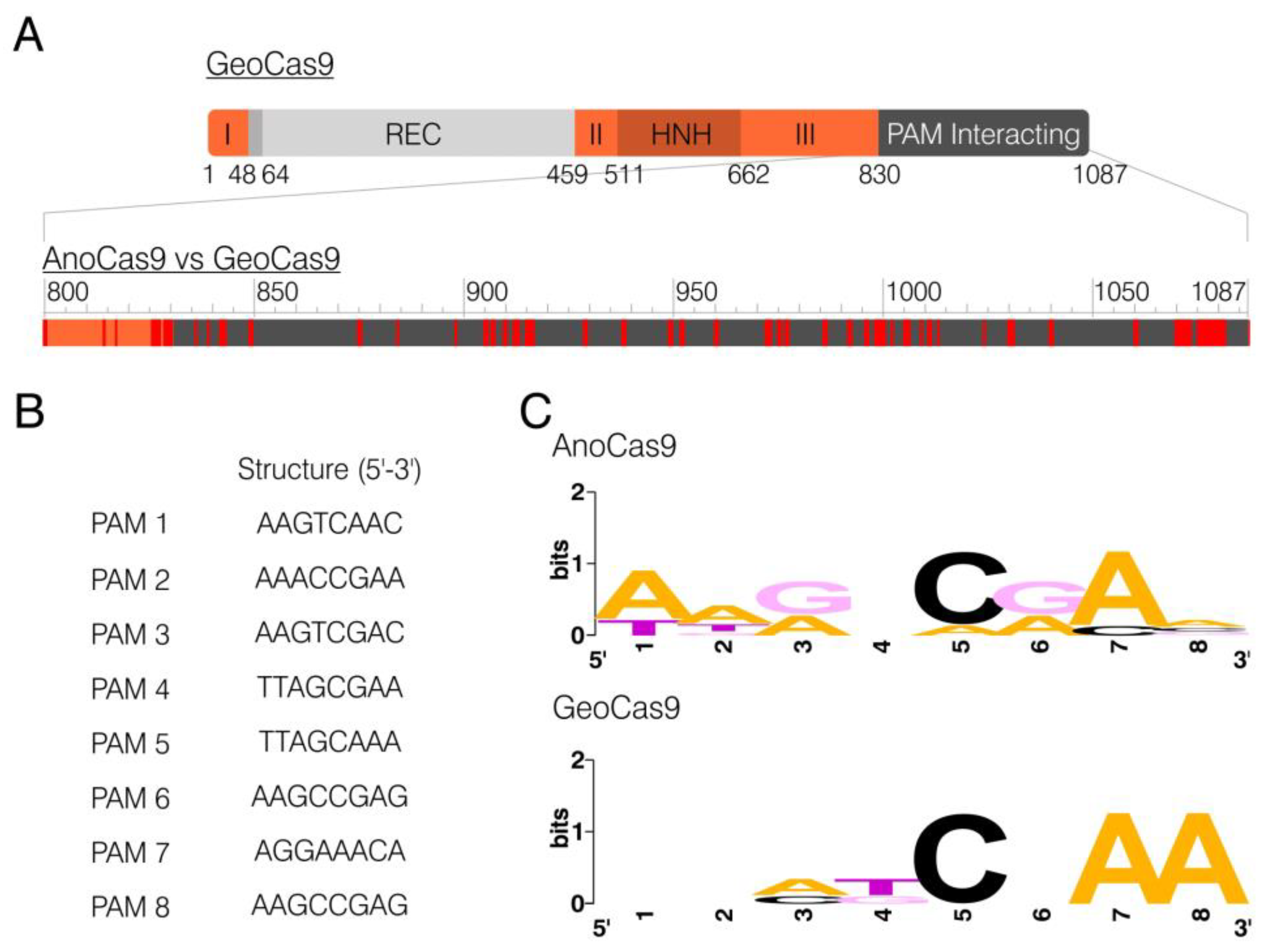
2.3. CRISPR Array Analysis
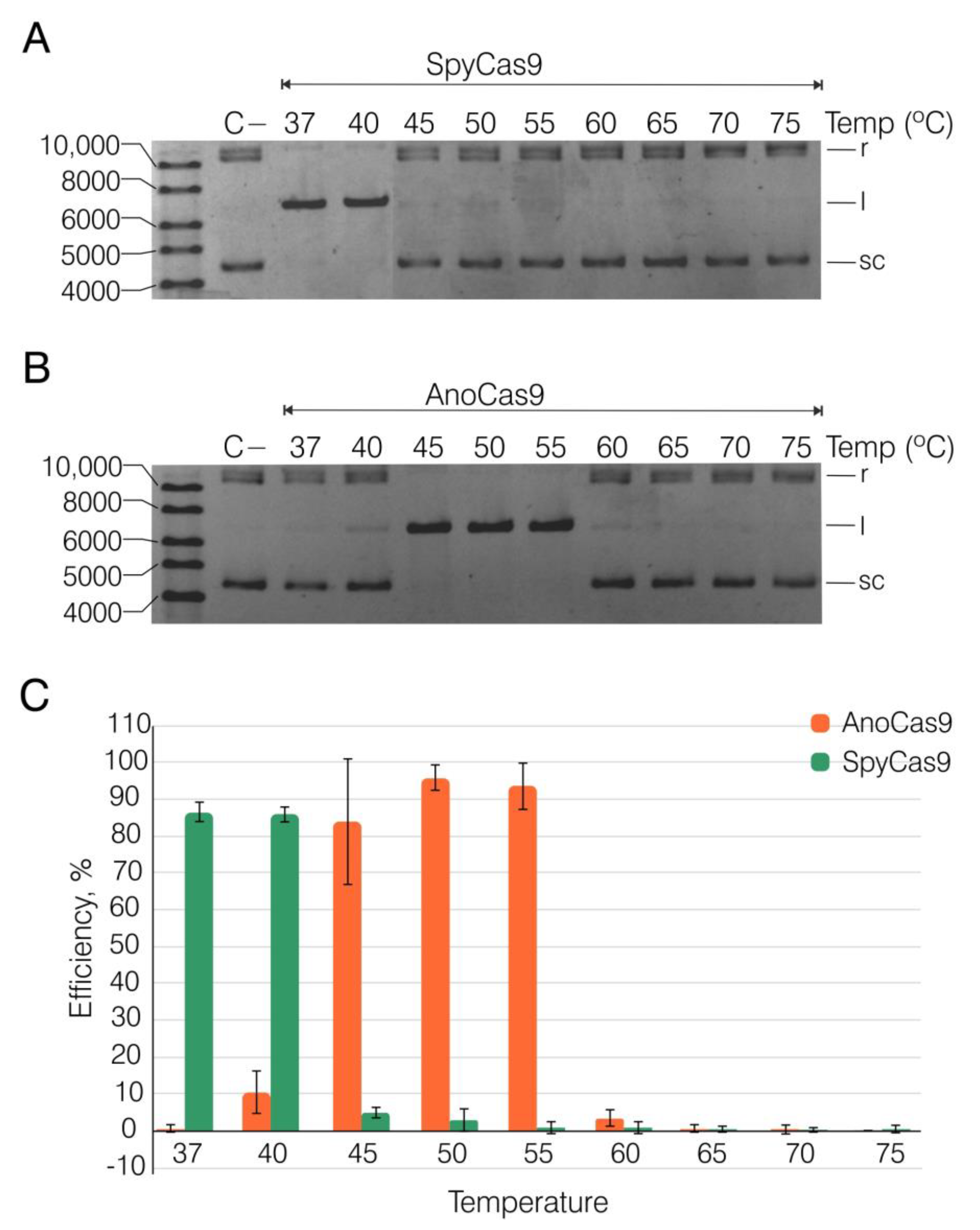
2.4. Recombinant AnoCas9 Nuclease Activity Assessment
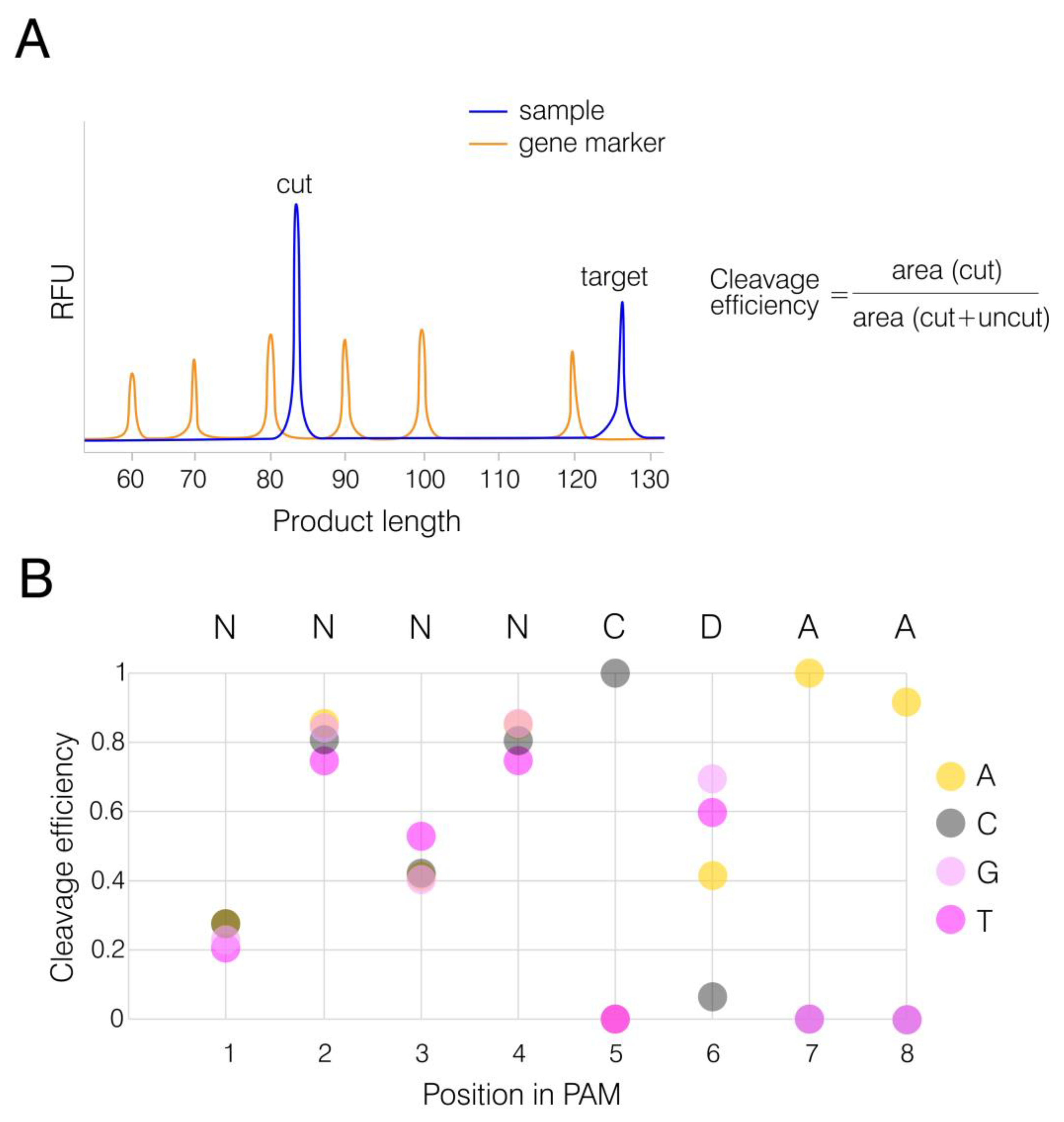
2.5. In Vitro PAM Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Whole Genome Sequencing and Data Analysis
4.3. Gene Sources and Cloning Strategy
4.4. Escherichia coli Cultivation Conditions
4.5. AnoCas9 Purification
4.5.1. Collection of the Cell Extract
4.5.2. IMAC-Sepharose Chromatography
4.5.3. SP-Sepharose Chromatography
4.6. CRISPR Array Analysis
4.7. CRISPR RNA Synthesis
4.8. In Vitro DNA Plasmid Cleavage with Cas9
4.9. In Vitro PAM Library Digestion with Cas9
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Swarts, D.C.; Staals, R.H.J.; Jore, M.M.; Brouns, S.J.J.; van der Oost, J. The CRISPRs, They Are A-Changin’: How Prokaryotes Generate Adaptive Immunity. Annu. Rev. Genet. 2012, 46, 311–339. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Ann Ran, F.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRIPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Fonfara, I.; Richter, H.; BratoviÄ, M.; Le Rhun, A.; Charpentier, E. The CRISPR-Associated DNA-Cleaving Enzyme Cpf1 Also Processes Precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-Wide Specificities of CRISPR-Cas Cpf1 Nucleases in Human Cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; Graaf, D.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A.; et al. Two Distinct RNase Activities of CRISPR-C2c2 Enable Guide-RNA Processing and RNA Detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Reardon, S. Welcome to the CRISPR Zoo. Nature 2016, 531, 160–163. [Google Scholar] [CrossRef] [PubMed]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Kordyś, M.; Sen, R.; Warkocki, Z. Applications of the Versatile CRISPR-Cas13 RNA Targeting System. WIREs RNA 2022, 13, e1694. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Eid, A.; Momin, A.A.; Bazin, J.; Crespi, M.; Arold, S.T.; Mahfouz, M.M. CRISPR Directed Evolution of the Spliceosome for Resistance to Splicing Inhibitors. Genome Biol. 2019, 20, 73. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Liu, D.; Jia, X.; Zheng, Y.; Liu, W.; Xiao, Y. CRISPR-Cas9/Cas12a Biotechnology and Application in Bacteria. Synth. Syst. Biotechnol. 2018, 3, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-Deployable Viral Diagnostics Using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Loutre, R.; Heckel, A.-M.; Smirnova, A.; Entelis, N.; Tarassov, I. Can Mitochondrial DNA Be CRISPRized: Pro and Contra. IUBMB Life 2018, 70, 1233–1239. [Google Scholar] [CrossRef]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.-H.; Heckl, D. Refined SgRNA Efficacy Prediction Improves Large- and Small-Scale CRISPR–Cas9 Applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef]
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 Genome Editing Technology for the Improvement of Crops Cultivated in Tropical Climates: Recent Progress, Prospects, and Challenges. Front. Plant Sci. 2018, 9, 617. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR Tool Kit for Genome Editing and Beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Lee, M. Deep Learning in CRISPR-Cas Systems: A Review of Recent Studies. Front. Bioeng. Biotechnol. 2023, 11, 1226182. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Jiang, T.; Yan, Y. The Expanded CRISPR Toolbox for Constructing Microbial Cell Factories. Trends Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.-H.; Suh, Y. In Vivo Epigenome Editing and Transcriptional Modulation Using CRISPR Technology. Transgenic Res. 2018, 27, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally Engineered Cas9 Nulceases with Improved Specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.C.; Tjian, R.; Doudna, J.A. Genomes in Focus: Development and Applications of CRISPR-Cas9 Imaging Technologies. Angew. Chem. Int. Ed. 2018, 57, 4329–4337. [Google Scholar] [CrossRef]
- Reeder, S.B.; Hu, H.H.; Sirlin, C.B.; Group, L.I.; Diego, S. Targeting Genomic Rearrangements in Tumor Cells Using Cas9- Mediated Insertion of a Suicide Gene. Nat. Biotechnol. 2017, 36, 1011–1014. [Google Scholar]
- Liszczak, G.P.; Brown, Z.Z.; Kim, S.H.; Oslund, R.C.; David, Y.; Muir, T.W. Genomic Targeting of Epigenetic Probes Using a Chemically Tailored Cas9 System. Proc. Natl. Acad. Sci. USA 2017, 114, 681–686. [Google Scholar] [CrossRef]
- Hirosawa, M.; Fujita, Y.; Parr, C.J.C.; Hayashi, K.; Kashida, S.; Hotta, A.; Woltjen, K.; Saito, H. Cell-Type-Specific Genome Editing with a MicroRNA-Responsive CRISPR-Cas9 Switch. Nucleic Acids Res. 2017, 45, e118. [Google Scholar] [CrossRef]
- Spencer, N.Y.; Yan, Z.; Cong, L.; Zhang, Y.; Engelhardt, J.F. Definitive Localization of Intracellular Proteins- Novel Approach Using CRISPR-Cas9 Genome Editing with Glucose 6-Phosphate Dehydrogenase as a Model. Anal. Biochem. 2017, 494, 55–67. [Google Scholar] [CrossRef]
- Burstein, D.; Harrington, L.B.; Strutt, S.C.; Probst, A.J.; Anantharaman, K.; Thomas, B.C.; Doudna, J.A.; Banfield, J.F. New CRISPR–Cas Systems from Uncultivated Microbes. Nature 2017, 542, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Young, J.; Bigelyte, G.; Silanskas, A.; Cigan, M.; Siksnys, V. Rapid Characterization of CRISPR-Cas9 Protospacer Adjacent Motif Sequence Elements. Genome Biol. 2015, 16, 253. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Gootenberg, J.S.; Abudayyeh, O.O. Discovery of Diverse CRISPR-Cas Systems and Expansion of the Genome Engineering Toolbox. Biochemistry 2023. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.; Edraki, A.; Lee, J.; Sontheimer, E.J. Type II-C CRISPR-Cas9 Biology, Mechanism, and Application. ACS Chem. Biol. 2018, 13, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.B.; Paez-Espino, D.; Staahl, B.T.; Chen, J.S.; Ma, E.; Kyrpides, N.C.; Doudna, J.A. A Thermostable Cas9 with Increased Lifetime in Human Plasma. Nat. Commun. 2017, 8, 1424. [Google Scholar] [CrossRef] [PubMed]
- da Rosa, D.F.; Macedo, A.J. The Genus Anoxybacillus: An Emerging and Versatile Source of Valuable Biotechnological Products. Extremophiles 2023, 27, 22. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Parise, M.T.D.; Parise, D.; Medeiros, L.G.; Sousa, T.J.; Kato, R.B.; Uetanabaro, A.P.T.; Araújo, F.; Ramos, R.T.J.; de Castro Soares, S.; et al. Unraveling the Genomic Potential of the Thermophilic Bacterium Anoxybacillus Flavithermus from an Antarctic Geothermal Environment. Microorganisms 2022, 10, 1673. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Ryabchenko, A.V.; Karavaev, V.S.; Beklemishev, A.B. Comparative Structural and Immunochemical Analysis of Recombinant OspC Antigens of Novosibirsk Borrelia Garinii and Borrelia Afzelii Isolates. Bull. SB RAMS 2010, 30, 6–12. [Google Scholar]
- Prokhorova, D.V.; Vokhtantsev, I.P.; Tolstova, P.O.; Zhuravlev, E.S.; Kulishova, L.M.; Zharkov, D.O.; Stepanov, G.A. Natural Nucleoside Modifications in Guide RNAs Can Modulate the Activity of the CRISPR-Cas9 System In Vitro. CRISPR J. 2022, 5, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.; Sun, J. CRISPR/Cas Genome Editing Systems in Thermophiles: Current Status, Associated Challenges, and Future Perspectives. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2022; pp. 1–30. [Google Scholar]
- Lau, M.S.H.; Sheng, L.; Zhang, Y.; Minton, N.P. Development of a Suite of Tools for Genome Editing in Parageobacillus thermoglucosidasius and Their Use to Identify the Potential of a Native Plasmid in the Generation of Stable Engineered Strains. ACS Synth. Biol. 2021, 10, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Raheja, Y.; Basotra, N.; Sharma, G.; Tsang, A.; Chadha, B.S. CRISPR/Cas9 Mediated Gene Editing of Transcription Factor ACE1 for Enhanced Cellulase Production in Thermophilic Fungus Rasamsonia Emersonii. Fungal Biol. Biotechnol. 2023, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Gallo, G.; Mougiakos, I.; Bianco, M.; Carbonaro, M.; Carpentieri, A.; Illiano, A.; Pucci, P.; Bartolucci, S.; van der Oost, J.; Fiorentino, G. A Hyperthermoactive-Cas9 Editing Tool Reveals the Role of a Unique Arsenite Methyltransferase in the Arsenic Resistance System of Thermus Thermophilus HB27. mBio 2021, 12, e02813-21. [Google Scholar] [CrossRef] [PubMed]
- Hand, T.H.; Das, A.; Li, H. Directed Evolution Studies of a Thermophilic Type II-C Cas9. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2019; pp. 265–288. [Google Scholar]
- Tsui, T.K.M.; Hand, T.H.; Duboy, E.C.; Li, H. The Impact of DNA Topology and Guide Length on Target Selection by a Cytosine-Specific Cas9. ACS Synth. Biol. 2017, 6, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Hand, T.H.; Smith, C.L.; Wickline, E.; Zawrotny, M.; Li, H. The Molecular Basis for Recognition of 5′-NNNCC-3′ PAM and Its Methylation State by Acidothermus Cellulolyticus Cas9. Nat. Commun. 2020, 11, 6346. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Mohanraju, P.; Bosma, E.F.; Vrouwe, V.; Finger Bou, M.; Naduthodi, M.I.S.; Gussak, A.; Brinkman, R.B.L.; van Kranenburg, R.; van der Oost, J. Characterizing a Thermostable Cas9 for Bacterial Genome Editing and Silencing. Nat. Commun. 2017, 8, 1647. [Google Scholar] [CrossRef]
- Ganguly, J.; Martin-Pascual, M.; van Kranenburg, R. CRISPR Interference (CRISPRi) as Transcriptional Repression Tool for Hungateiclostridium thermocellum DSM 1313. Microb. Biotechnol. 2020, 13, 339–349. [Google Scholar] [CrossRef]
- Trasanidou, D.; Barendse, P.; Bouzetos, E.; de Haan, L.; Bouwmeester, H.; Staals, R.H.J.; Mougiakos, I.; van der Oost, J. Efficient Genome and Base Editing in Human Cells Using ThermoCas9. CRISPR J. 2023, 6, 278–288. [Google Scholar] [CrossRef]
- Schmidt, S.T.; Yu, F.B.; Blainey, P.C.; May, A.P.; Quake, S.R. Nucleic Acid Cleavage with a Hyperthermophilic Cas9 from an Uncultured Ignavibacterium. Proc. Natl. Acad. Sci. USA 2019, 116, 23100–23105. [Google Scholar] [CrossRef]
- Xie, L.; Hu, Y.; Li, L.; Jiang, L.; Jiao, Y.; Wang, Y.; Zhou, L.; Tao, R.; Qu, J.; Chen, Q.; et al. Expanding PAM Recognition and Enhancing Base Editing Activity of Cas9 Variants with Non-PI Domain Mutations Derived from XCas9. FEBS J. 2022, 289, 5899–5913. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Legut, M.; Daniloski, Z.; Xue, X.; McKenzie, D.; Guo, X.; Wessels, H.-H.; Sanjana, N.E. High-Throughput Screens of PAM-Flexible Cas9 Variants for Gene Knockout and Transcriptional Modulation. Cell Rep. 2020, 30, 2859–2868.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, B. SpRY: Engineered CRISPR/Cas9 Harnesses New Genome-Editing Power. Trends Genet. 2020, 36, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Leenay, R.T.; Maksimchuk, K.R.; Slotkowski, R.A.; Agrawal, R.N.; Gomaa, A.A.; Briner, A.E.; Barrangou, R.; Beisel, C.L. Identifying and Visualizing Functional PAM Diversity across CRISPR-Cas Systems. Mol. Cell 2016, 62, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Siksnys, V. Methods for Decoding Cas9 Protospacer Adjacent Motif (PAM) Sequences: A Brief Overview. Methods 2017, 121–122, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Siksnys, V. Harnessing the Natural Diversity and in Vitro Evolution of Cas9 to Expand the Genome Editing Toolbox. Curr. Opin. Microbiol. 2017, 37, 88–94. [Google Scholar] [CrossRef]
- Ma, D.; Xu, Z.; Zhang, Z.; Chen, X.; Zeng, X.; Zhang, Y.; Deng, T.; Ren, M.; Sun, Z.; Jiang, R.; et al. Engineer Chimeric Cas9 to Expand PAM Recognition Based on Evolutionary Information. Nat. Commun. 2019, 10, 560. [Google Scholar] [CrossRef]
- Adalsteinsson, B.T.; Kristjansdottir, T.; Merre, W.; Helleux, A.; Dusaucy, J.; Tourigny, M.; Fridjonsson, O.; Hreggvidsson, G.O. Efficient Genome Editing of an Extreme Thermophile, Thermus Thermophilus, Using a Thermostable Cas9 Variant. Sci. Rep. 2021, 11, 9586. [Google Scholar] [CrossRef]
- Shi, Y.-J.; Duan, M.; Ding, J.-M.; Wang, F.-Q.; Bi, L.-L.; Zhang, C.-X.; Zhang, Y.-Z.; Duan, J.-Y.; Huang, A.-H.; Lei, X.-L.; et al. DNA Topology Regulates PAM-Cas9 Interaction and DNA Unwinding to Enable near-PAMless Cleavage by Thermophilic Cas9. Mol. Cell 2022, 82, 4160–4175.e6. [Google Scholar] [CrossRef]
- Chen, J.; Qiu, T.; Mauk, M.G.; Su, Z.; Fan, Y.; Yuan, D.J.; Zhou, Q.; Qiao, Y.; Bau, H.H.; Ying, J.; et al. Programmable Endonuclease Combined with Isothermal Polymerase Amplification to Selectively Enrich for Rare Mutant Allele Fractions. Chin. Chem. Lett. 2022, 33, 4126–4132. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.S.; Gavrilov, M.; Liu, Y.; Rasoloson, D.; Conte, M.; Hardick, J.; Shen, L.; Chen, S.; Pekosz, A.; Seydoux, G.; et al. Improving the Specificity of Nucleic Acid Detection with Endonuclease-Actuated Degradation. Commun. Biol. 2022, 5, 290. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Qiu, T.; Mauk, M.G.; Fan, Y.; Jiang, Y.; Ying, J.; Zhou, Q.; Qiao, Y.; Bau, H.H.; Song, J. CRISPR Cas9-Mediated Selective Isothermal Amplification for Sensitive Detection of Rare Mutant Alleles. Clin. Chem. 2021, 67, 1569–1571. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Jinek, M. Chapter One—In Vitro Enzymology of Cas9. In Methods in Enzymology; Doudna, J.A., Sontheimer, E.J., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 546, pp. 1–20. ISBN 0076-6879. [Google Scholar]
- Daniel, E.; Onwukwe, G.U.; Wierenga, R.K.; Quaggin, S.E.; Vainio, S.J.; Krause, M. ATGme: Open-Source Web Application for Rare Codon Identification and Custom DNA Sequence Optimization. BMC Bioinform. 2015, 16, 303. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein Production by Auto-Induction in High-Density Shaking Cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Biswas, A.; Gagnon, J.N.; Brouns, S.J.J.; Fineran, P.C.; Brown, C.M. CRISPRTarget. RNA Biol. 2013, 10, 817–827. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator: Figure 1. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Wright, A.V.; Sternberg, S.H.; Taylor, D.W.; Staahl, B.T.; Bardales, J.A.; Kornfeld, J.E.; Doudna, J.A. Rational Design of a Split-Cas9 Enzyme Complex. Proc. Natl. Acad. Sci. USA 2015, 112, 2984–2989. [Google Scholar] [CrossRef]
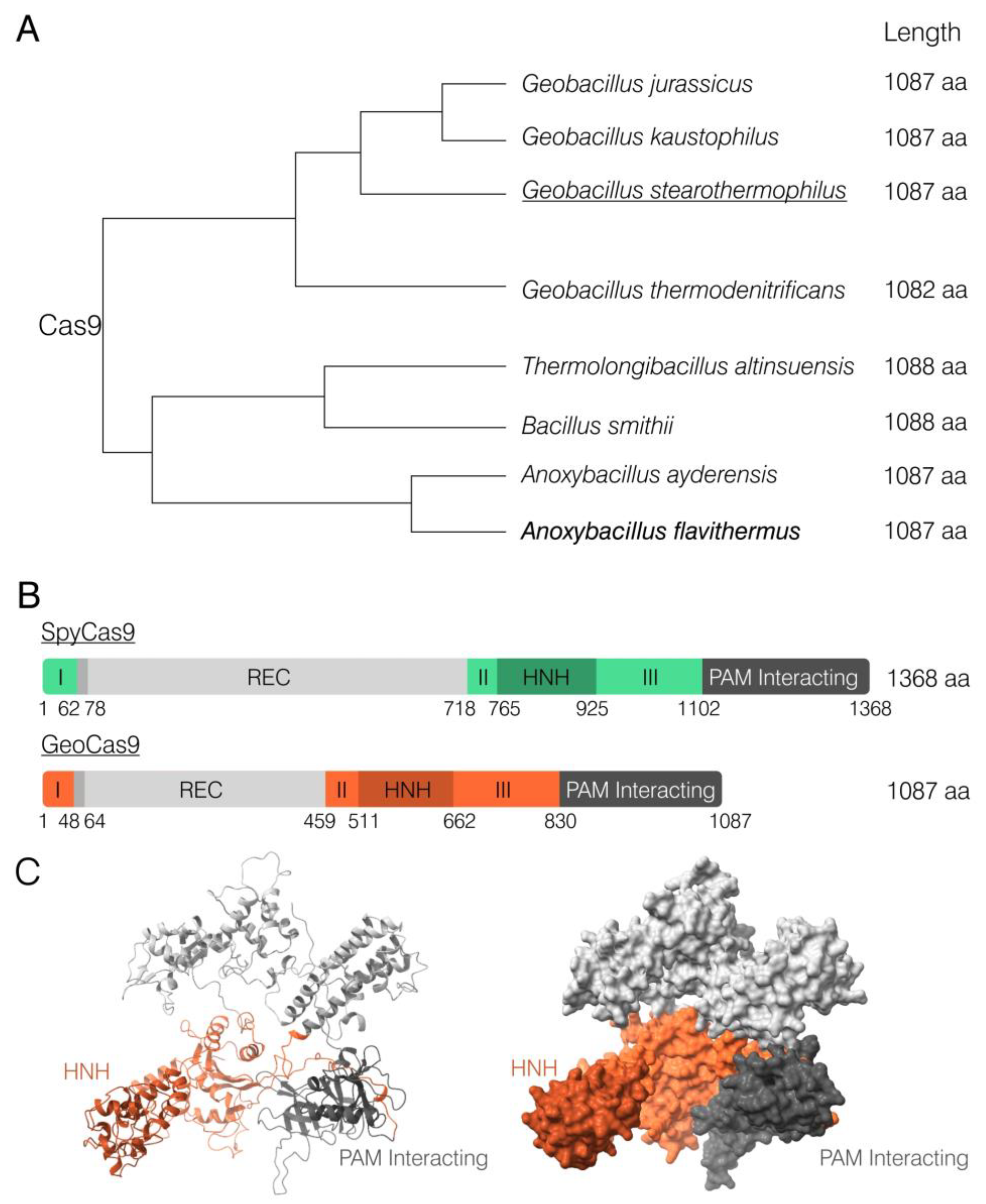


| Name | Source | Size, aa | PAM Sequence (5′-3′) | References |
|---|---|---|---|---|
| ThermoCas9 | Geobacillus thermodenitrificans T12 | 1082 | NNNNCVAA NNNNCCCA | [49] |
| GeoCas9 | Geobacillus stearothermophilus | 1087 | NNNNCRAA | [35] |
| CaldoCas9 | Geobacillus sp. LC300 | 1087 | NNNNGNMA | [61] |
| AceCas9 | Acidothermus cellulolyticus | 1138 | NNNCC | [46,47] |
| IgnaviCas9 | Ignavibacteriae phylum | 1241 | NRRNAT | [52] |
| AtCas9 | Alicyclobacillus tengchongensis | 1147 | NNNNCNNN NNNNRNNA | [62] |
| AnoCas9 | Anoxybacillus flavithermus | 1087 | NNNNCDAA |
| Name | Structure (5′–3′) |
|---|---|
| Cas9-4025_ext_F | cgcagcggtcacccgaagcttatgagatataaaataggattagacatag |
| Cas9-4025_ext_R | tggtggtgctcgagcagaagcttatgactaattgattgtaacgaatg |
| 1F | cgaaaacctgtattttcagggcggtaccatgcgttataaaattggtttagacatc |
| 2R | cgttaaaacaccttcacggactaaaagacggcgaatacgttctaaacgatgtttg |
| 3F | cgtcttttagtccgtgaaggtg |
| 4R | tcagtcgaaagactgggcctttcgcccgggctaatttcaatgactaattgattgtaacgaatg |
| 268F | atgcgtccggcgtaga |
| 269R | gctagttattgctcagcggtg |
| 5F | agactgtcgatgaagccctg |
| 5R | tgccgaactcagaagtgaaac |
| Seq_Cas9(Anoav)_1 | actccatcgcagaagctgc |
| Seq_Cas9(Anoav)_2 | gcaaatgtcttgaacaagtttatgg |
| Seq_Cas9(Anoav)_3 | cgtcttttagtccgtgaaggtg |
| Seq_Cas9(Anoav)_4 | ccagctgagtatttaggattcg |
| Seq_Cas9(Anoav)_5 | ggtccaatcattcgtactgtg |
| Seq_Cas9(Anoav)_6 | cagtaccgttacaccataattcac |
| Seq_Cas9(Anoav)_7 | acgcatcaacacgcaactgc |
| Seq_Cas9(Anoav)_8 | ctctttgaattcttgttcttctgc |
| Seq_Cas9(Anoav)_9 | caagttctttttccatcagcacg |
| Seq_Cas9(Anoav)_10 | tcaaaaacatcggtgcgaacg |
| Strains | Relevant Genotype | Source |
|---|---|---|
| Anoxybacillus flavithermus | Wild type | CEMTC (ICBFM SB RAS) |
| E. coli TOP10 | F– mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Thermo Scientific™ (Waltham, MA, USA) |
| E. coli BL21 (DE3) | F– ompT hsdSB (rB–, mB–) gal dcm (DE3) | Thermo Scientific™ |
| E. coli Rosetta 2 (DE3) | F– ompT hsdSB (rB–, mB–) gal dcm (DE3) pRARE2 (CamR) | Novagen (Sigma-Aldrich, St. Louis, MO, USA) |
| Plasmids | Description | Source |
| pET36b(+) | PT7lac, KanR | Novagen |
| pETm-Cas9 | PT7lac, KanR | This study |
| pUC19-210Cas9 | pBR233ori-F, AmpR | ICBFM SB RAS |
| pD441-HMBP | PT5lac, KanR | DNA TwoPointO Inc. (Menlo Park, CA, USA) («ATUM») |
| pD441-HMBP-Cas9 | PT5lac, KanR | This study |
| Name | Structure (5′-3′) |
|---|---|
| Ano_tracrRNA_T7 | atgcagctaatacgactcactataggtcagggttactatgataagg |
| Ano_tracrRNA_R | atggggagtgcccttaaagg |
| Ano_ANXA_cr | cagggaugcauuuguggccauguaauaguuccccuga |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveeva, A.; Ryabchenko, A.; Petrova, V.; Prokhorova, D.; Zhuravlev, E.; Zakabunin, A.; Tikunov, A.; Stepanov, G. Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease. Int. J. Mol. Sci. 2023, 24, 17121. https://doi.org/10.3390/ijms242317121
Matveeva A, Ryabchenko A, Petrova V, Prokhorova D, Zhuravlev E, Zakabunin A, Tikunov A, Stepanov G. Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease. International Journal of Molecular Sciences. 2023; 24(23):17121. https://doi.org/10.3390/ijms242317121
Chicago/Turabian StyleMatveeva, Anastasiya, Alexander Ryabchenko, Viktoria Petrova, Daria Prokhorova, Evgenii Zhuravlev, Alexander Zakabunin, Artem Tikunov, and Grigory Stepanov. 2023. "Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease" International Journal of Molecular Sciences 24, no. 23: 17121. https://doi.org/10.3390/ijms242317121
APA StyleMatveeva, A., Ryabchenko, A., Petrova, V., Prokhorova, D., Zhuravlev, E., Zakabunin, A., Tikunov, A., & Stepanov, G. (2023). Expression and Functional Analysis of the Compact Thermophilic Anoxybacillus flavithermus Cas9 Nuclease. International Journal of Molecular Sciences, 24(23), 17121. https://doi.org/10.3390/ijms242317121








