Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål)
Abstract
:1. Introduction
2. BPH-Resistance Gene Mapping
3. Cloning and Mechanisms of BPH-Resistance Genes
3.1. CC–NB–LRR Gene
3.2. Atypical CC–NB–LRR Genes
3.3. LRD Genes
3.4. LecRK Genes
3.5. Other Types of BPH-Resistance Genes
4. Responses of Rice to BPH Infection
4.1. MAPK Signal Transduction
4.2. Phytohormones
4.3. Transcription Factors
4.4. Metabolites
4.5. Calcium Signaling
4.6. MicroRNAs
5. BPH-Secreted Proteins That Involved in Rice–BPH Interactions
5.1. BPH Elicitors
5.2. BPH Effectors
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, X.; Zhu, L.; He, G. Genetic and molecular understanding of host rice resistance and Nilaparvata lugens adaptation. Curr. Opin. Insect Sci. 2021, 45, 14–20. [Google Scholar] [CrossRef]
- Shi, S.; Wang, H.; Nie, L.; Tan, D.; Zhou, C.; Zhang, Q.; Li, Y.; Du, B.; Guo, J.; Huang, J.; et al. Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol. Plant 2021, 14, 1714–1732. [Google Scholar] [CrossRef] [PubMed]
- Muduli, L.; Pradhan, S.K.; Mishra, A.; Bastia, D.N.; Samal, K.C.; Agrawal, P.K.; Dash, M. Understanding brown planthopper resistance in rice: Genetics, biochemical and molecular breeding approaches. Rice Sci. 2021, 28, 532–546. [Google Scholar] [CrossRef]
- Chang, X.; Wang, F.; Fang, Q.; Chen, F.; Yao, H.; Gatehouse, A.; Ye, G. Virus-induced plant volatiles mediate the olfactory behaviour of its insect vectors. Plant Cell Environ. 2021, 44, 2700–2715. [Google Scholar] [CrossRef]
- Sarao, P.S.; Sahi, G.K.; Neelam, K.; Mangat, G.S.; Patra, B.C.; Singh, K. Donors for resistance to brown planthopper Nilaparvata lugens Stål from wild rice species. Rice Sci. 2016, 23, 219–224. [Google Scholar] [CrossRef]
- Feng, C.; Wan, Z.; Lan, L.; Le, K. Rice responses and resistance to planthopper-borne viruses at transcriptomic and proteomic levels. Curr. Issues Mol. Biol. 2015, 19, 43–52. [Google Scholar]
- Min, S.; Lee, S.W.; Choi, B.R.; Lee, S.H.; Kwon, D.H. Insecticide resistance monitoring and correlation analysis to select appropriate insecticides against Nilaparvata lugens (Stål), a migratory pest in Korea. J. Asia-Pac. Entomol. 2014, 17, 711–716. [Google Scholar] [CrossRef]
- Lu, K.; Chen, X.; Liu, W.; Zhang, Z.; Wang, Y.; You, K.; Li, Y.; Zhang, R.; Zhou, Q. Characterization of heat shock protein 70 transcript from Nilaparvata lugens Stål: Its response to temperature and insecticide stresses. Pestic. Biochem. Phys. 2017, 42, 102–110. [Google Scholar] [CrossRef]
- Alam, M.J.; Das, G. Toxicity of insecticides to predators of rice brown planthopper: Wolf spider and carabid beetle. J. Sci. Food Agric. 2020, 3, 9–13. [Google Scholar] [CrossRef]
- Wu, J.; Ge, L.; Liu, F.; Song, Q.; Stanley, D. Pesticide-induced planthopper population resurgence in rice cropping systems. Annu. Rev. Entomol. 2020, 65, 409–429. [Google Scholar] [CrossRef]
- Ge, L.Q.; Huang, L.J.; Yang, G.Q.; Song, Q.S.; Stanley, D.; Gurr, G.M.; Wu, J.C. Molecular basis for insecticide-enhanced thermotolerance in the brown planthopper Nilaparvata lugens Stål (Hemiptera: Delphacidae). Mol. Ecol. 2013, 22, 5624–5634. [Google Scholar] [CrossRef]
- Wu, S.; Zeng, B.; Zheng, C.; Mu, X.; Zhang, Y.; Hu, J.; Zhang, S.; Gao, C.; Shen, J. The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012–2016. Sci. Rep. 2018, 8, 4586. [Google Scholar] [CrossRef]
- Zeng, Q.; Yu, C.; Chang, X.; Wan, Y.; Ba, Y.; Li, C.; Lv, H.; Guo, Z.; Cai, T.; Ren, Z.; et al. CeO2 nanohybrid as a synergist for insecticide resistance management. Chem. Eng. J. 2022, 446, 137074. [Google Scholar] [CrossRef]
- Jin, R.; Wang, Y.; He, B.; Zhang, Y.; Cai, T.; Wan, H.; Jin, B.R.; Li, J. Activator protein-1 mediated CYP6ER1 overexpression in the clothianidin resistance of Nilaparvata lugens (Stål). Pest Manag. Sci. 2021, 77, 4476–4482. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Jin, R.; Zhang, X.; Ali, E.; Mao, K.; Xu, P.; Li, J.; Wan, H. Characterization of sulfoxaflor resistance in the brown planthopper, Nilaparvata lugens (Stål). Pest Manag. Sci. 2019, 75, 1646–1654. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Mao, K.; Liao, X.; Xu, P.; Li, Z.; Ali, E.; Wan, H.; Li, J. Overexpression of CYP6ER1 associated with clothianidin resistance in Nilaparvata lugens (Stål). Pestic. Biochem. Physiol. 2019, 154, 39–45. [Google Scholar] [CrossRef]
- Sharma, H.C. Host plant resistance to insects: Modern approaches and limitations. Plant Prot. Assoc. India 2007, 35, 179–184. [Google Scholar]
- Gurr, G.M.; Liu, J.; Read, D.; Catindig, J.; Cheng, J.A.; Lan, L.P.; Heong, K.L. Parasitoids of Asian rice planthopper (Hemiptera: Delphacidae) pests and prospects for enhancing biological control by ecological engineering. Ann. Appl. Biol. 2015, 158, 149–176. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Pathak, M.; Cheng, C.; Fortuno, M. Resistance to Nephotettix impicticeps and Nilaparvata lugens in varieties of rice. Nature 1969, 223, 502–504. [Google Scholar] [CrossRef]
- Hirabayashi, H.; Ogawas, T. RFLP mapping of Bph-1 (brown planthopper resistance gene) in rice. Jpn. J. Breed. 1995, 45, 369–371. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, Q.; Chen, Y.; Huang, J.; Guo, Q.; Li, Y.; Wang, W.; Qiu, Y.; Guan, W.; Zhang, J.; et al. Balancing selection and wild gene pool contribute to resistance in global rice germplasm against planthopper. J. Integr. Plant Biol. 2021, 63, 1695–1711. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.V.; Hechanova, S.L.; Hernandez, J.E.; Li, C.P.; Tülek, A.; Ahn, E.K.; Jairin, J.; Choi, I.R.; Sundaram, R.M.; Jena, K.K.; et al. Available cloned genes and markers for genetic improvement of biotic stress resistance in rice. Front. Plant Sci. 2023, 14, 1247014. [Google Scholar] [CrossRef] [PubMed]
- Kiswanto, I.; Soetopo, L.; Adiredjo, A.L. Identification of novel candidate of brown planthopper resistance gene Bph44 in rice (Oryza sativa L.). Genome 2022, 65, 505–511. [Google Scholar] [CrossRef]
- Liu, M.; Fan, F.; He, S.; Guo, Y.; Chen, G.; Li, N.; Li, N.; Yuan, H.; Si, F.; Yang, F.; et al. Creation of elite rice with high-yield, superior-quality and high resistance to brown planthopper based on molecular design. Rice 2022, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Cheng, L.; Yan, L.; Shu, W.; Wang, X.; Qiu, Y. Mapping and characterization of a quantitative trait locus resistance to the brown planthopper in the rice variety IR64. Hereditas 2019, 156, 22. [Google Scholar] [CrossRef] [PubMed]
- Balachiranjeevi, C.H.; Prahalada, G.D.; Mahender, A.; Jamaloddin, M.; Sevilla, M.A.L.; Marfori-Nazarea, C.M.; Vinarao, R.; Sushanto, U.; Baehaki, S.E.; Li, Z.; et al. Identification of a novel locus, BPH38(t), conferring resistance to brown planthopper (Nilaparvata lugens Stål.) using early backcross population in rice (Oryza sativa L.). Euphytica 2019, 215, 185. [Google Scholar] [CrossRef]
- Naik, S.B.; Divya, D.; Sahu, N.; Sundaram, R.M.; Sarao, P.S.; Singh, K.; Lakshmi, V.J.; Bentur, J.S. A new gene Bph33(t) conferring resistance to brown planthopper (BPH), Nilaparvata lugens (Stål) in rice line RP2068-18-3-5. Euphytica 2018, 214, 53. [Google Scholar] [CrossRef]
- Renganayaki, K.; Fritz, A.K.; Sadasivam, S.; Pammi, S.; Harrington, S.E.; McCouch, S.R.; Kumar, S.M.; Reddy, A.S. Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice, O. sativa. Crop Sci. 2002, 42, 2112–2117. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Pang, X.; Pan, Q. Genetic analysis and fine mapping of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene bph19(t). Mol. Genet. Genom. 2006, 275, 321–329. [Google Scholar] [CrossRef]
- Prahalada, G.D.; Shivakumar, N.; Lohithaswa, H.C.; Gowda, D.K.S.; Ramkumar, G.; Kim, S.R.; Ramachandra, C.; Hittalmani, S.; Mohapatra, T.; Jena, K.K. Identification and fine mapping of a new gene, BPH31 conferring resistance to brown planthopper biotype 4 of India to improve rice, Oryza sativa L. Rice 2017, 10, 41. [Google Scholar] [CrossRef]
- Hirabayashi, H.; Angeles, E.R.; Kaji, R.; Ogawa, T.; Brar, D.S.; Khush, G.S. Identification of a brown planthopper resistance gene derived from O. officinalis using molecular markers in rice. Breed. Sci. 1998, 48, 82. [Google Scholar]
- Hu, J.; Xiao, C.; Cheng, M.; Gao, G.; Zhang, Q.; He, Y. Fine mapping and pyramiding of brown planthopper resistance genes QBph3 and QBph4 in an introgression line from wild rice O. officinalis. Mol. Breed. 2015, 35, 3. [Google Scholar] [CrossRef]
- Du, B.; Zhang, W.; Liu, B.; Hu, J.; Wei, Z.; Shi, Z.; He, R.; Zhu, L.; Chen, R.; Han, B.; et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. USA 2009, 106, 22163–22168. [Google Scholar] [CrossRef]
- Mohanty, S.K.; Panda, R.S.; Mohapatra, S.L.; Nanda, A.; Behera, L.; Jena, M.; Sahu, R.K.; Sahu, S.C.; Mohapatra, T. Identification of novel quantitative trait loci associated with brown planthopper resistance in the rice landrace Salkathi. Euphytica 2017, 213, 38. [Google Scholar] [CrossRef]
- Hu, J.; Chang, X.; Zou, L.; Tang, W.; Wu, W. Identification and fine mapping of Bph33, a new brown planthopper resistance gene in rice (Oryza sativa L.). Rice 2018, 11, 55. [Google Scholar] [CrossRef]
- Tan, H.; Palyam, S.; Gouda, J.; Kumar, P.P.; Chellian, S.K. Identification of two QTLs, BPH41 and BPH42, and their respective gene candidates for brown planthopper resistance in rice. Sci. Rep. 2022, 12, 18538. [Google Scholar] [CrossRef] [PubMed]
- Kamolsukyeunyong, W.; Ruengphayak, S.; Chumwong, P.; Kusumawati, L.; Chaichoompu, E.; Jamboonsri, W.; Saensuk, C.; Phoonsiri, K.; Toojinda, T.; Vanavichit, A. Identification of spontaneous mutation for broad-spectrum brown planthopper resistance in a large, long-term fast neutron mutagenized rice population. Rice 2019, 12, 16. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2015, 33, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qin, G.; Ma, Q.; Wei, M.; Yang, X.; Ma, Z.; Liang, H.; Liu, C.; Li, Z.; Liu, F.; et al. Identification of a major resistance locus Bph35 to brown planthopper in rice (Oryza sativa L.). Rice Sci. 2020, 27, 237–245. [Google Scholar]
- Hu, J.; Xiao, C.; Cheng, M.; Gao, G.; Zhang, Q.; He, Y. A new finely mapped Oryza australiensis-derived QTL in rice confers resistance to brown planthopper. Gene 2015, 561, 132–137. [Google Scholar] [CrossRef]
- Li, Z.; Xue, Y.; Zhou, H.; Li, Y.; Usman, B.; Jiao, X.; Wang, X.; Liu, F.; Qin, B.; Li, R.; et al. High-resolution mapping and breeding application of a novel brown planthopper resistance gene derived from wild rice (Oryza rufipogon Griff). Rice 2019, 12, 41. [Google Scholar] [CrossRef]
- Rahman, M.L.; Jiang, W.Z.; Chu, S.H.; Qiao, Y.L.; Ham, T.H.; Woo, M.O.; Lee, J.; Khanam, M.S.; Chin, J.H.; Jeung, J.U.; et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor. Appl. Genet. 2009, 119, 1237–1246. [Google Scholar] [CrossRef]
- Sun, L.; Su, C.; Wang, C.; Zhai, H.; Wan, J. Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breed. Sci. 2005, 55, 391–396. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Liu, Y.; Jiang, L.; Wu, H.; Kang, H.; Liu, S.; Chen, L.; Liu, X.; Cheng, X.; et al. High-resolution mapping of brown planthopper (BPH) resistance gene Bph27(t) in rice (Oryza sativa L.). Mol. Breed. 2013, 31, 549–557. [Google Scholar] [CrossRef]
- Guo, J.; Xu, C.; Wu, D.; Zhao, Y.; Qiu, Y.; Wang, X.; Ouyang, Y.; Cai, B.; Liu, X.; Jing, S.; et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018, 50, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Sarao, P.S.; Bhatia, D.; Neelam, K.; Kaur, A.; Mangat, G.S.; Brar, D.S.; Singh, K. High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. × Oryza nivara (Sharma & Shastry) derived interspecific F2 population. Theor. Appl. Genet. 2018, 131, 1163–1171. [Google Scholar] [PubMed]
- Myint, K.K.M.; Fujita, D.; Matsumura, M.; Sonoda, T.; Yoshimura, A.; Yasui, H. Mapping and pyramiding of two major genes for resistance to the brown planthopper (Nilaparva talugens [Stål]) in the rice cultivar ADR52. Theor. Appl. Genet. 2012, 124, 495–504. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, L.; Zhang, Y.; Cao, C.; Liu, F.; Huang, F.; Qiu, Y.; Li, R.; Luo, X. Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J. Exp. Bot. 2015, 66, 6035–6045. [Google Scholar] [CrossRef]
- Ren, J.; Gao, F.; Wu, X.; Lu, X.; Zeng, L.; Lv, J.; Su, X.; Luo, H.; Ren, G. Bph32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice. Sci. Rep. 2016, 6, 37645. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Murata, K.; Ishii, T.; Takumi, S.; Mori, N.; Nakamura, C. Assignment of a brown planthopper (Nilaparvata lugens Stål) resistance gene bph4 to the rice chromosome 6. Breed. Sci. 2001, 51, 13–18. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Y.; He, J.; Liu, Y.; Jiang, L.; Liu, L.; Wang, C.; Cheng, X.; Wan, J. Fine mapping of brown planthopper (Nilaparvata lugens Stål) resistance gene Bph28(t) in rice (Oryza sativa L.). Mol. Breed. 2014, 33, 909–918. [Google Scholar] [CrossRef]
- Sun, L.; Wang, C.; Su, C.; Liu, Y.; Zhai, H.; Wan, J. Mapping and marker-assisted selection of a brown planthopper resistance gene bph2 in rice (Oryza sativa L.). Acta Genet. Sin. 2006, 33, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, J.; Wang, Z.; Jing, S.; Wang, Y.; Ouyang, Y.; Cai, B.; Xin, X.; Liu, X.; Zhang, C.; et al. Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc. Natl. Acad. Sci. USA 2016, 113, 12850–12855. [Google Scholar] [CrossRef]
- Ishii, T.; Brar, D.S.; Multani, D.S.; Khush, G.S. Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice O. sativa. Genome 1994, 37, 217–221. [Google Scholar] [CrossRef]
- Su, C.; Zhai, H.; Wang, C.; Sun, L.; Wan, J. SSR mapping of brown planthopper resistance gene Bph9 in Kaharamana, an indica rice (Oryza sativa L.). Acta Genet. Sin. 2006, 33, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.N.; Ketipearachchi, Y.; Murata, K.; Torii, A.; Takumi, S.; Mori, N.; Nakamura, C. RFLP/AFLP mapping of a brown planthopper (Nilaparvata lugens Stål) resistance gene Bph1 in rice. Euphytica 2002, 129, 109–117. [Google Scholar] [CrossRef]
- Tamura, Y.; Hattori, M.; Yoshioka, H.; Yoshioka, M.; Takahashi, A.; Wu, J.; Sentoku, N.; Yasui, H. Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci. Rep. 2014, 4, 5872. [Google Scholar]
- Ji, H.; Kim, S.R.; Kim, Y.H.; Suh, J.P.; Park, H.M.; Sreenivasulu, N.; Misra, G.; Kim, S.M.; Hechanova, S.L.; Kim, H.; et al. Map-based cloning and characterization of the BPH18 gene from wild rice conferring resistance to brown planthopper (BPH) insect pest. Sci. Rep. 2016, 6, 34376. [Google Scholar] [CrossRef]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef]
- Mishra, A.; Barik, S.R.; Pandit, E.; Yadav, S.S.; Das, S.R.; Pradhan, S.K. Genetics, mechanisms and deployment of brown planthopper resistance genes in rice. Crit. Rev. Plant Sci. 2022, 41, 91–127. [Google Scholar] [CrossRef]
- Yang, L.; Li, R.; Li, Y.; Huang, F.; Chen, Y.; Huang, S.; Huang, L.; Liu, C.; Ma, Z.; Huang, D.; et al. Genetic mapping of bph20(t) and bph21(t) loci conferring brown planthopper resistance to Nilaparvata lugens Stål in rice (Oryza sativa L.). Euphytica 2012, 183, 161–171. [Google Scholar] [CrossRef]
- Hettenhausen, C.; Schuman, M.C.; Wu, J. MAPK signaling: A key element in plant defense response to insects. Insect Sci. 2015, 22, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Wang, Q.; Huangfu, J.; Schuman, M.C.; Lou, Y. A group D MAPK protects plants from autotoxicity by suppressing herbivore-induced defense signaling. Plant Physiol. 2019, 179, 1386–1401. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Chen, M.; Zhang, Y.; Gao, Q.; Noman, A.; Wang, Q.; Li, H.; Chen, L.; Zhou, P.; Lu, J.; et al. OsMKK3, a stress-responsive protein kinase, positively regulates rice resistance to Nilaparvata lugens via phytohormone dynamics. Int. J. Mol. Sci. 2019, 20, 3023. [Google Scholar] [CrossRef] [PubMed]
- Nanda, S.; Wan, P.; Yuan, S.; Lai, F.; Wang, W.; Fu, Q. Differential responses of OsMPKs in IR56 rice to two BPH populations of different virulence levels. Int. J. Mol. Sci. 2018, 19, 4030. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Sun, Z.; Wang, R.; Du, Y.; Zhang, Z.; Lan, T.; Song, Y.; Zeng, R. Integration of transcriptome and metabolome analyses reveals the role of OsSPL10 in rice defense against brown planthopper. Plant Cell Rep. 2023, 42, 2023–2038. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Lin, X.; Yuan, X.; Zou, J.; Zhang, H.; Zhang, Y.; Liu, Z. The salivary chaperone protein NlDNAJB9 of Nilaparvata lugens activated plant immune responses. J. Exp. Bot. 2023, 27, erad154. [Google Scholar]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Zu, H.; Zeng, X.; Baldwin, I.T.; Lou, Y.; Li, R. Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore. New Phytol. 2021, 230, 1639–1652. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.; Ju, H.; Liu, X.; Erb, M.; Wang, X.; Lou, Y. Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol. Plant 2014, 7, 1670–1682. [Google Scholar] [CrossRef]
- Chen, L.; Cao, T.; Zhang, J.; Lou, Y. Overexpression of OsGID1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2018, 19, 2744. [Google Scholar] [CrossRef]
- Pan, G.; Liu, Y.; Ji, L.; Zhang, X.; He, J.; Huang, J.; Qiu, Z.; Liu, D.; Sun, Z.; Xu, T.; et al. Brassinosteroids mediate susceptibility to brown planthopper by integrating with the salicylic acid and jasmonic acid pathways in rice. J. Exp. Bot. 2018, 69, 4433–4442. [Google Scholar] [CrossRef]
- Liu, J.; Du, H.; Ding, X.; Zhou, Y.; Xie, P.; Wu, J. Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae). Pest Manag. Sci. 2017, 73, 2559–2568. [Google Scholar] [CrossRef]
- Shi, S.; Zha, W.; Yu, X.; Wu, Y.; Li, S.; Xu, H.; Li, P.; Li, C.; Liu, K.; Chen, J.; et al. Integrated transcriptomics and metabolomics analysis provide insight into the resistance response of rice against brown planthopper. Front. Plant Sci. 2023, 14, 1213257. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; de Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef]
- Huangfu, J.; Li, J.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.; Lou, Y. The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ye, M.; Li, R.; Lou, Y. OsWRKY53, a versatile switch in regulating herbivore-induced defense responses in rice. Plant Signal. Behav. 2016, 11, e1169357. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ju, H.; Zhou, G.; Zhu, C.; Erb, M.; Wang, X.; Wang, P.; Lou, Y. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, H.; Li, H.; Zhang, H.; Miao, X. Identification of transcription factors potential related to brown planthopper resistance in rice via microarray expression profiling. BMC Genom. 2012, 13, 687. [Google Scholar] [CrossRef]
- Lu, H.; Luo, T.; Fu, H.; Wang, L.; Tan, Y.; Huang, J.; Wang, Q.; Ye, G.; Gatehouse, A.M.R.; Lou, Y.; et al. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 2018, 4, 338–344. [Google Scholar] [CrossRef]
- Hao, P.; Feng, Y.; Zhou, Y.; Song, X.; Li, H.; Ma, Y.; Ye, C.; Yu, X. Schaftoside interacts with NlCDK1 protein: A mechanism of rice resistance to brown planthopper, Nilaparvata lugens. Front. Plant Sci. 2018, 9, 710. [Google Scholar] [CrossRef]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef]
- Ye, W.; Yu, H.; Jian, Y.; Zeng, J.; Ji, R.; Chen, H.; Lou, Y. A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci. Rep. 2017, 7, 40498. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, L.; Zheng, J.; Chen, F.; Wang, T.; Wang, M.; Tao, Y.; Wang, H.; Hong, Z.; Huang, Y.; et al. A missense mutation in large grain size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 2019, 70, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Kryovrysanaki, N.; James, A.; Tselika, M.; Bardani, E.; Kalantidis, K. RNA silencing pathways in plant development and defense. Int. J. Dev. Biol. 2022, 66, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lv, W.; Hu, L.; Rao, W.; Zeng, Y.; Zhu, L.; He, Y.; He, G. Identification and analysis of brown planthopper-responsive microRNAs in resistant and susceptible rice plants. Sci. Rep. 2017, 7, 8712. [Google Scholar] [CrossRef]
- Nanda, S.; Yuan, S.; Lai, F.; Wang, W.; Fu, Q.; Wan, P. Identification and analysis of miRNAs in IR56 rice in response to BPH infestations of different virulence levels. Sci. Rep. 2020, 10, 19093. [Google Scholar] [CrossRef]
- Lü, J.; Liu, J.; Chen, L.; Sun, J.; Su, Q.; Li, S.; Yang, J.; Zhang, W. Screening of brown planthopper resistant miRNAs in rice and their roles in regulation of brown planthopper fecundity. Rice Sci. 2022, 29, 559–568. [Google Scholar]
- Ge, Y.; Han, J.; Zhou, G.; Xu, Y.; Ding, Y.; Shi, M.; Guo, C.; Wu, G. Silencing of miR156 confers enhanced resistance to brown planthopper in rice. Planta 2018, 248, 813–826. [Google Scholar] [CrossRef]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Yang, F.; Zhang, S.; Sun, S.; Miao, X.; Shi, Z. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Will, T.; Furch, A.C.; Zimmermann, M.R. How phloem-feeding insects face the challenge of phloem-located defenses. Front. Plant Sci. 2013, 4, 336. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lu, J.; Li, Q.; Bao, Y.; Zhang, C. Combined transcriptomic/proteomic analysis of salivary gland and secreted saliva in three planthopper species. J. Proteom. 2018, 172, 25–35. [Google Scholar] [CrossRef]
- Hu, C.; Li, Y.; Liu, Y.; Hao, G.; Yang, X. Molecular interaction network of plant-herbivorous insects. Adv. Agrochem 2023, in press. [CrossRef]
- Tufail, M.; Takeda, M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008, 54, 1447–1458. [Google Scholar] [CrossRef]
- Gong, G.; Yuan, L.; Li, Y.; Xiao, H.; Li, Y.; Zhang, Y.; Wu, W.; Zhang, Z. Salivary protein 7 of the brown planthopper functions as an effector for mediating tricin metabolism in rice plants. Sci. Rep. 2022, 12, 3205. [Google Scholar] [CrossRef]
- Snoeck, S.; Guayazán-Palacios, N.; Steinbrenner, A.D. Molecular tug-of-war: Plant immune recognition of herbivory. Plant Cell 2022, 34, 1497–1513. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef]
- Zeng, J.; Ye, W.; Hu, W.; Jin, X.; Kuai, P.; Xiao, W.; Jian, Y.; Turlings, T.C.J.; Lou, Y. The N-terminal subunit of vitellogenin in planthopper eggs and saliva acts as a reliable elicitor that induces defenses in rice. New Phytol. 2023, 238, 1230–1244. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Ji, R.; Ye, W.; Chen, H.; Zeng, J.; Li, H.; Yu, H.; Li, J.; Lou, Y. A salivary endo-β-1,4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant Physiol. 2017, 173, 1920–1932. [Google Scholar] [CrossRef]
- Zhou, J.M.; Zhang, Y. Plant Immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Guo, J.; Wang, H.; Guan, W.; Guo, Q.; Wang, J.; Yang, J.; Peng, Y.; Shan, J.; Gao, M.; Shi, S.; et al. A tripartite rheostat controls self-regulated host plant resistance to insects. Nature 2023, 618, 799–807. [Google Scholar] [CrossRef]
- Jeevanandham, N.; Raman, R.; Ramaiah, D.; Senthilvel, V.; Mookaiah, S.; Jegadeesan, R. Rice: Nilaparvata lugens Stal interaction—Current status and future prospects of brown planthopper management. J. Plant Dis. Prot. 2023, 130, 125–141. [Google Scholar] [CrossRef]
- Claridge, M.F.; Hollander, D. The “biotypes” of the rice brown planthopper, Nilaparvata lugens. Entomol. Exp. Appl. 1980, 27, 23–30. [Google Scholar] [CrossRef]
- Saxena, R.C.; Barrion, A.A. Biotypes of the brown planthopper Nilaparvata lugens (Stål) and strategies in deployment of host plant resistance. Int. J. Trop. Insect Sci. 1985, 6, 271–289. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Rechavi, O. Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 2022, 23, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Hamby, R.; Sanchez, J.N.; Cai, Q.; Yan, Q.; Jin, H. RNAs—A new frontier in crop protection. Curr. Opin. Biotechnol. 2021, 70, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, M.; Ying, J.; Shen, J.; Dou, D.; Yin, M.; Whisson, S.C.; Birch, P.R.J.; Yan, S.; Wang, X. High-efficiency green management of potato late blight by a self-assembled multicomponent nano-bioprotectant. Nat. Commun. 2023, 14, 5622. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Li, X.; Li, J.; Yu, C.; Zeng, Q.; Ning, G.; Wan, H.; Li, J.; Ma, K.; He, S. Overcoming resistance in insect pest with a nanoparticle-mediated dsRNA and insecticide co-delivery system. Chem. Eng. J. 2023, 475, 146239. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, H. Trans-kingdom RNA interactions drive the evolutionary arms race between hosts and pathogens. Curr. Opin. Genet. Dev. 2019, 58–59, 62–69. [Google Scholar] [CrossRef]
- Yan, S.; Yin, M.; Shen, J. Nanoparticle-based nontransformative RNA insecticides for sustainable pest control: Mechanisms, current status and challenges. Entomolol. Gen. 2023, 43, 21–30. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.; Thomma, B.P.; Huang, H.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, H. RNA silencing: From discovery and elucidation to application and perspectives. J. Integr. Plant Biol. 2022, 64, 476–498. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Gao, J.; Huang, G.; Chen, X.; Zhu, Y. Protein S-acyl transferase 13/16 modulate disease resistance by S-acylation of the nucleotide binding, leucine-rich repeat protein R5L1 in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1789–1802. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Zhang, B.; Gao, X.; Shi, M.; Zhang, S.; Zhong, S.; Zheng, Y.; Liu, X. Functionalized carbon dot-delivered RNA nano fungicides as superior tools to control phytophthora pathogens through plant RdRP1 mediated spray-induced gene silencing. Adv. Funct. Mater. 2023, 33, 2213143. [Google Scholar] [CrossRef]
- Yu, C.; Li, J.; Zhang, Z.; Zong, M.; Qin, C.; Mo, Z.; Sun, D.; Yang, D.; Zeng, Q.; Wang, J.; et al. Metal-organic framework-based insecticide and dsRNA codelivery system for insecticide resistance management. ACS. Appl. Mater. Interfaces 2023, 15, 48495–48505. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Chu, Y.; Yin, M.; Müllen, K.; An, C.; Shen, J. Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv. Mater. 2013, 25, 4580–4584. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhou, H.; Wei, Y.; Yan, S.; Shen, J. A novel plasmid–Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef]
- Sharma, A.; Shrestha, G.; Reddy, G.V.P. Trap crops: How far we are from using them in cereal crops? Ann. Entomol. Soc. Am. 2019, 112, 330–339. [Google Scholar] [CrossRef]
- Vlahova, V. Trap cropping: A useful approach in farming systems. Sci. Pap-Ser. A-Agron. 2021, 64, 342–349. [Google Scholar]
- Sarkar, S.C.; Wang, E.; Wu, S.; Lei, Z. Application of trap cropping as companion plants for the management of agricultural pests: A review. Insects 2018, 9, 128. [Google Scholar] [CrossRef]
- Fahad, S.; Nie, L.; Hussain, S.; Khan, F.; Khan, F.A.; Saud, S.; Muhammad, H.; Li, L.; Liu, X.; Tabaassum, A.; et al. Rice pest management and biological control. In Sustainable Agriculture Reviews: Cereals; Lichtfouse, E.A., Goyal, A.B., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 85–106. [Google Scholar]
- Krishnaiah, N. A global perspective of rice brown planthopper management II-after green revolution era. Mol. Entomol. 2014, 5, 46–55. [Google Scholar]
- Lou, Y.; Zhang, G.; Zhang, W.; Hu, Y.; Zhang, J. Biological control of rice insect pests in China. Biol. Control 2013, 67, 8–20. [Google Scholar] [CrossRef]
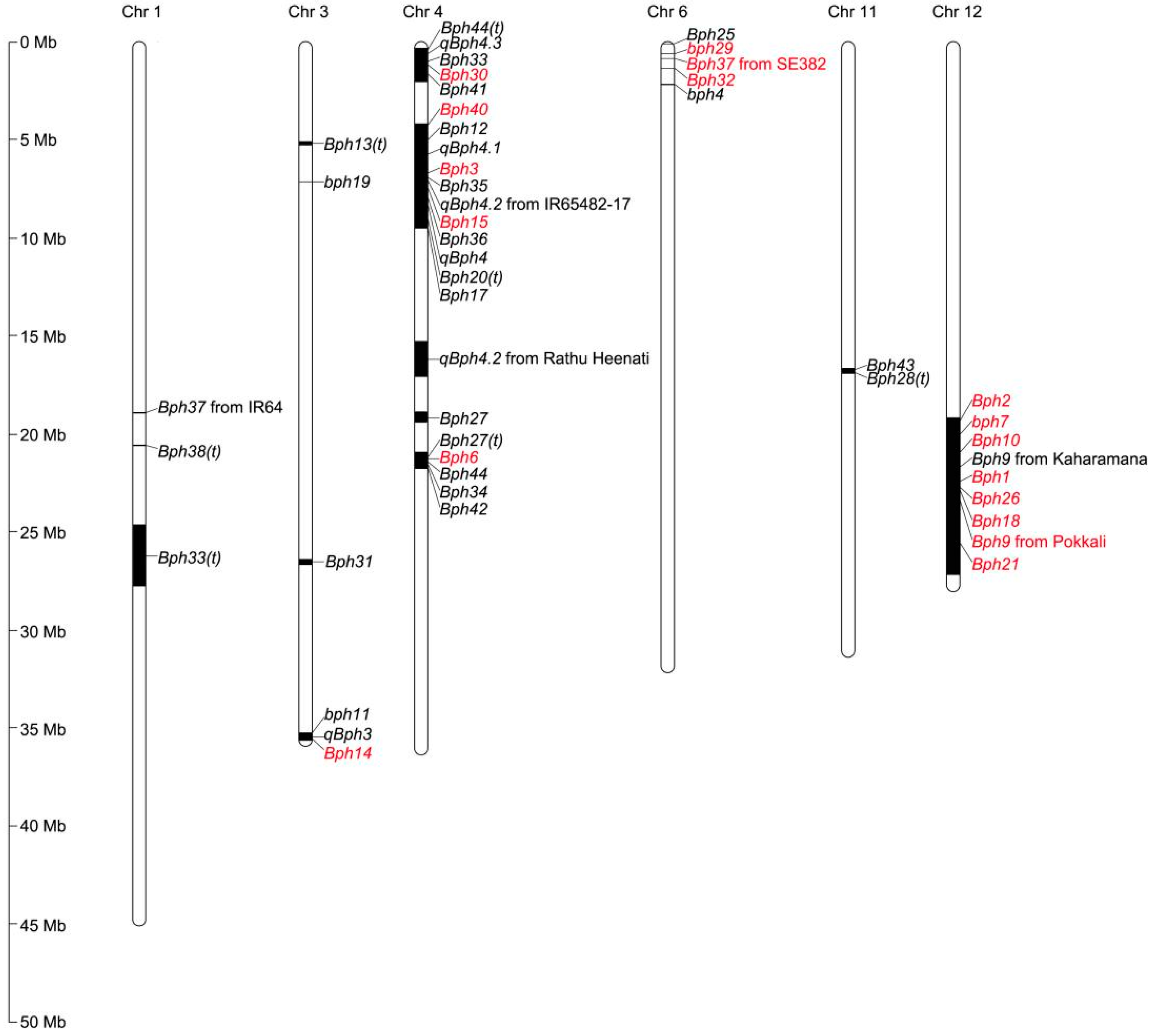

| Gene | Germplasm | Chromosome | Linked Markers | Reference |
|---|---|---|---|---|
| Bph37 | IR64 | 1L | RM302, YM35 | [26] |
| Bph38(t) | Khazar | 1L | 693369, id1012165 | [27] |
| Bph33(t) | RP2068 | 1L | RM488, RM11522 | [28] |
| Bph13(t) | O. officinalis | 3S | AJ09b, AJ09c | [29] |
| bph19 | AS20-1 | 3S | RM6308, RM3134 | [30] |
| Bph31 | CR2711-76 | 3L | PA26, RM2334 | [31] |
| bph11 | O. officinalis | 3L | G1318 | [32] |
| qBph3 | IR02W101 (O. officinalis) | 3L | t6, f3, c3-14 | [33] |
| Bph14 | B5 (O. officinalis) | 3L | SM1, G1318 | [34] |
| Bph44(t) | IRGC 15344 | 4S | 344-0-6, 344-1-2 | [24] |
| qBph4.3 | Salkathi | 4S | RM551, RM335 | [35] |
| Bph33 | Kolayal, Poliyal | 4S | H99, H101 | [36] |
| Bph30 | AC-1613 | 4S | SSR28, SSR69 | [2] |
| Bph41 | SWD10 | 4S | SWRm_01617, SWRm_01522 | [37] |
| Bph40 | SE232, SE67, C334 | 4S | - | [2] |
| Bph12 | O. latifolia | 4S | RM16459, RM1305 | [3] |
| qBph4.1 | Rathu Heenati | 4S | - | [38] |
| Bph3 | Rathu Heenati | 4S | RHD9, RHC10 | [39] |
| Bph35 | RBPH660 (O. rufipogon) | 4S | PSM16, R4M13 | [40] |
| qBph4.2 | IR65482-17 | 4S | RM261, S1, XC4-27 | [41] |
| Bph15 | B5 (O. officinalis) | 4S | RG1, RG2 | [3] |
| qBph4.4 | Salkathi | 4S | RM335, RM5633 | [35] |
| Bph36 | O. rufipogon | 4L | S13, X48 | [42] |
| qBph4 | IR02W101 (O.officinalis) | 4S | p17, xc4-27 | [33] |
| Bph20(t) | O. minuta | 4S | B42, B44 | [43] |
| Bph17 | Rathu Heenati | 4S | RM8213, RM5953 | [44] |
| qBph4.2 | Rathu Heenati | 4L | - | [38] |
| Bph27 | O. rufipogon | 4L | RM16766, RM17033 | [42] |
| Bph27(t) | Balamawee | 4L | Q52, Q20 | [45] |
| Bph6 | Swarnalata | 4L | H, Y9 | [46] |
| Bph44 | Balamawee | 4L | Q31, RM17007 | [24] |
| Bph34 | O. nivara | 4L | RM16994, RM17007 | [47] |
| Bph42 | SWD10 | 4L | SWRm_01695, SWRm_00328 | [37] |
| Bph25 | ADR52 | 6S | S00310 | [48] |
| bph29 | O. rufipogon | 6S | BYL8, BID2 | [49] |
| Bph37 | SE382 | 6S | - | [22] |
| Bph32 | Ptb33 | 6S | RM19291, RM8072 | [50] |
| bph4 | Babawee | 6S | RM190, C76A | [51] |
| Bph43 | IRGC 8678 | 11L | 16-22, 16-30 | [23] |
| Bph28(t) | DV85 | 11L | Indel55, Indel66 | [52] |
| bph2 | ASD7 | 12L | RM7102, RM463 | [53] |
| bph7 | T12 | 12L | RM3448, RM313 | [54] |
| Bph10 | O. australiensis | 12L | RG457 | [55] |
| Bph9 | Kaharamana | 12L | RM463, RM5341 | [56] |
| Bph1 | Mudgo | 12L | em5814N, em2802N | [57] |
| Bph26 | ADR52 | 12L | DS72B4, DS173B | [58] |
| Bph18 | O. australiensis | 12L | BIM3, BN162 | [59] |
| Bph9 | Pokkali | 12L | InD2, RsaI | [54] |
| Bph21 | O. minuta | 12L | S12094A, B122 | [43] |
| Gene | Germplasm | Chromosome | Encoded Protein | Defense Mechanism | Reference |
|---|---|---|---|---|---|
| Bph14 | B5 | 3L | CC-NB-LRR | SA↑, callose deposition | [34] |
| Bph9 | Pokkali | 12L | CC-NB-NB-LRR | SA↑ | [54] |
| Bph1 | Mudgo | 12L | CC-NB-NB-LRR | - | [54] |
| Bph2 | ASD7 | 12L | CC-NB-NB-LRR | - | [54] |
| bph7 | T12 | 12L | CC-NB-NB-LRR | - | [54] |
| Bph10 | IR65482-4-136-2-2 | 12L | CC-NB-NB-LRR | - | [54] |
| Bph18 | IR65482-7-216-1-2 | 12L | CC-NB-NB-LRR | - | [59] |
| Bph21 | IR71033-121-15 | 12L | CC-NB-NB-LRR | - | [54] |
| Bph26 | ADR52 | 12L | CC-NB-NB-LRR | - | [58] |
| Bph37 | SE382 | 6S | CC-NB | - | [22] |
| Bph6 | Swarnalata | 4L | Atypical LRR | SA↑, JA↑, CK↑, enhanced cell walls | [46] |
| Bph30 | AC-1613 | 4S | LRD | enhanced cell walls, IAA↓ | [2] |
| Bph40 | SE232, SE67, C334 | 4S | LRD | enhanced cell walls | [2] |
| Bph15 | B5 | 4S | Lectin receptor kinase | OsPR1a↑, OsLOX↑, OsCHS↑ | [60] |
| Bph3 | Rathu Heenati | 4S | Lectin receptor kinase | - | [39] |
| bph29 | RBPH54 | 6S | B3 DNA-binding | SA↑, JA/ET↓ | [49] |
| Bph32 | PTB33 | 6S | SCR | - | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; Wang, H.; Zha, W.; Wu, Y.; Liu, K.; Xu, D.; He, G.; Zhou, L.; You, A. Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål). Int. J. Mol. Sci. 2023, 24, 16959. https://doi.org/10.3390/ijms242316959
Shi S, Wang H, Zha W, Wu Y, Liu K, Xu D, He G, Zhou L, You A. Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål). International Journal of Molecular Sciences. 2023; 24(23):16959. https://doi.org/10.3390/ijms242316959
Chicago/Turabian StyleShi, Shaojie, Huiying Wang, Wenjun Zha, Yan Wu, Kai Liu, Deze Xu, Guangcun He, Lei Zhou, and Aiqing You. 2023. "Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål)" International Journal of Molecular Sciences 24, no. 23: 16959. https://doi.org/10.3390/ijms242316959
APA StyleShi, S., Wang, H., Zha, W., Wu, Y., Liu, K., Xu, D., He, G., Zhou, L., & You, A. (2023). Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål). International Journal of Molecular Sciences, 24(23), 16959. https://doi.org/10.3390/ijms242316959








