Biochemical Characterization of Parsley Glycosyltransferases Involved in the Biosynthesis of a Flavonoid Glycoside, Apiin
Abstract
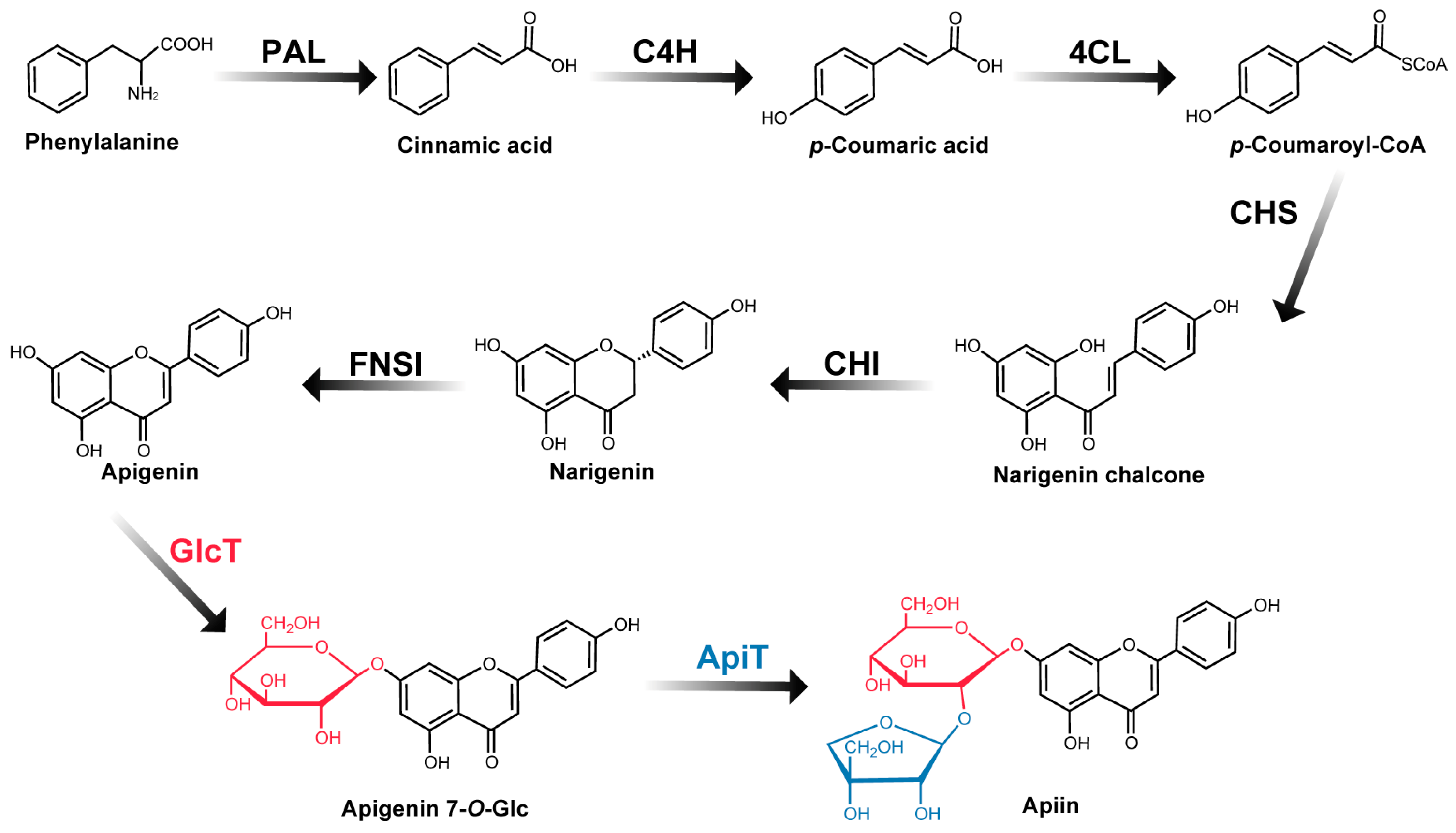
:1. Introduction
2. Results and Discussion
2.1. Gene Identification of PcGlcT and PcApiT
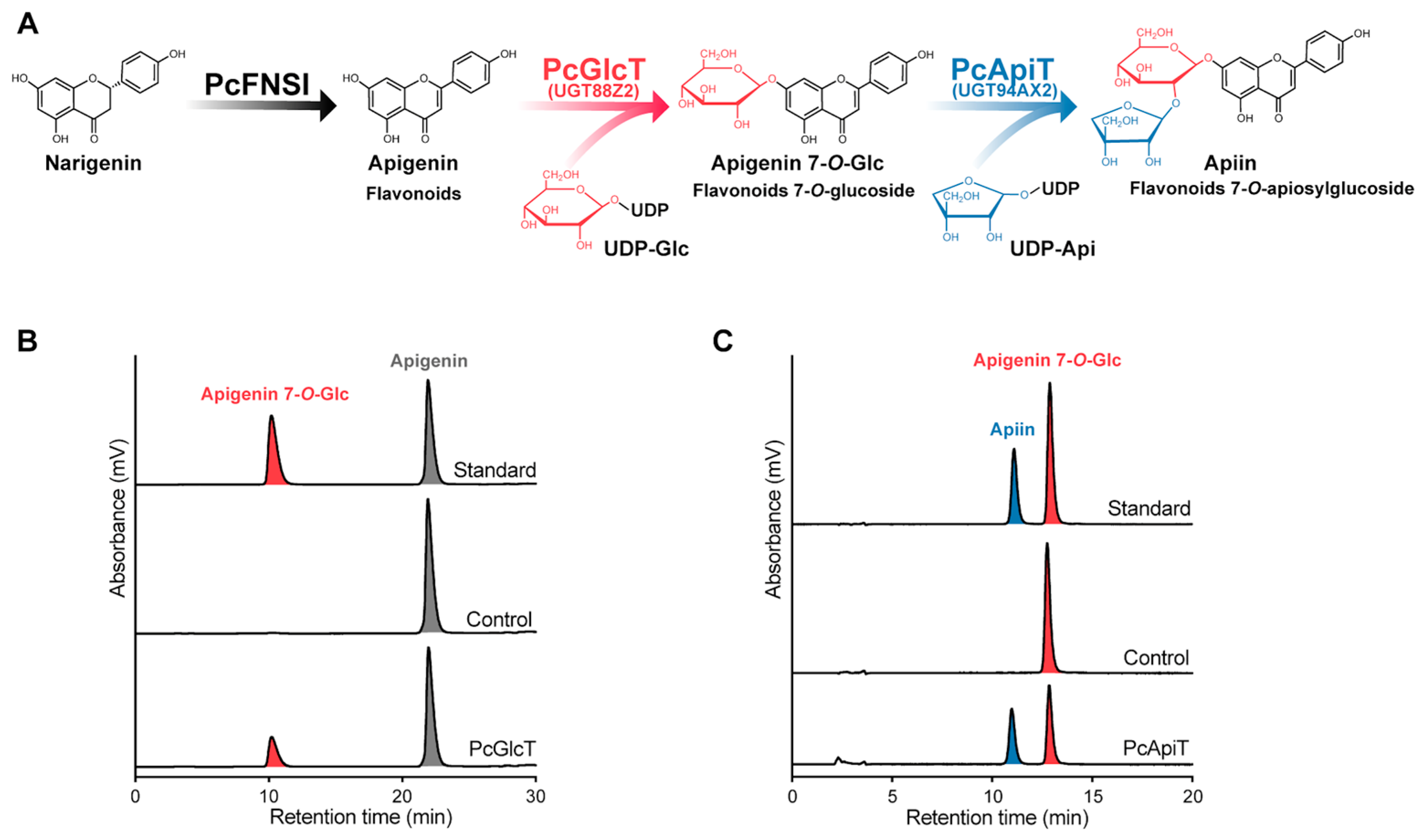
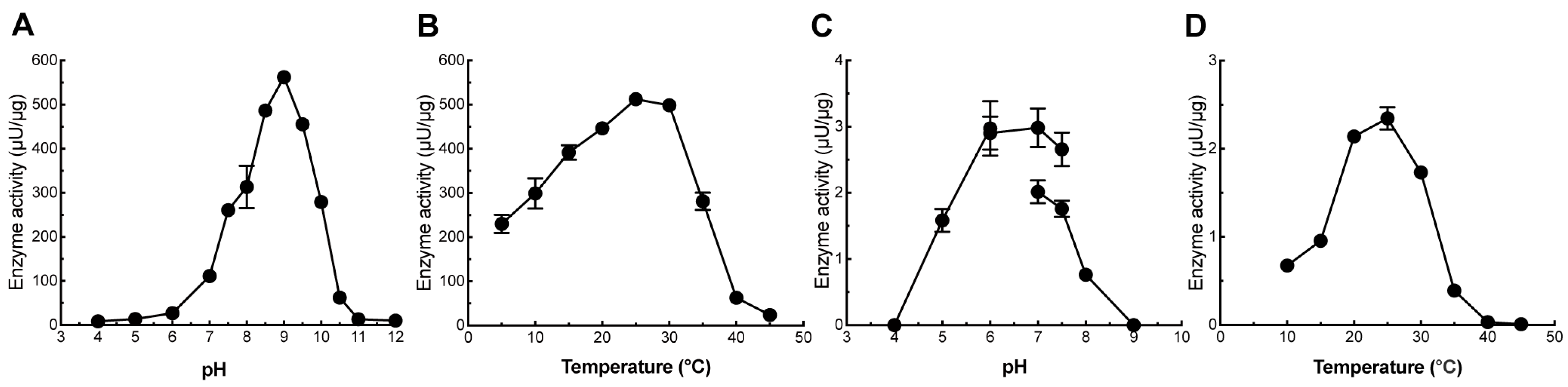
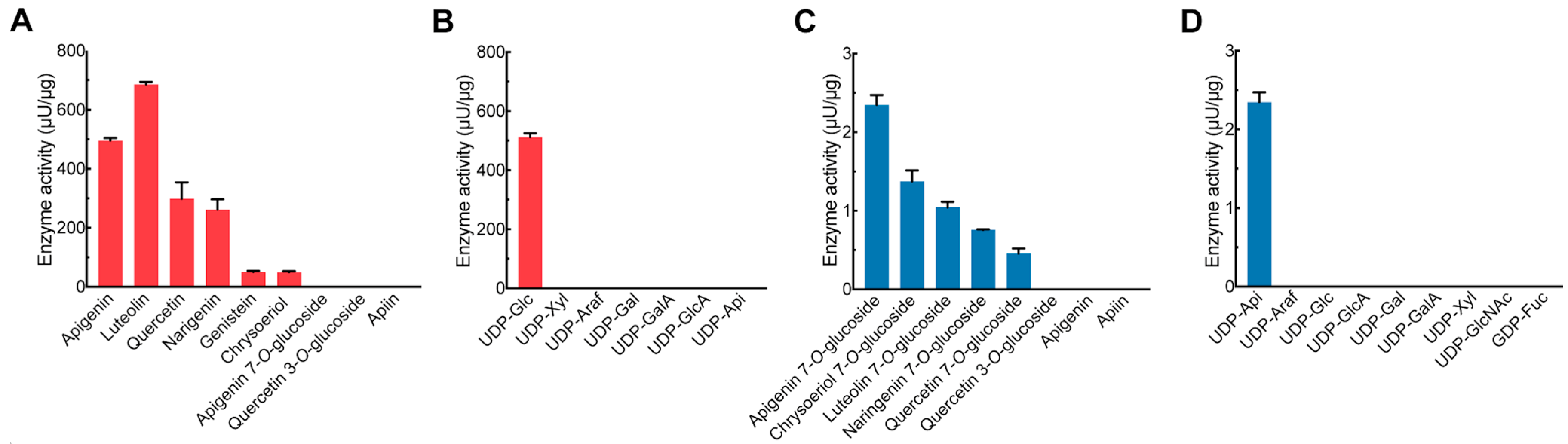
2.2. Biochemical Characterization of PcGlcT and PcApiT
2.3. PcGlcT and PcApiT Expression and Flavonoid Glucosides Accumulation Profiles in Parsley
3. Materials and Methods
3.1. Plant Cultivation
3.2. PcGlcT and PcApiT Candidate Genes
3.3. Heterologous Expression of PcGlcT and PcApiT
3.4. Enzymatic Assays of PcGlcT and PcApiT
3.5. Gene Expression Analysis
3.6. Quantification of Flavonoid 7-O-β-Apioglucosides in Parsley
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mahmood, S.; Hussain, S.; Malik, F. Critique of medicinal conspicuousness of parsley (Petroselinum crispum): A culinary herb of Mediterranean region. Pak. J. Pharm. Sci. 2014, 27, 193–202. [Google Scholar]
- Chomchalow, N. Production of herbs in Asia: An overview. AU JT 2002, 6, 95–108. [Google Scholar]
- Farzaei, M.H.; Abbasabadi, Z.; Ardekani, M.R.S.; Rahimi, R.; Farzaei, F. Parsley: A review of ethnopharmacology, phytochemistry and biological activities. J. Tradit. Chin. Med. 2013, 33, 815–826. [Google Scholar] [CrossRef]
- Pollastro, F.; Gaeta, S. Apiaceae, a family of species rich in secondary metabolites: Aromatic compounds and medicinal attributes. In Carrots and Related Apiaceae Crops; CABI: Wallingford, UK, 2020; pp. 35–46. [Google Scholar]
- Chaves, D.S.A.; Frattani, F.S.; Assafim, M.; de Almeida, A.P.; Zingali, R.B.; Costa, S.S. Phenolic chemical composition of Petroselinum crispum extract and its effect on haemostasis. Nat. Prod. Commun. 2011, 6, 961–964. [Google Scholar] [CrossRef]
- Kreuzaler, F.; Hahlbrock, K. Flavonoid glycosides from illuminated cell suspension cultures of Petroselinum hortense. Phytochemistry 1973, 12, 1149–1152. [Google Scholar] [CrossRef]
- Hudson, C.S. Apiose and the glycosides of the parsley plant. Adv. Carbohydr. Chem. 1949, 4, 57–74. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, F.; Wang, X.; Yao, H.-Y. Evaluation of antioxidant activity of parsley (Petroselinum crispum) essential oil and identification of its antioxidant constituents. Food Res. 2006, 39, 833–839. [Google Scholar] [CrossRef]
- Boutsika, A.; Sarrou, E.; Cook, C.M.; Mellidou, I.; Avramidou, E.; Angeli, A.; Martens, S.; Ralli, P.; Letsiou, S.; Selini, A.; et al. Evaluation of parsley (Petroselinum crispum) germplasm diversity from the greek gene bank using morphological, molecular and metabolic markers. Ind. Crops Prod. 2021, 170, 113767. [Google Scholar] [CrossRef]
- Eckey-Kaltenbach, H.; Heller, W.; Sonnenbichler, J.; Zetl, I.; Schäfer, W.; Ernst, D.; Sandermann, H. Oxidative stress and plant secondary metabolism: 6″-O-malonylapiin in parsley. Phytochemistry 1993, 34, 687–691. [Google Scholar] [CrossRef]
- Mara De Menezes Epifanio, N.; Rykiel Iglesias Cavalcanti, L.; Falcão Dos Santos, K.; Soares Coutinho Duarte, P.; Kachlicki, P.; Ozarowski, M.; Jorge Riger, C.; Siqueira De Almeida Chaves, D. Chemical characterization and in vivo antioxidant activity of parsley (Petroselinum crispum) aqueous extract. Food Funct. 2020, 11, 5346–5356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, M.; Jiang, B.; Yu, J.; Hao, Q.; Liu, W.; Hu, Z.; Zhang, Y.; Song, C. Extraction optimization, structural characterization and potential alleviation of hyperuricemia by flavone glycosides from celery seeds. Food Funct. 2022, 13, 9832–9846. [Google Scholar] [CrossRef]
- Adem, Ş.; Eyupoglu, V.; Ibrahim, I.M.; Sarfraz, I.; Rasul, A.; Ali, M.; Elfiky, A.A. Multidimensional in silico strategy for identification of natural polyphenols-based SARS-CoV-2 main protease (Mpro) inhibitors to unveil a hope against COVID-19. Comput. Biol. Med. 2022, 145, 105452. [Google Scholar] [CrossRef]
- Husain, I.; Ahmad, R.; Siddiqui, S.; Chandra, A.; Misra, A.; Srivastava, A.; Ahamad, T.; Khan, M.F.; Siddiqi, Z.; Trivedi, A.; et al. Structural interactions of phytoconstituent(s) from cinnamon, bay leaf, oregano, and parsley with SARS-CoV-2 nucleocapsid protein: A comparative assessment for development of potential antiviral nutraceuticals. J. Food Biochem. 2022, 46, e14262. [Google Scholar] [CrossRef]
- Roviello, V.; Gilhen-Baker, M.; Vicidomini, C.; Roviello, G.N. The healing power of clean rivers: In silico evaluation of the antipsoriatic potential of apiin and hyperoside plant metabolites contained in river waters. Int. J. Environ. Res. Public Health 2022, 19, 2502. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Takahashi, S.; Waki, T. Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 2019, 10, 821. [Google Scholar] [CrossRef]
- Ono, E.; Homma, Y.; Horikawa, M.; Kunikane-Doi, S.; Imai, H.; Takahashi, S.; Kawai, Y.; Ishiguro, M.; Fukui, Y.; Nakayama, T. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 2010, 22, 2856–2871. [Google Scholar] [CrossRef]
- Sasaki, N.; Nakayama, T. Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol. 2015, 56, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Isayenkova, J.; Lim, E.-K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef]
- Salas, J.A.; Méndez, C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007, 15, 219–232. [Google Scholar] [CrossRef]
- Thorson, J.S.; Vogt, T. Glycosylated natural products. In Carbohydrate-Based Drug Discovery; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 685–711. [Google Scholar]
- Yang, Q.; Zhang, Y.; Qu, X.; Wu, F.; Li, X.; Ren, M.; Tong, Y.; Wu, X.; Yang, A.; Chen, Y.; et al. Genome-wide analysis of UDP-glycosyltransferases family and identification of ugt genes involved in abiotic stress and flavonol biosynthesis in nicotiana tabacum. BMC Plant Biol. 2023, 23, 204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Feng, Y.; Ke, D.; Teng, Y.; Chen, Y.; Langjia, R. Molecular identification and characterization of UDP-glycosyltransferase (UGT) multigene family in pomegranate. Horticulturae 2023, 9, 540. [Google Scholar] [CrossRef]
- Watson, R.R.; Orenstein, N.S. Chemistry and biochemistry of apiose. Adv. Carbohydr. Chem. Biochem. 1975, 31, 135–184. [Google Scholar]
- Kashiwagi, T.; Horibata, Y.; Mekuria, D.B.; Tebayashi, S.; Kim, C.S. Ovipositional deterrent in the sweet pepper, Capsicum annuum, at the mature stage against Liriomyza trifolii (Burgess). Biosci. Biotechnol. Biochem. 2005, 69, 1831–1835. [Google Scholar] [CrossRef]
- Sutter, A.; Ortmann, R.; Grisebach, H. purification and properties of an enzyme from cell suspension cultures of parsley catalyzing the transfer of D-glucose from UDP-D-glucose to flavonoids. Biochim. Biophys. Acta Enzymol. 1972, 258, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, A.; Horikawa, M.; Fukui, Y.; Fukuchi-Mizutani, M.; Iuchi-Okada, A.; Ishiguro, M.; Kiso, Y.; Nakayama, T.; Ono, E. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in lamiales. Plant Cell 2009, 21, 1556–1572. [Google Scholar] [CrossRef]
- Ohgami, S.; Ono, E.; Toyonaga, H.; Watanabe, N.; Ohnishi, T. Identification and characterization of Camellia sinensis glucosyltransferase, UGT73A17: A possible role in flavonol glucosylation. Plant Biotechnol. 2014, 31, 573–578. [Google Scholar] [CrossRef]
- Hirotani, M.; Kuroda, R.; Suzuki, H.; Yoshikawa, T. Cloning and expression of UDP-Glucose: Flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 2000, 210, 1006–1013. [Google Scholar] [CrossRef]
- Su, X.; Wang, W.; Xia, T.; Gao, L.; Shen, G.; Pang, Y. Characterization of a heat responsive UDP: Flavonoid glucosyltransferase gene in tea plant (Camellia sinensis). PLoS ONE 2018, 13, e0207212. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, B.G.; Park, Y.; Ko, J.H.; Lim, C.E.; Lim, J.; Lim, Y.; Ahn, J.-H. Characterization of flavonoid 7-O-glucosyltransferase from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2006, 70, 1471–1477. [Google Scholar] [CrossRef]
- Zhu, T.-T.; Liu, H.; Wang, P.-Y.; Ni, R.; Sun, C.-J.; Yuan, J.-C.; Niu, M.; Lou, H.-X.; Cheng, A.-X. Functional characterization of UDP-glycosyltransferases from the liverwort Plagiochasma appendiculatum and their potential for biosynthesizing flavonoid 7-O-glucosides. Plant Sci. 2020, 299, 110577. [Google Scholar] [CrossRef]
- Noguchi, A.; Saito, A.; Homma, Y.; Nakao, M.; Sasaki, N.; Nishino, T.; Takahashi, S.; Nakayama, T. A UDP-glucose:isoflavone 7-O-glucosyltransferase from the roots of soybean (glycine max) seedlings. Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J. Biol. Chem. 2007, 282, 23581–23590. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shi, X.; Yang, C.; Zhao, X.; Zhuang, J.; Liu, Y.; Gao, L.; Xia, T. Two UDP-glycosyltransferases catalyze the biosynthesis of bitter flavonoid 7-O-neohesperidoside through sequential glycosylation in tea plants. J. Agric. Food Chem. 2022, 70, 2354–2365. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shen, G.; Di, S.; Dixon, R.A.; Pang, Y. Characterization of UGT716A1 as a multi-substrate UDP:flavonoid glucosyltransferase gene in Ginkgo Biloba. Front. Plant Sci. 2017, 8, 2085. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Hughes, M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 1994, 5, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Ortmann, R.; Sandermann, H.; Grisebach, H. Transfer of apiose from UDP-Apiose to 7-O-(β-D-glucosyl)-apigenin and 7-O-(β-D-glucosyl)-chrysoeriol with an enzyme preparation from parsley. FEBS Lett. 1970, 7, 164–166. [Google Scholar] [CrossRef]
- Ortmann, R.; Sutter, A.; Grisebach, H. Purification and properties of UDP-apiose: 7-O-(β-D-glucosyl)-flavone apiosyltransferase from cell suspension cultures of parsley. Biochim. Biophys. Acta Enzymol. 1972, 289, 293–302. [Google Scholar] [CrossRef]
- Fujimori, T.; Matsuda, R.; Suzuki, M.; Takenaka, Y.; Kajiura, H.; Takeda, Y.; Ishimizu, T. Practical preparation of UDP-apiose and its applications for studying apiosyltransferase. Carbohydr. Res. 2019, 477, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Fujimori, T.; An, S.; Iguchi, S.; Takenaka, Y.; Kajiura, H.; Yoshizawa, T.; Matsumura, H.; Kobayashi, M.; Ono, E.; et al. The apiosyltransferase celery UGT94AX1 catalyzes the biosynthesis of the flavone glycoside apiin. Plant Physiol. 2023, 193, 1758–1771. [Google Scholar] [CrossRef]
- Wang, H.-T.; Wang, Z.-L.; Chen, K.; Yao, M.-J.; Zhang, M.; Wang, R.-S.; Zhang, J.-H.; Ågren, H.; Li, F.-D.; Li, J.; et al. Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants. Nat. Commun. 2023, 14, 6658. [Google Scholar] [CrossRef]
- Griesser, M.; Vitzthum, F.; Fink, B.; Bellido, M.L.; Raasch, C.; Munoz-Blanco, J.; Schwab, W. Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria×ananassa) achene and receptacle. J. Exp. Bot. 2008, 59, 2611–2625. [Google Scholar] [CrossRef] [PubMed]
- Durren, R.L.; McIntosh, C.A. Flavanone-7-O-glucosyltransferase activity from Petunia hybrida. Phytochemistry 1999, 52, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, J.; Yue, J.; Huang, Z.; Zhang, L.; Yao, W.; Guan, R.; Wu, J.; Liang, J.; Duan, L.; et al. Functional characterization of a novel glycosyltransferase (UGT73CD1) from Iris tectorum Maxim. for the substrate promiscuity. Mol. Biotechnol. 2021, 63, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Y.; Liu, Y.; Zhao, L.; Pei, J. Screening and characterizing flavone synthases and its application in biosynthesizing vitexin from naringenin by a one-pot enzymatic cascade. Enzym. Microb. Technol. 2022, 160, 110101. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization by One Table (TvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W58–W592. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Ishimizu, T.; Uchida, T.; Sano, K.; Hase, S. Chemical synthesis of uridine 5′-diphospho-α-D-xylopyranose. Tetrahedron Asymmetry 2005, 16, 309–311. [Google Scholar] [CrossRef]
- Ohashi, T.; Cramer, N.; Ishimizu, T.; Hase, S. Preparation of UDP-galacturonic acid using UDP-sugar pyrophosphorylase. Anal. Biochem. 2006, 352, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yu, L.; Xu, S.; Gu, W.; Zhu, W. Apigenin accumulation and expression analysis of apigenin biosynthesis relative genes in celery. Sci. Hortic. 2014, 165, 218–224. [Google Scholar] [CrossRef]
- Li, M.Y.; Song, X.; Wang, F.; Xiong, A.S. Suitable reference genes for accurate gene expression analysis in parsley (Petroselinum crispum) for abiotic stresses and hormone stimuli. Front. Plant Sci. 2016, 7, 1481. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Lin, L.Z.; Lu, S.; Harnly, J.M. Detection and quantification of glycosylated flavonoid malonates in celery, Chinese celery, and celery seed by LC-DAD-ESI/MS. J. Agric. Food Chem. 2007, 55, 1321–1326. [Google Scholar] [CrossRef]
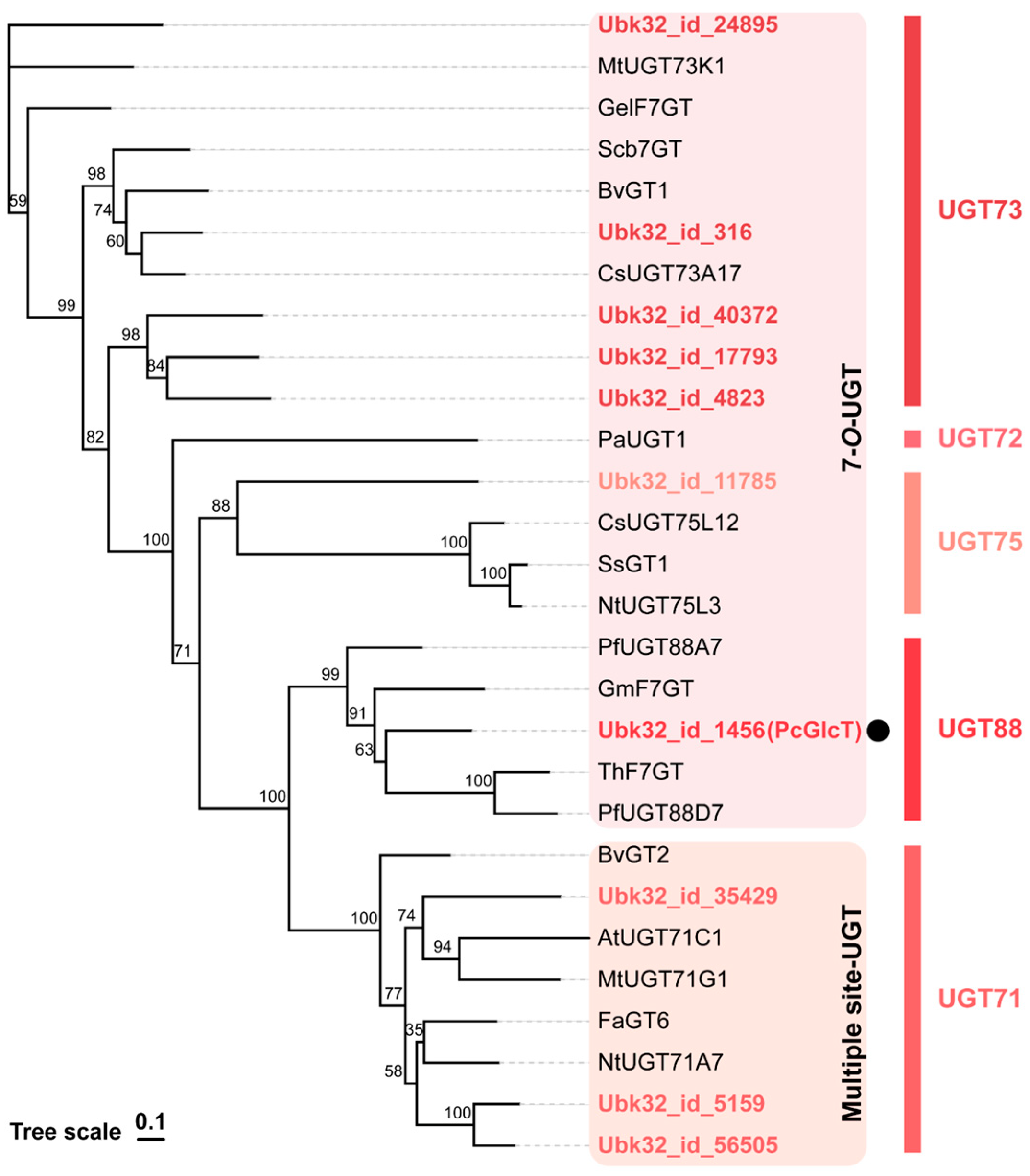
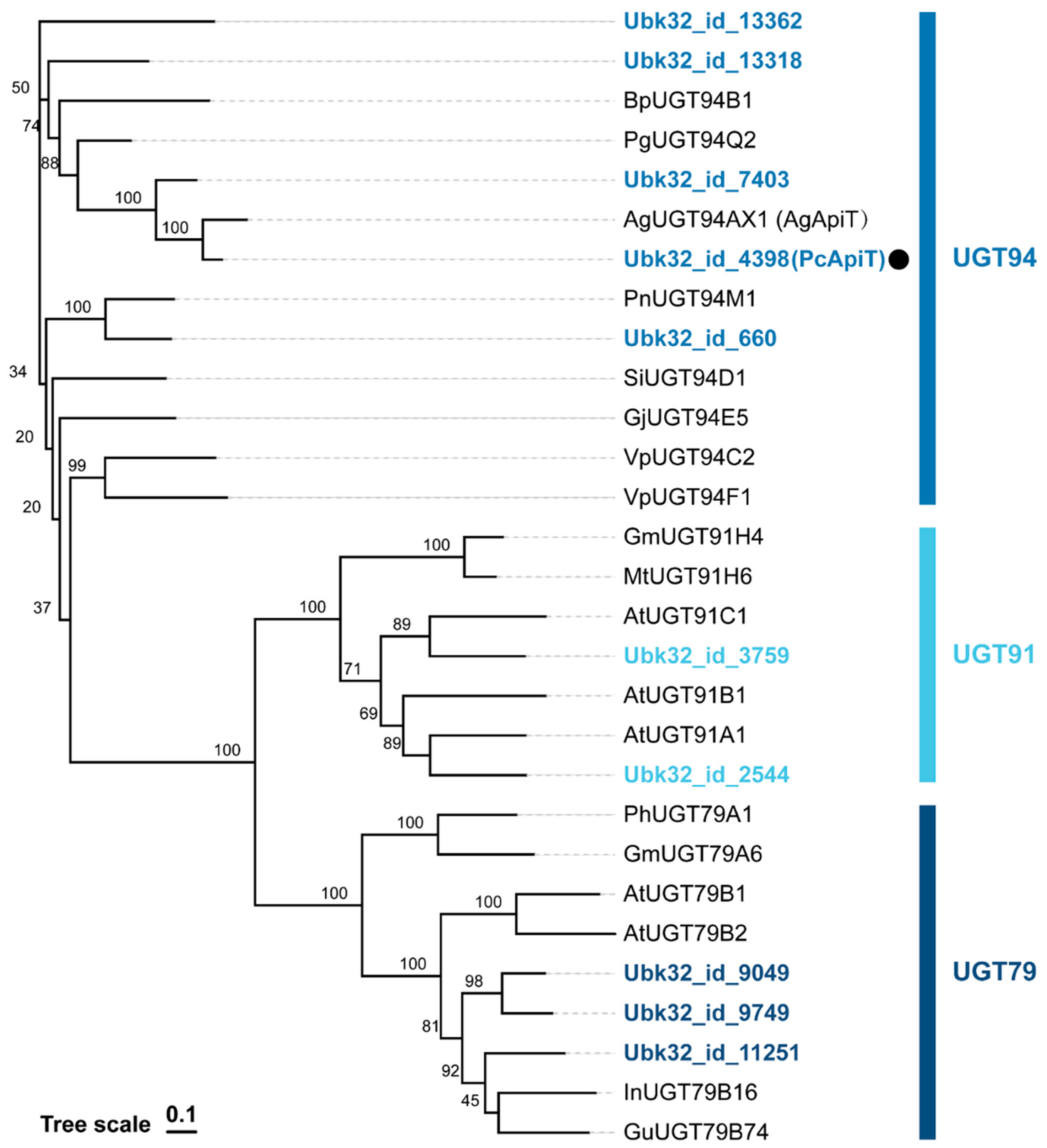

| Protein ID | UGT Family | Regioselectivity | TPM |
|---|---|---|---|
| Ubk32_id_1456 (PcGlcT) | UGT88 | 7-O-UGT | 59.7 |
| Ubk32_id_316 | UGT73 | 7-O-UGT | 16.1 |
| Ubk32_id_5159 | UGT71 | Multiple site-UGT | 9.8 |
| Ubk32_id_11785 | UGT75 | 7-O-UGT | 9.1 |
| Ubk32_id_35429 | UGT71 | Multiple site-UGT | 4.2 |
| Ubk32_id_4823 | UGT73 | 7-O-UGT | 1.8 |
| Ubk32_id_17793 | UGT73 | 7-O-UGT | 1.6 |
| Ubk32_id_24895 | UGT73 | 7-O-UGT | 1.4 |
| Ubk32_id_40372 | UGT73 | 7-O-UGT | 1.1 |
| Ubk32_id_56505 | UGT71 | Multiple site-UGT | 1.0 |
| Protein ID | UGT Family | TPM | Identity to AgApiT (%) |
|---|---|---|---|
| Ubk32_id_4398 | UGT94 | 147.1 | 78 |
| Ubk32_id_2544 | UGT91 | 61.4 | 28 |
| Ubk32_id_11251 | UGT79 | 25.8 | 26 |
| Ubk32_id_9049 | UGT79 | 6.7 | 27 |
| Ubk32_id_660 | UGT94 | 4.9 | 41 |
| Ubk32_id_9749 | UGT79 | 3.5 | 24 |
| Ubk32_id_7403 | UGT94 | 3.1 | 62 |
| Ubk32_id_13362 | UGT94 | 2.1 | 39 |
| Ubk32_id_3759 | UGT91 | 1.6 | 30 |
| Ubk32_id_13318 | UGT94 | 1.5 | 44 |
| Source | Enzyme | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1/mM) |
|---|---|---|---|---|---|
| Parsley | PcGlcT | Apigenin | 320 ± 70 | (6.2 ± 0.5) × 10−1 | 1.97 ± 0.69 |
| Lamiales | UGT88D7 [30] | Apigenin | 8.5 ± 1.9 | (6.0 ± 0.3) ×10−3 | 0.71 |
| Liverwort | PaUGT1 [35] | Apigenin | 22.2 ± 3.3 | (2.2 ± 0.1) ×10−3 | 0.098 |
| Tea | UGT73A17 [33] | Apigenin | 5.5 ± 1.5 | 2.3 × 10−3 | 0.54 |
| Tea | UGT75L12 [37] | Apigenin | 4.6 ± 0.2 | 2.1 × 10−2 | 4.54 |
| Ginkgo biloba | UGT716A1 [38] | Apigenin | 230 ± 0.0 | 1.7 × 10−2 | 0.073 |
| Parsley | PcGlcT | UDP-Glc | 610 ± 110 | (6.2 ± 0.5) × 10−1 | 1.01 ± 0.44 |
| Lamiales | UGT88D7 [30] | UDP-Glc | 540 ± 20 | 0.011 | |
| Tea | UGT75L12 [37] | UDP-Glc | 233.3 ± 41.7 | 14.9 × 10−2 | 0.64 |
| Source | Enzyme | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1/mM) |
|---|---|---|---|---|---|
| Parsley | PcApiT | Apigenin 7-O-Glc | 81 ± 20 | 3.2 ± 0.3 | 40 ± 17 |
| Celery | AgApiT [44] | Apigenin 7-O-Glc | 15 ± 3 | 0.88 ± 0.05 | 58 ± 15 |
| Parsley | PcApiT | UDP-Api | 360 ± 40 | 4.5 ± 0.2 | 12 ± 5 |
| Celery | AgApiT [44] | UDP-Api | 8.6 ± 0.6 | 0.65 ± 0.01 | 76 ± 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, S.; Yamashita, M.; Iguchi, S.; Kihara, T.; Kamon, E.; Ishikawa, K.; Kobayashi, M.; Ishimizu, T. Biochemical Characterization of Parsley Glycosyltransferases Involved in the Biosynthesis of a Flavonoid Glycoside, Apiin. Int. J. Mol. Sci. 2023, 24, 17118. https://doi.org/10.3390/ijms242317118
An S, Yamashita M, Iguchi S, Kihara T, Kamon E, Ishikawa K, Kobayashi M, Ishimizu T. Biochemical Characterization of Parsley Glycosyltransferases Involved in the Biosynthesis of a Flavonoid Glycoside, Apiin. International Journal of Molecular Sciences. 2023; 24(23):17118. https://doi.org/10.3390/ijms242317118
Chicago/Turabian StyleAn, Song, Maho Yamashita, Sho Iguchi, Taketo Kihara, Eri Kamon, Kazuya Ishikawa, Masaru Kobayashi, and Takeshi Ishimizu. 2023. "Biochemical Characterization of Parsley Glycosyltransferases Involved in the Biosynthesis of a Flavonoid Glycoside, Apiin" International Journal of Molecular Sciences 24, no. 23: 17118. https://doi.org/10.3390/ijms242317118
APA StyleAn, S., Yamashita, M., Iguchi, S., Kihara, T., Kamon, E., Ishikawa, K., Kobayashi, M., & Ishimizu, T. (2023). Biochemical Characterization of Parsley Glycosyltransferases Involved in the Biosynthesis of a Flavonoid Glycoside, Apiin. International Journal of Molecular Sciences, 24(23), 17118. https://doi.org/10.3390/ijms242317118






